Abstract
Jute fibre is the second most important fibre next to cotton. It is obtained from the bark of plant through microbial retting process. Here we report optimized microbial retting protocol that can lower retting period and produce high fibre quality. A total of 451 bacterial colonies have been isolated from five jute retting water samples in Bangladesh. Higher pectinolytic bacterial isolates were predominant in the later stage of jute retting. Out of these, 168 isolates have been screened by both semi-quantitative and quantitative pectinase, xylanase and cellulase enzyme assay. Among them, 144 isolates have been selected on the basis of extra cellular enzyme activity of these three enzymes. 16 s ribosomal gene sequencing analysis identified 2 phyla- Firmicutis (80.55%) and Proteobacteria (19.45%). To check the synergistic and antagonistic effect 10 selected isolates were tested in 167 different combinations. Three best combinations were identified that lowered retting period from 18–21 days to 10 days producing high quality fibre in both laboratory and field trial. This improved retting technology can be adopted in industrial scale for the production of quality jute fibre in a controlled condition in reduced water quantity without polluting the environment.
Similar content being viewed by others
Introduction
Jute, second most important natural fibre after cotton, is cultivated in East Asia and some parts of Latin America1,2,3. Jute is a bast or phloem fibre in the bark of stems, cemented together by pectin and gummy substances4. Commercial extraction of jute fibre is water based microbiological retting where jute bundles are submerged into slow running river water and subjected to decomposition of pectin, hemicelluloses, and other mucilaginous substances2,5. In this process, pectin is depolymerized by pectinases, primarily comprising four enzymes: Polygalacturonase (PG), Pectin Lyase (PNL), Pectate lyase and Pectin esterase. However, PG6, and PNL7 are primary retting enzymes. In addition, xylanase makes jute fibre softer by selective removal of non-fibrous hemicelluloses without affecting strength of cellulosic fibre. Pectinolytic microorganisms having xylanase activity devoid of cellulase activity is an additional beneficial aspect to improve fibre quality8. The quality of fibre is largely determined by the efficiency of retting process1,2,9,10 and various factors are responsible for proper retting as well as improved fibre quality. Most promising water based microbiological retting process mostly involves bacteria along with various fungi, protozoa, algae and diatoms10,11,12,13. Main aerobic retting bacteria belong to genus Bacillus viz., B. subtilis, B. polymyxa, B. mesentericus, B.pumilus, B. cereus, B. megaterium and B. macerans, initiate retting14,15,16,17,18 along with large numbers of gram-negative genera such as Erwinia and Pseudomonas9,19. At the later stage of retting some anaerobic bacteria from genus Clostridium - Clostridium acetobutylicum, Clostridium stercorarium, Clostridium tertium come to carry on the retting process20. As microorganisms are the main pectinolytic agent during retting, affecting the process and end product quality in depth knowledge of microbial community is essential. Moreover, retting water varies from place to place with respect to its physico-chemical, microbial and biochemical properties which affect fibre quality21,22. Due to scarcity of water, farmers are compelled to use water bodies repeatedly for retting, leading to poor quality jute fibre production21,23. Acute shortage of water and the environmental pollution created from conventional system of retting has demanded for improving the retting process. Aim of this work was to optimize microbial retting protocol that will lower retting period and increase fibre quality. Our strategy involved analysis of pectinolytic retting bacteria with selection of best synergistic effect producing combinations of microbes. In summary, we had to optimize retting process with the best bacterial consortium to yield high quality fibre with less volume of water by reducing retting time from 18–21 days to 10 days.
Results and Discussion
Isolation and screening of isolates
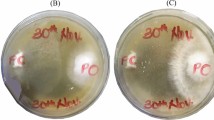
Four hundred and fifty one bacterial isolates were selected based on the colony morphology (Fig. S1). Most of the isolates were found to produce cream color colonies with dry or moist surface. Majority of them were rod shaped and spore former. However some colonies with different color and shape were also observed (Fig. S1). The colony morphology of isolates, as found here in, is comparable with the previous report by Das et al.3. Qualitatively these bacteria were screened on the basis of zone of hydrolysis around the bacterial growth. Out of 451 isolates, 168 having high pectinolytic and xylanolytic and low or no cellulolytic activities were selected (Supplementary Table 1). In primary screening, the colonies forming clear zone, after flooding the substrate with iodine solution indicated to be the enzyme producers (Fig. S2). Efficiency of these isolates was further assessed for extra cellular enzymatic activities through liquid culture (Fig. S3). Finally 144 potential isolates were obtained as pectinase, xylanase and low cellulase producer by forming clear zone within the range of 7.5–25.5 mm, 8.5–25.5 mm and 7.0–25.0 mm diameter respectively (Supplementary Table 2 and Fig. 1). In this study, microorganisms with high pectinolytic, xylanolytic and low cellulolytic activities have been focused because quality of jute fibre mostly depends on retting process and the retting process mainly depends on these enzymatic activities of microorganisms9,15,17. Polygalacturonase is considered to be the most important enzyme in this process6,24,25 and xylanase is required for the partial removal of hemicellulose that makes the jute fibre softer for finer spinning26,27. So it is prime need to have these enzymatic activities of isolates for efficient use as retting inocula.
Identification and phylogenic analysis
A molecular approach based on 16S rRNA gene sequencing was used to identify and distinguish closely related bacterial strains (Supplementary Table 3) and the retrieved sequences were used to construct phylogenetic trees to show relative positions of the isolates at genus level. The 16s rDNA sequence analysis and estimation of phylogenetic relationships (Fig. 2) assigned all 144 strains into two predominant phyla-Firmicutes (most abundant components ~80.55%) and Proteobacteria (~19.45%) which corroborated with the findings of Munshi and Chattoo2. The comparative sequence analysis revealed a rich spectrum of bacterial diversity. The total of 144 bacterial isolates belonged to 14 phylogenetically related species- Bacillus aryabhattai (29%), Bacillus subtilis (19.44%), Bacillus cereus (17.36%), Bacillus megaterium (4.86%), Bacillus koreensis (3.47%), Bacillus xiamenensis (2.78%), Staphylococcus arlettae (0.69%), Clostridium aurantibutyricum (2.78%), Aeromonas jandaei (12.5%), Proteus mirabilis (3.47%), Serratia nematodiphila (1.39%), Kosakonia sacchari (0.69%), Kosakonia oryzae (0.69%) and Enterobacter tabaci (0.69%) under 8 genera (Fig. 3). Comparative sequence analysis revealed that 144 isolates belong to phylogenetically related 14 species and the Bacillus spp. was dominant. Many Bacillus sp. are also widely reported to produce enzymes of industrial application in paper industry28,29,30,31,32,33 and bast fibre degumming34,35. The abundance of these species in our study is in accordance with the earlier culture-based studies, which implicate their role in the retting of jute3,13. Interestingly, we have identified seven phylogenetically related bacterial species namely Aeromonas jandaei, Proteus mirabilis, Bacillus xiamenensis, Bacillus koreensis, Serratia nematodiphila, Kosakonia oryzae and Enterobacter tabaci as retting bacteria which were not reported in previous study as jute retting bacteria.
Selection of best retting bacterial strains
Out of 144, only 10 bacterial strains were selected from retting niche based on extracellular enzymatic activities for preparing retting consortia (Table 1). The selected 10 bacterial isolates comprised namely Bacillus megaterium, Bacillus subtilis, Bacillus cereus, Bacillus xiamenensis, Bacillus koreensis, Proteus mirabilis, Enterobacter tabaci, Kosakonia oryzae, Serratia nematodiphila and Aeromonas jandaei. Among them, Bacillus megaterium, Bacillus subtilis and Bacillus cereus were previously reported to play role in retting process2,3,23. Rest of seven phylogenetically related species, Bacillus xiamenensis, Aeromonas jandae, Proteus mirabilis, Serratia nematodiphila, Bacillus koreensis, Kosakonia oryzae and Enterobacter tabaci were not previously studied as retting bacteria. Bacillus megaterium showed highest pectinolytic and xylanolytic activity.
Comparison of individual and consortial enzyme activity
Strains within various species usually differ in their capacity to ret jute due to varying in enzyme activities. From 10 selected isolates, the highest pectinase activity was noted in Bacillus megaterium (ID-417; 25 mm clear zone, 14.59 U/ml) while lowest was in Kosakonia oryzae (ID-105; 16 mm clear zone, 6.22 U/ml). Aeromonas jandae (ID-191; 23.5 mm clear zone, 6.03 U/ml) and Proteus mirabilis (ID-104; 23.5 mm clear zone, 6.38 U/ml) recorded as the highest xylanase activity while it was lowest in Kosakonia oryzae (ID-105; 15 mm clear zone, 1.63 U/ml) (Table 1). We observed that the pectinolytic and xylanolytic activities associated with cellulolytic activity. Every pectinolytic microbes found in our study had cellulolytic activity. Brühlmann, and his colleague36 reported that cellulolytic activities were always found to be associated with pectinolytic and xylanolytic activities.
Based on pectinolytic and xylanolytic activities 10 bacterial isolates were selected for retting consortia. Because, a mixture of microbes secreting different enzymes are more effective for retting of jute plants than a single microbe37. Microbial consortia included 10 phylogenetically related species namely, Bacillus megaterium, Bacillus subtilis, Bacillus cereus, Bacillus xiamenensis, Bacillus koreensis, Proteus mirabilis, Enterobacter tabaci, Kosakonia oryzae, Serratia nematodiphila and Aeromonas jandaei. One hundred and sixty seven combinations were evaluated for their enzyme activities (Supplementary Table 4). Finally, three consortia namely C-51, C-67 and C-90 were (having highest pectinolytic and xylanase activities however lower cellulolytic activity) selected as potential for use in large scale jute retting process after screened for antagonism. In most of the cases, all the enzyme activities of the consortia were higher than the individual organisms. For example, consortium C-67 showed higher pectinase (31.5 mm clear zone, 21.4 U/ml) and xylanase (30.5 mm clear zone, 8.56 U/ml) activities (Table 2) corroborating the potentiality of using microbial consortia instead of single microbial strain (Fig. 4).
Effectiveness of bacterial consortia in retting
Three bacterial consortia, C-51, C-67 and C-90 with an inoculum of 1 × 108 CFU/ml concentrations and a 5:1 water-inocula ratio reduced the retting period to 10–11 days from 21days (Table 3). It also made remarkable improvement in jute fibre strength, twisting force and luster over control. The results showed that the fibre strength, twisting force and luster increased when microbial consortia were used for retting compared with positive control. The increment of fibre strength and luster could amount to about 20% and fibre twisting force could amount to 9%. It indicated that retting with microbial consortia could results in the effectively removal of pectinolytic material linking gum component within a short time from the fibre. It also confirmed that the microbial consortia did not damage jute fibre. Tenkanen and his colleague38 have shown that the residual lignin linking with xylan in birch kraft pulp could be removed by xylanase. Higher fibre strength, twisting force and luster could be achieved by using C-67 consortium compared with C-90 and C-51. The results clearly demonstrated that microbial consortia significantly reduced the retting period as well as improved the fibre quality in respect of strength, twisting force and luster. Among them, C-67 consortium gave the best result that reduced the retting time from 18–21days to 10 days with better fibre quality. Thus water-retting process and fibre quality were substantially improved by simultaneously inoculating water tanks with six selected pectinolytic strains. Saha et al.39 used a microbial consortia for jute retting comprising three strain of Bacillus pumilus which retted jute plants in 13–15 days. But in the current study, bacterial consortia comprising more than one species has been used to reduce retting period from 18–21 days to 10 days. It is also worth mentioning here that the bacterial consortium is capable of improving fibre strength, color and fineness without using any chemical.
Conclusion
Jute cultivation in water scarce environment is under threat due to higher water requirements for separating fibre from jute stem through retting process. The present study indicated that selective inoculation with efficient microorganisms could be an alternative method of jute retting to obtain quality fibre in water scarce environment. The consortium C-67 could be adopted after commercial field trial to ret jute in minimum water within short time.
Methods
Source of plant material
Defoliated 120 days aged green jute stem (Corchorus olitorius variety 04) samples were collected from Jute Experimental Research Station, Manikganj, Bangladesh (23°52′N, 90°1.4′E), and sub-merged in slow moving Dhaleswari River Water (DRW).
Retting sample collection
Five retting liquor samples namely- DRW-1, DRW-2, DRW-3, DRW-4 and DRW-5, were collected at four days interval within 4–20 days of retting from close proximity of submerged mat of jute bundles to get maximum coverage of retting microbial population. Three sub-samples of retting water were collected from evenly spaced parts (upper, middle and lower) of the jute bundle for each sample at a depth of 20 cm from water surface. All the samples were immediately placed on ice for transportation. The three sub-samples were pooled to create a single composite sample and filtered using Cheese cloth (50–100 μm) followed by Mira cloth (25 μm) repeatedly to separate the rough debris. The temperature and pH of the retting water during the retting season was 25–30 °C and 5.04–7.75, respectively.
Isolation and purification of bacterial strains
The composite samples were diluted up to 10−5 in peptone water (Peptone 10 g/L, NaCl 5 g/L; pH 7.2 ± 0.2). Then 100 μl of samples from each dilution were spread on Nutrient agar (NA) (Peptone 5 g/L, Yeast extract 2 g/L, Meat extract 1 g/L, NaCl 5 g/L, Agar 15 g/L; pH 7.2 ± 0.2) and incubated at 34 °C for 24 hour in aerobic and anaerobic condition (AnaeroGen™ AN0035 kit). All the media were collected from Sigma-Aldrich. Triplicate set of plates were used for all the treatments throughout the study. Bacterial colonies were picked up based on different appearance and colony characteristics were determined according to Holtz40.
Enzyme activity of the isolates
Solid culture extra-cellular enzyme assay
Primary screening of the isolates having pectinase, xylanase and cellulase activities was done by single touch inoculation of each colony onto 0.5% pectin, xylan and Carboxymethyl cellulose (CMC) agar respectively and incubated at 34 °C for 24 hours. Plates were observed for enzyme activity by flooding them with iodine solution (Iodine 1 g and Potassium Iodide 5 g for 330 ml iodine solution)41. Presence of clear zone developed through hydrolysis around the growth indicated positive enzyme activity and the extent of clear zone indicated the capacity.
Liquid culture extra-cellular enzyme assay
Bacteria were cultured in 100 ml Nutrient Broth (Peptone 5 g/L, Yeast extract 2 g/L, Meat extract 1 g/L, NaCl 5 g/L; pH 7.2 ± 0.2) and incubated at 34 °C in shaking incubator with 200 rpm for 20 hours. After incubation, a suitable volume of bacterial cultures were centrifuged at 22,000 × g rpm for 20 min and filtered the supernatant through Millipore filter (0.22 μm). The filtrate was subjected to the screening of three (pectinase, xylanase and cellulose) enzymatic activities by cup-plate diffusion technique42. The 4.0 mm diameter wells were prepared in agar plates containing pectin, xylan and CMC with a sterile cork borer to inoculate 25 μl of filtrate. After 24 hours of incubation at 34 °C, the plates were flooded with iodine solution. Presence of clear zone around the well indicated the enzymatic activity of the isolates. The average diameter (mm) of clear zones of 4 replications for each isolate was recorded.
Quantitative extra-cellular enzyme assay
Quantitative enzyme assay of retting isolates was based on the determination of reducing sugars produced as a result of enzymatic hydrolysis of pectin, xylan and cellulose by dinitrosalicylic acid reagent (DNS) method43. For the assay, 5 ml bacterial culture grown in modified MS medium containing 0.3% KH2PO4, 0.6% Na2HPO4, 0.2% NH4Cl, 0.5% NaCl, 0.01% MgSO4.7H2O supplemented with 0.5% yeast extract and 0.5% pectin (for pectinase production) or xylan (for xylanase production) or carboxymethyl cellulose (for cellulase production) was centrifuged at 10,000 rpm for 10 min at 4 °C and the clear supernatant was used as crude enzyme. Beechwood xylan 1% and carboxymethyl cellulose (CMC) 1% (Sigma-Aldrich) were prepared in 0.05M Na-citrate buffer of pH 5.3 and pH 4.8 respectively. Whereas Polygalacturonic acid 0.5% was prepared in 0.1M phosphate buffer (pH 7.5) and used as substrates for xylanase, cellulase and pectinase. The reaction mixture contained 900 μl of respective substrate (pectin and xylan) and 100 μl of appropriately diluted enzyme (pectinase, and xylanase), whereas, 100 μl of distilled water was used for enzyme blank. In case of cellulase, 1 ml of substrate (CMC) and 1 ml of appropriately diluted crude enzyme were used and were incubated at 50 °C for 10 min and the reaction was terminated by adding 1 mL of 3,5-dinitrosalicylic acid (DNS) and boiling (92 °C) for 5 min. Finally, the tubes were cooled and optical density (OD) was measured using spectrophotometer (Bio-Rad SmartSpec Plus) at 540 nm. For pectin, xylan and CMC, reducing sugar concentrations were estimated as D-galacturonic acid D-xylose and D-glucose equivalents, using a calibration curve constructed with D-galacturonic acid (Sigma-Aldrich, ≥98.0% purity) D-xylose (Sigma-Aldrich, ≥99% purity) and D-glucose (Sigma-Aldrich, ≥99.5% purity) respectively. The enzyme unit (U) was defined as the amount of enzyme that catalyzes the formation of 1 μmol of reducing sugar per minute under the specified assay conditions.
Isolation of genomic DNA
Genomic DNA was isolated following the method of Sambrook and Russel44 and treated with RNase A. The quantity and purity of DNA was assessed using a NanoDrop 2000 spectrophotometer (NanoDrop, Thermo Scientific). The integrity of DNA was evaluated by 1% agarose gel electrophoresis.
Amplification of 16S rDNA and PCR product purification
PCR amplification of 16S rDNA gene was carried out in GeneAmp PCR System 9700 (Applied Biosystems, Life Technologies, USA) by using forward primer 27 F (5′-AGAGTTTGATCMTGGCTCAG-3′) and the reverse primer 1492 R (5′-GGTTACCTTGTTACGACTT-3′). Final volume of PCR mixture was 50 μl containing 5 μl of 50 ng/μl genomic DNA, 0.25 μl of Platinum Taq DNA Polymerase (Invitrogen, Life Technologies, USA), 5 μl of 10x PCR buffer with 2 μl of 20 mM MgSO4, 1 μl of 10 mM dNTP (Invitrogen, Life Technologies, USA), 2 μl Dimethyl sulfoxide (DMSO), 2 μl of 10 μM each primer and 32.75 μl of nuclease free water. The cycling parameter consisted of 25 cycles: denaturation at 94 °C, 30 s; primer annealing at 55 °C, 30 s; extension at 68 °C, 1 min. Before amplification cycle, DNA was denatured for 5 min at 94 °C and after amplification an extension step for 7 min at 68 °C was performed. All the amplified PCR products were eluted from agarose gel using Qiagen QuickSpin PCR purification columns (Qiagen,Catalog No. 28706).
Sequencing of 16S rDNA fragment, assembly and BLAST search
The amplified and purified PCR fragments were sequenced in ABI 3730XL DNA Analyzer (Applied Biosystems, Life Technologies, USA) with the primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), 533R (5′-TTACCGCGGCTGCTGGCAC-3′), 981R (5′-GGGTTGCGCTCGTTGCGGG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). Sequencing reaction was performed by using Big Dye V3.1 sequencing reagents (Applied Biosystems, Life Technologies, USA) following the manufacturer’s protocol. The four sequences for each sample were assembled using CAP3 Sequence Assembly Program, to obtain the full 16S rRNA genes. The 16S rDNA sequences of the isolated strains were compared to public databases available in NCBI. Identification to the species level was determined as a 16S rDNA sequence similarity of >99% with that of the prototype strain sequences in the GenBank.
Phylogenetic analysis
The 16S rDNA sequences of isolates were aligned with the MUSCLE and phylogenetic trees were inferred using the neighbor-joining method45. The software MEGA, version 6.0, was used to construct trees46. Bootstrap analysis (100 replicates) was used to test the topology of the neighbor-joining method.
Extracellular enzymatic assay of bacterial consortia
A total of 167 consortia of different combinations containing 3 to 10 bacterial isolates, were selected to test synergistic effects in extracellular enzyme production. For each treatment, primary inoculum of individual strain was prepared by growing in 20 ml NB at 30 °C for 24 hours which was used to inoculate 99 ml NB in 1:99 ratio after adjustment of Optical Density (OD) at 6–7(±0.4) (A = 600). After attaining mid-log phase (0.4–0.6 OD) of growth, bacterial cultures were mixed equally and OD of mixed culture was adjusted at 6.5–7.0 to inoculate 99 ml NB in 1:99 ratio and incubated for their respective incubation period (Optimization). Enzyme activity was checked according to same procedure as extracellular enzyme assay. The bacterial isolates were further screened for enzyme activity by quantitative assay. To study the pectinase, xylanase and cellulase activities in various consortia, the respective enzyme producing media were simultaneously inoculated with the bacterial strains and the cell free supernatants were used for the assays. The enzyme producing media composition and quantitative enzyme assay procedure were as same as described above in Quantitative extra-cellular enzyme assay section.
Retting efficiency test
Small scale
Artificial plastic retting tanks (Length 150 inch, width 14 inch and depth 7 inch) were used for jute retting trial. Green ribbons were extracted from 120 days aged defoliated jute plant (Corchorus olitorius, variety O-4) with an average height 3.30 m and diameter 23 mm by a ribbon extractor machine (CRIJAF, 2011). Two kg of green ribbon was submerged in 10 L of distilled water at a substrate liquor ratio of 1:5. Three consortia C-51, C-67 and C-90 with 5, 6 and 7 bacterial strains respectively were used as inocula in each treatment. The bacterial inocula were added with water of 10:1 at 1 × 108 cells/ml. Plastic tanks were covered with cork sheet and kept at green house to maintain the temperature at 30–32 °C until completion of retting. Each treatment was carried out in three replicated tanks. A positive and negative control was maintained using 10 L natural river water and 10 L distilled water without inocula, respectively.
Large scale
Large scale retting experiment was also carried out with 10 kg and 50 kg of 120 days aged ribbon in cemented retting concrete tank. We have used combination C-67 containing 6 bacterial isolates that produced best results in plastic tank retting experiment. Ribbon and inocula and water were added in 1:1:5. Number of days required for retting, average day temperature, OD and pH of retting liquor during progress of retting was periodically recorded. After completion of retting, fibre was extracted, washed and sun dried.
Fibre quality test method
Bundle strength of jute fibre was determined by taking 20 randomly selected jute samples in each set and following the method described by Bandyopadhyay and Mokhapadhyay47 on a Pressly fibre bundle strength tester (Model-215, USA). 100 reading from each set of yarn were taken to measure Twisting force and luster of the fibre on a Goodbrand Manual Twist Tester (Goodbrand and Company Ltd, UK 80600) and Reflection meter (Model-577, USA) respectively.
Statistical analysis
All experiments were laid out a complete randomized design with four replications. Analysis of variance and comparison of means were calculated with the statistical package Mstat-C v2.10. Means were compared by using the Least Significance Differences (LSD) test (P < 0.05).
Data availability
All 16S rRNA partial gene sequences are publicly available from the GenBank under the accession number MH010052 to MH010195, with specific numbers listed in Supplementary Table 3.
References
Jarman, C. G. The retting of Jute. FAO Agriculture Organization of the United Nations, Rome, Italy, pp 1–5 (1985).
Munshi, T. K. & Chattoo, B. B. Bacterial Population Structure of the Jute Retting Environment. Microb. Ecol. 56, 270–282 (2008).
Das, B. et al. Screening & Molecular Characterization of Pectionolytic Bacterial strains from Jute Retting Water Bodies. Research Journal of Microbiology. 7, 50–58 (2012).
Ali, M. H., Samad, M. A. & Sarker, N. N. Comparative study on the physico-mechanical properties of Jute, kenaf and Mesta-Part-V. Bangladesh J. Jute Fibre Res. 21, 71–80 (1996).
Haque, M. S., Zakaria, A., Adhir, K. B. & Firoza, A. Identification of Micrococcus sp. Responsible for the Accelaration of Jute Retting. Pakistan Journal of Biological Sciences 6, 686–687 (2003).
Zhang, J., Henriksson, G. & Johansson, G. Polygalacturonase is the key component in enzymatic retting of flax. J. Biotechnol. 81, 85–89 (2000).
Soriano, M., Diaz, P. & Pastor, F. I. J. Pectinolytic systems of two aerobic sporogenous bacterial strains with high activity on pectin. Curr. Microbiol. 50, 114–118 (2005).
Gomes, I., Saha, R. K., Mohiuddin, G. & Hoq, M. M. Isolation and characterization of a cellulase-free pectinolytic and hemicellulolytic thermophilic fungus. World J. Microbiol. Biotechnol. 8, 589–592 (1992).
Ahmed, Z. & Akhter, F. Jute retting: An overview. Online J Biol Sci. 1, 685–688 (2001).
Ahmed, Z. & Nizam, S. A. Jute-Microbiological and Biochemical Research. Plant Tissue Cult. & Biotech. 18, 197–220 (2008).
Donaghy, J. A., Levette, P. N. & Haylock, R. W. Changes in microbial populations during anaerobic flax retting. J Appl Bacterio. l69, 634–641 (1990).
Islam, M. M. & Rahman, M. M. Advances in Jute and Allied fibres post-harvest processing technologies in Bangladesh: Adoption constraints, Prospect and Future Thrust. Research Web Pub 1, 20–30 (2013).
Tamburini, E., Gordillo, L. A., Perito, B. & Mastromei, G. Characterization of bacterial pectinolytic strains involved in the water retting process. Environ Microbio. 15, 730–736 (2003).
Majumdar, A. K. & Day, A. Chemical constituents of jute ribbon and the materials removed by retting. Food farming and agriculture 21, 25–26 (1977).
Nath, M. et al. Optimization of Fermentation Conditions for pectin degrading enzyme production by pectinolytic microbial consortia used for jute retting. Int. J. Curr. Microbiol. App. Sci 6, 925–931 (2017).
Das, S., Majumdar, B. & Saha, A. R. Biodegradation of plant pectin and hemicelluloses with three novel Bacillus pumilus strains and their combined application for quality jute fibre production. Agric. Res. 4, 354–364 (2015).
Cheng, L. et al. Screening a bacterium and its effect on the biological degumming of ramie and kenaf. Sci. Agric. 75, 375–380 (2018).
Ge, J. et al. Production of Pectinolytic Enzymes by Two Bacillus spp. Strains and Their Application in Flax Degumming. Trans. Tianjin Univ. 25, 413–419 (2019).
Wang, Q. et al. Isolation of Bacillus cereus P05 and Pseudomonas sp. X12 and their application in the ramie retting. Industrial Crops and Products 97, 518–524 (2017).
Duan, S. et al. Diversity and Characteristics of Kenaf Bast Degumming Microbial Resources. Journal of Natural Fibres 15, 799–807 (2018).
Das, B., Chakraborty, A., Majumder, B. & Chakraborti, A. Studies on physic-chemical and microbial parameters of jute growing soil, retting water and jute fibre quality. Bangladesh J. Agric. Environ. 6, 1–13 (2010).
Das, B. et al. Assessment of changes in community level physiological profile and molecular diversity of bacterial communities in different stages of jute retting. Pak. J. Biol. Sci. 16, 1722–1727 (2003).
Banik, S., Basak, M. K. & Sil, S. C. Effect of Inoculation of Pectinolytic Mixed Bacterial Culture on Improvement of Ribbon Retting of Jute and Kenaf. Journal of Natural Fibres 4, 33–50 (2007).
Anand, G., Yadav, S. & Yadav, D. Purification and characterization of polygalacturonase from Aspergillus fumigatus MTCC 2584 and elucidating its application in retting of Crotalaria juncea fibre. 3 Biotech 6, 201–207 (2016).
De Prez, J., Van Vuure, A. W., Ivens, J., Aerts, G. & Van de Voorde, I. Effect of enzymatic treatment of flax on chemical composition and the extent of fibre separation. Bio. Res. 14, 3012–3030 (2019).
Haque, M. S. et al. Upgradation of SMR and seed-cut low grade barky jute fibre through the application of fungi. Bangladesh J. Jute Fib. Res. 14, 53–58 (1989).
Das, B. et al. Effect of efficient pectinolytic bacterial isolates on retting and fibre quality of jute. Industrial Crops and Products 36, 415–419 (2012).
Kulkarni, N. & Rao, M. Application of xylanase from alkaliphilic thermophilic Bacillus sp.NCIM 59 in biobleaching of bagasse pulp. J. Biotechnol. 51, 167–173 (1996).
Khanongnuch, C. et al. β-Mannanase and xylanase of Bacillus subtilis 5H active for bleaching of crude pulp. J. Fermentation Bioeng. 86, 461–466 (1998).
Shah, A. K., Sidid, S. S., Ahmad, A. & Rele, M. V. Treatment of bagasse pulp with cellulase-free xylanases from an alkalophilic Bacillus sp. Sam-3. Bioresour. Technol. 68, 133–140 (1999).
Bim, M. A. & Franco, T. T. Extraction in aqueous two-phase systems of alkaline xylanase produced by Bacillus pumilus and its application in kraft pulp bleaching. J. Chromatogr. B Biomed. Sci. Appl. 743, 349–356 (2000).
Chauhan, S., Choudhury, B., Singh, S. N. & Ghosh, P. Application of xylanase enzyme of Bacillus coagulans as a pre-bleaching agent on non-woody pulps. Process Biochem. 41, 226–231 (2006).
Battan, B., Sharma, J., Dhiman, S. S. & Kuhad, R. C. Enhanced production of cellulase-free thermostable xylanase by Bacillus pumilus ASH and its potential application in paper industry. Enzyme Microb.Technol. 41, 733–739 (2007).
Zhao, D. et al. Bacterial succession and metabolite changes during flax (Linum usitatissimum L.) retting with Bacillus cereus HDYM-02. Sci. Rep. 6, 31812 (2016).
Das, S. et al. Comparative Study of Conventional and Improved Retting of Jute with Microbial Formulation. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 88, 1351 (2017).
Brühlmann, F., Kim, K. S., Zimmerman, W. & Fiechter, A. Pectinolytic Enzymes from Actinomycetes for the Degumming of Ramie Bast Fibres. Applied and Environmental Microbiology 60, 2107–2112 (1994).
Banik, S. et al. Ribbon retting of jute-a prospective and eco-friendly method for improvement of fibre quality. Industrial Crops and Products. 17, 183–190 (2003).
Tenkanen, M., Tamminen, B. & Hortling, B. Investigation of lignin-carbohydrate complexes in kraft pulps by selective enzymatic treatments. Appl. Microbiol. Biotechnol. 51, 241–248 (1999).
Mahapatra, B. S. et al. A microbial consortium used in faster retting of jute and mesta. Application no. 418/KOL/2011 (2011).
Holtz, J. D. Bergey’s Manual of Determinative Bacteriology. (Williams and Wilkins, Baltimore, 1993).
Beg, Q. K., Bhushan, B., Kapoor, M. & Hoondal, G. S. Production and characterization of thermostable xylanase and pectinase from Streptomyces sp. QG-11-3. Journal of Industrial Microbiology and Biotechnology 24, 396–402 (2000).
Dingle, J., Reid, W. W. & Solomons, G. L. The enzymatic degradation of pectin and other polysaccharides. II. Application of the cup-plate assay to the estimation of enzymes. J. Sci. Food Agric. 4, 149–155 (1953).
Miller, G. H. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–429 (1959).
Sambrook, J. & Russel, D.W. Molecular Cloning: A Laboratory Manual. (CSH Laboratory Press, Cold Spring Harbor, NY. 2001).
Saitou, N. & Nei, M. The Neighbor-Joining method-a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Tamura, K. et al. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Bandyopadhyay, S. B. & Mokhapadhyay, S. K. Assessment of jute fibre strength. Jute Bull. 27, 193–202 (1964).
Acknowledgements
A portion of this research (16S rRNA gene sequencing) was performed under the collaborations of The Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB), University of Hawai‘i at Mānoa, USA. The authors would like to thank Jennifer Saito, Ph.D, Senior Researcher, ASGPB for critically reviewed the experiment and Shaobin Hou, Ph. D, Genomics Leader, ASGPB for the sequencing of 16S rRNA. This work was funded by the grants from Ministry of Agriculture, Government of Bangladesh.
Author information
Authors and Affiliations
Contributions
M.S.H., M.S.I. and M.M.A. conceived the study, and R.H., N.A., U.H. S.M.T.K. and R.I. performed the investigations. A.H. contributed to collect sample and M.D.H.S. performed field experiment. N.A., R.H. and M.S.I. performed the formal analyses and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hasan, R., Aktar, N., Kabir, S.M.T. et al. Pectinolytic Bacterial Consortia Reduce Jute Retting Period and Improve Fibre Quality. Sci Rep 10, 5174 (2020). https://doi.org/10.1038/s41598-020-61898-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61898-z
This article is cited by
-
Innovations in papermaking using enzymatic intervention: an ecofriendly approach
Cellulose (2023)
-
Comparative metagenomics and microbial dynamics of jute retting environment
International Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.