Abstract
Volatile organic compounds (VOCs) produced by Aureobasidium pullulans were investigated for antagonistic actions against Alternaria alternata and Botrytis cinerea. Conidia germination and colony growth of these two phytopathogens were suppressed by A. pullulans VOCs. A novel experimental setup was devised to directly extract VOCs using solid-phase microextraction-gas chromatography-mass spectrometry (SPME-GC-MS) from antagonist-pathogen culture headspace. The proposed system is a robust method to quantify microbial VOCs using an internal standard. Multivariate curve resolution-alternating least squares deconvolution of SPME-GC-MS spectra identified fourteen A. pullulans VOCs. 3-Methyl-1-hexanol, acetone, 2-heptanone, ethyl butyrate, 3-methylbutyl acetate and 2-methylpropyl acetate were newly identified in A. pullulans headspace. Partial least squares discriminant analysis models with variable importance in projection and selectivity ratio identified four VOCs (ethanol, 2-methyl-1-propanol, 3-methyl-1-butanol and 2-phenylethanol), with high explanatory power for discrimination between A. pullulans and pathogen. The antifungal activity and synergistic interactions of the four VOCs were evaluated using a Box-Behnken design with response surface modelling. Ethanol and 2-phenylethanol are the key inhibitory A. pullulans VOCs against both B. cinerea and A. alternata. Our findings introduce a novel, robust, quantitative approach for microbial VOCs analyses and give insights into the potential use of A. pullulans VOCs to control B. cinerea and A. alternata.
Similar content being viewed by others
Introduction
Microbial antagonists have been widely explored as more environmentally friendly disease management alternatives to reduce the excessive use of synthetic fungicides1,2. Among a wide array of mechanisms exploited by microbial antagonists, the production of volatile organic compounds (VOCs) with antimicrobial properties3 has become popular in the recent research due to their low risk of toxic residues, biodegradability4 and enormous spectrum of activity regardless of a physical contact of the commodity5.
Aureobasidium pullulans is a yeast-like saprophytic fungus, naturally inhabiting plant and fruit surfaces6. This yeast is a well-known biocontrol agent against a range of pathogenic fungi7,8 and strains of the organism have been formulated as commercial preparations for the practical management of fungal phytopathogens of a range of horticultural crops9. Production of antifungal VOCs has been recently identified as a potential mode of action for its biocontrol10,11. One of these studies has confirmed the identity of its four VOCs, 2-methyl-1-butanol, 3-methyl-1-butanol, 2-methyl-1-propanol and 2-phenylethanol11. These compounds were shown to suppress conidia germination and mycelium growth of Botrytis cinerea, Colletotrichum acutatum, Penicillium expansum, Penicillium digitatum and Penicillium italicum under both in vitro and in vivo conditions11. A similar study identified over 20 VOCs from A. pullulans isolated from grapevine12. However, none of these compounds were tested for antifungal properties12. Thus, further investigations are necessary to identify the antifungal properties of A. pullulans VOCs. In the absence of this knowledge, it is difficult to fully understand the mode of action of A. pullulans as a biological control agent. Four A. pullulans isolates were used in the current study. All four isolates have been previously proven to have antifungal properties against Greeneria uvicola in a dual culture system which reduced the radial growth of the pathogen compared to controls8. However their ability to produce VOCs has not been investigated.
The present study investigated A. pullulans antifungal VOCs against two necrotrophic fungal pathogens; Botrytis cinerea and Alternaria alternata. The two pathogens infect a number of horticultural crops causing both pre and postharvest yield losses worldwide13,14. Black mould and grey mould are two plant diseases attributed to A. alternata and B. cinerea respectively13,14. In addition to yield losses, the production of deleterious off flavours leads to a loss of wine quality when wine is made with grey mould infected grapes15.
To our knowledge, a robust automated technique for quantitative analysis of microbial VOCs has not previously been reported to assess antagonist and pathogen interaction and the inhibitory actions of VOCs. We propose a novel set up for the quantitative analysis of VOCs by automated solid phase-microextraction-gas chromatography-mass spectrometry (SPME-GC-MS). The setup facilitates extraction of VOCs from the headspace of A. pullulans and pathogen interaction system. Such a system allows variations in VOCs production by antagonists and the influence of target pathogens to be monitored. Previous studies manually extracted VOCs from a double Petri dish system11,16 without incorporation of suitable internal standards (IS). Thus, studies on the efficacy of mixtures of microbial VOCs have based their findings on the proportions of relative peak areas, derived from GC-MS chromatograms17,18. However, the degradation of SPME fibre sensitivity between injections, different affinities of different analytes to SPME fibres19 and detector sensitivity to different analytes11 make the interpretation of volatile profiles challenging. The employment of appropriate ISs for VOCs peak area normalisation generally overcomes these constraints and is widely practised in areas such as wine and food metabolomics20. The current study proposes a method to introduce an IS to the antagonist-pathogen culture system without disturbing the headspace and thereby perform quantitative sampling of VOCs.
Multivariate chemometrics which uses a combination of mathematical and statistical approaches21, have become important in data resolution obtained from analytical platforms such as gas chromatography-mass spectrometry (GC-MS)22. A range of unsupervised and supervised approaches to resolve complex multivariate data sets allows identification of compositional differences between samples22. Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS), one of the popular chemometric methods used in the resolution of multiple component responses23, was employed in this study in an association with untargeted, SPME-GC-MS analysis, to characterise A. pullulans VOCs.
The antifungal activity of VOCs has been suggested to be synergistic or additive rather than a sole inhibitory action11,24. An inability to mimic the antagonists’ antimicrobial activity by VOCs when applied individually24 and production of VOCs at very low concentrations by antagonists25 suggest that a synergistic or additive effect may occur. In this context, our study aimed to identify A. pullulans VOCs by a robust, direct, quantitative approach based on untargeted metabolomics and chemometric analysis. Also the antifungal properties of VOCs were evaluated against B. cinerea and A. alternata. Lastly, the synergistic behavior of VOCs to cause antifungal effects were studied using a response surface modelling (RSM) approach.
Methods
Antagonist and pathogen isolates
A. pullulans (KR605651.1(A1), KR605650.1(A2), KR605653.1(A3), KR605665.1(A4), B. cinerea TN080 (all isolated from wine grapes)8 and B. cinerea DAR69764 (isolated from tomato) were obtained from the culture collection of the laboratory of National Wine and Grape Industry Centre, Charles Sturt University, Wagga Wagga, Australia. A. alternata (MH931372) was isolated from a tomato fruit showing characteristic black mould symptoms. Cultures were maintained on Potato Dextrose Agar (PDA) at 25 °C for routine use.
Evaluation of the antifungal activity of A. pullulans VOCs
Mycelium growth inhibition
A double Petri dish method26 was adopted to evaluate the effect of VOCs on mycelium growth. A cell suspension of A. pullulans (108 cells/mL, 100 µL), harvested in sterile distilled water (SDW), was spread on PDA, sealed and incubated at 25 °C for 48 h. Mycelial agar discs (7 mm diameter) from 7-day-old fungal pathogen cultures were placed at the centre of the PDA base plate. Antagonist and pathogen plates were sealed together with Parafilm and incubated at 25 °C. Control plates devoid of A. pullulans, were treated with SDW (100 µL). The diameter of the pathogen colony was measured after three days of incubation at four radial distances and averaged. Six replicates were performed for each antagonist-pathogen combination and controls.
Inhibition of conidia germination
A conidial suspension of either B. cinerea or A. alternata (106 spores/mL, 30 µL), in potato dextrose broth (PDB), was placed on a glass microscope slide, in a Petri dish lined with a moistened filter paper. A second Petri dish containing a 48 h culture of A. pullulans growing on PDA was placed face-down on the base plate containing the microscope slide with the fungal pathogen. Control plates were prepared as described above. Plates were sealed and incubated for 5 h at 25 °C in darkness. Two hundred conidia were assessed for germination and the results expressed as a percentage. Conidia were considered as germinated when the germ tube length was equal to or greater than the transverse diameter of conidia and there was no observable disruption. Three replicates were performed.
Analysis of A. pullulans VOCs by headspace SPME-GC-MS
Experimental set up for SPME automated sampling of VOCs
PDA (1.5 mL) was poured into a 20 mL headspace vial laying on its side such that the agar solidified on the vertical wall of the vial. The vial was then rotated and a further volume of PDA (1.5 mL) was poured on to the opposite side of the vial, in the same way the next day ensuring that the two layers were not mixed. A cell suspension of A. pullulans in SDW (108 cells/mL, 10 µL) was spread on to one of the PDA layers of the 20 mL headspace vial. The vial was sealed with a screw cap with PTFE septum and incubated in a horizontal position at 25 °C for 48 h. The opposite PDA layer was then inoculated with a spore suspension of either B. cinerea or A. alternata (106 spores/mL, 10 µL). A paper disc (10 mm, Sigma-Aldrich, MO, USA) glued (PVA based glue) on a strip of aluminium foil and heated at 120 °C in an oven for 2 h to volatilise any compounds present in the adhesive, was hung on the wall of the headspace vial. The disc was used to add the IS immediately before the GC-MS analysis (Fig. 1). Care was taken to keep the strip away from the PDA layer and the length was kept to approximately 1 cm in order to prevent contamination of the SPME fibre. After pathogen inoculation, the vials were incubated for 72 h at 25 °C. Each pathogen-antagonist combination consisted of four types of growth each with four replicates; negative antagonist (A. pullulans only), negative pathogen (either B. cinerea or A. alternata only) interaction (inoculated with both A. pullulans and respective pathogen), and the blank (non-inoculated media). The following antagonist-pathogen combinations were grown in interaction vials, A. pullulans A1-B. cinerea TN080, A. pullulans A2-B. cinerea DAR69764, A. pullulans A2-A. alternata and A. pullulans A3 with either B. cinerea TN080, B. cinerea DAR69764 or A. alternata.
An IS comprising 2-methyl-4-pentanol in methanol (40 mg/L, 5 µL) was added to the disc, using a glass syringe (Hamilton syringe®) with Chaney adapter through the septum of the headspace vial cap, immediately before the GC-MS analysis. The concentration, volume and type of IS were decided and adjusted from a series of preliminary injections (data not shown), in order to produce a peak height in the chromatogram at approximately the midpoint of peaks height for the unknown VOCs.
Headspace vials containing samples were placed in a Peltier cooled sample tray at 10 °C which minimised further growth of organisms and reduced the impact on changing headspace composition, and were sampled in a random order. Vials were transferred into a heated (40 °C) block and incubated for 1 min. A 2 cm SPME fibre with 50/30 µm DVB/CAR/PDMS (Supelco, PA, USA) coating was inserted into the headspace of the sample vial and exposed to the headspace for 15 min at 40 °C.
GC-MS analysis
VOCs analysis was performed with a 7890B gas chromatography system (Agilent, Palo Alto, CA, USA) equipped with a CombiPal auto-sampler (CTC Analytics, Zwingen, Switzerland) coupled to an Agilent 5975C triple quadrapole mass detector. The SPME fibre was desorbed into an ultra-inert straight SPME liner (0.75 mm, Agilent Technologies Inc., USA) at 250 °C in splitless mode for 2 min, and separation of compounds achieved through a DB-Waxetr column (60 m × 250 µm inner diameter × 0.25 µm film thickness; Agilent Technologies Ltd., USA) with helium at a flow rate of 1 mL min−1. The oven temperature was set to 40 °C for 3 min and then ramped from 40 to 90 °C at 10 °C min−1, 90 to 180 °C at 5 °C min−1, 180 to 250 °C at 20 °C min−1 and held for 2 min, resulting in a total run time of 31.5 min. The mass spectrometer was operated in an electron impact (EI) ionization at 70 eV with an ion source temperature of 250 °C, to scan a mass range from 35 m/z to 350 m/z.
Data processing
The total ion chromatogram (TIC) of all samples were overlaid in Agilent MassHunter Qualitative analysis B.07.00 software to identify appropriate time windows for automated processing. Each time window was selected along a stable base line including two to three peaks with a profile height not less than 106 abundance. The GC-MS data files (.d) were converted into excel (.xlsx) files with Openchrom (community edition 1.2.0.) and imported into Matlab (version R.2018b, The Mathworks Inc., MI, USA). Files were processed according to previously published MCR-ALS analysis procedures23,27.
Siloxane related mass ions, including 73, 133, 147, 149, 191, 193, 207, 208, 209, 221, 249, 265, 267, 268, 269, 281, 282, 283, 295, 325, 327, 341, 34227 were excluded from the data matrix as they were due to SPME fibre breakdown products. Spectra extracted during the chromatogram deconvolution by MCR-ALS were exported from Matlab in a format compatible with the National Institute of Standards and Technology (NIST) Mass Spectral Search Program, 2014, version 2.2 (USA), and a peak table with areas associated with extracted variables including the IS, used for subsequent multivariate analysis.
Identification of compounds
The spectra imported in to NIST−14 library and variables were tentatively identified by comparing with those reference compounds from the library. Any known artificial peaks were excluded from the data set. The compounds with more than 80% of similarity with reference spectra were selected. The production of these selected compounds by A. pullulans were confirmed using box-plots (Supplementary material Fig. S5) of feature (VOCs) abundances after normalisation to the IS from MCR-ALS extracted data. The identity of those selected compounds were confirmed by comparison with the retention time of authentic standards run in identical GC-MS conditions. Linear retention indices (LRI) were calculated using n-alkane series (C8−C20) run in similar oven ramp and gas flow conditions and the equation of van den Dool and Kratz and compared with literature28.
Sample classification by PCA and PLS-DA
An offset of 1 was added to all m/z channels of the normalised data matrix of variables followed by log 10 transformation, mean-centering and Pareto-scaling prior to principal component analysis (PCA). Scores were plotted with samples coded for negative antagonist, negative pathogen, interaction and blank.
Subsequently, data from A. pullulans A1-B. cinerea TN080, A. pullulans A2-B. cinerea DAR69764 and A. pullulans A2-A. alternata (assigned to class interaction) and combined data from negative pathogen and blanks (assigned to class blank) were subjected to a partial least squares-discriminant analysis (PLS-DA) as a supervised classification method. PLS-DA was conducted using two latent variables. PLS-DA model efficacy was evaluated using a twofold evaluation. Firstly, model cross-validation29 was performed using a subset resampling methodology by omitting random subsets of the samples with ten data splits. Cross-validation was followed with a Monte Carlo permutation of the data class with the null hypothesis being no predictive acuity. Metrics of true model performance (Q2, area under the receiver operator curve (AUROC) and number of misclassifications) were compared to the permuted models (1,000 iterations) and empirical p-values determined from the proportion of test statistics from the random models equal to or higher than the true model30. Results of the permutation testing are presented in the supplementary material (Supplementary material Figs. S1,S2).
Selectivity ratios (SR)31 and variable importance in projection (VIP)31 scores from the PLS-DA model were used to determine the most important variables responsible for the discrimination between sample groups. Peaks with a VIP score of greater than one and with high SR scores were considered important target compounds for quantification and interaction studies.
Quantification of A. pullulans VOCs
Calibration curves (R2 > 0.99) were prepared for ethanol, 2-methyl-1-propanol, 3-methyl-1-butanol and 2-phenylethanol. A vial of target compounds in a water matrix (5 mL) was spiked with IS (2-methyl-4-pentanol, 0.04 mg/L) and analysed using SPME-GC-MS conditions as described above. At least six concentrations of each compound were used with two replicates. Concentrations of the four analytes in the interaction sample headspace were calculated.
Antifungal activity and synergistic effects of VOCs
The concentrations of compounds needed to be added to the mixture used for RSM were determined using a series of preliminary experiments as the PDA potentially absorb compounds which would confound the analytical results reliant on headspace concentration. Each compound mixture (500 µL) prepared in water, was added to a filter paper (Whatmann No. 1, 85 mm diameter) placed on one side of a Petri dish and covered immediately with a base Petri dish containing PDA (10 mL). The two dishes were immediately sealed with Parafilm and incubated at 25 °C. After three days of incubation, the headspace was manually sampled by SPME-GC-MS. The IS (2-methyl-4-pentanol, 330 mg/L, 5 µL) was spotted onto a paper disc glued on aluminium foil as described above and inserted into the double Petri dish setup immediately before sampling through a small pre-cut open of the Parafilm, keeping any loss of compounds to a minimum. The SPME fiber was manually inserted into the double Petri dish setup through a previously prepared hole on a side of the Petri dish and kept for 15 mins. The fibre was retracted, desorbed into the GC/MS inlet and VOCs were analysed using the same oven ramp conditions described above. The normalised peak areas of the four compounds in the double Petri dish headspace were compared with the normalised peak areas obtained from the original A. pullulans culture headspace in vials as the IS concentration in the two headspaces were identical. When the two normalised peak areas share a similar range, those concentrations added to double Petri dish setups were selected for the following RSM experiment.
Response surface modelling (RSM)
A Box–Behnken experimental design was applied to investigate the synergistic inhibitory effects of the four compounds (ethanol, 3-methyl-1-butanol, 2-methyl-1-propanol and 2-phenylethanol) on the growth of B. cinerea and A. alternata. A total of twenty seven experiments were performed, including three centre points. All variables were in three-coded factor levels: −1, 0, +1, corresponding to the low, mid and high levels respectively (Supplementary material Table S1). The top PDA plate of the double Petri dish was inoculated with a pathogen mycelial plug. The compound mixture (500 µL) was added to a filter paper in the bottom Petri dish. Colony diameter was measured after three days of incubation at 25 °C.
The following quadratic polynomial equation was fitted to model the relationship between four variables and the response,
where A, B, C and D represent the coded values of ethanol, 3-methyl-1-butanol, 2-methyl-1-propanol and 2-phenylethanol respectively. Y is the predicted response, β0 is the intercept term, βi is the linear coefficient, βij is the interaction coefficient and βii is the quadratic coefficient. Depending on the significant terms and other statistical parameters of validation, models were simplified and fitted by suppressing quadratic terms and certain interaction terms which were not statistically significant (P > 0.05). Simplified final models are presented in the results section. Design generation, analysis and optimisation were carried out using Design Expert STAT Ease software 7.0 version (Minneapolis, USA).
Statistical analysis
The data from the mycelium growth inhibition assay and the conidia germination assay were subjected to an analysis of variance (ANOVA) with Tukey’s mean separation test to evaluate significant differences among treatments at a 95% confidence level. Analyses were conducted using SPSS ver. 24.0 (IBM SPSS Statistics 24).
Results
Reduced mycelium growth and conidia germination of B. cinerea and A. alternata by A. pullulans VOCs
Mycelium growth
VOCs of the four A. pullulans isolates significantly inhibited (P < 0.05) the colony growth of the three pathogens. The degree of inhibition after three days of incubation varied between isolates (Fig. 2a–c). A. pullulans A1 VOCs resulted in the highest inhibition of B. cinerea TN080, whereas B. cinerea DAR69764 and A. alternata were highly inhibited by A. pullulans A2 VOCs. In comparison with controls, these inhibitions were 74%, 84% and 47% for B. cinerea TN080, B. cinerea DAR69764 and A. alternata respectively. Among the four A. pullulans isolates examined in this study, A. pullulans A3 headspace inhibited the growth of B. cinerea TN080, B. cinerea DAR69764 and A. alternata the least,(Fig. 2a–c).
Conidia germination and morphological deformations
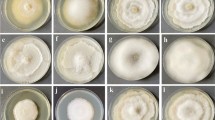
VOCs of the four A. pullulans isolates supressed conidia germination of the three pathogens to various degrees (Fig. 2d–f). These results were consistent with the mycelium growth inhibition assay. Conidia germination of B. cinerea TN080 was highly inhibited by A. pullulans A1 headspace whereas the highest inhibition of conidia germination of B. cinerea DAR69764 and A. alternata were found when exposed to A. pullulans A2 VOCs. The least inhibition of conidia germination was observed in the presence of A. pullulans A3 against all three pathogen isolates. Light microscopic images of pathogen conidia exposed to headspace of the highest inhibitory A. pullulans isolates; B. cinerea TN080 with A. pullulans A1 VOCs (Fig. 3a), B. cinerea DAR69764 with A. pullulans A2 VOCs (Fig. 3b), and A. alternata with A. pullulans A2 VOCs (Fig. 3c), revealed that exposure to A. pullulans VOCs may cause damage to the cell wall and cell membrane, resulting in the release of cellular content.
The effect of VOCs produced by isolates of A. pullulans (A1–A4) on the mycelium growth and conidia germination. Mycelium growth of (a) B. cinerea TN080, (b) B. cinerea DAR69764 and (c) A. alternata after three days of incubation. Error bars indicate standard error of mean of the six replicates. Suppression of conidia germination of (d) B. cinerea TN080, (e) B. cinerea DAR69764 and (f) A. alternata by exposure to VOCs of A. pullulans. Error bars indicate standard error of mean of the three replicates. In every graph, ‘C’ represents the control. Data of the mycelium growth and conidia germination were subjected to analysis of variance. Different letters represent significant differences (P < 0.05) explained by one way ANOVA with Tukey’s mean separation test.
Deformed germ tubes and conidia upon exposure to A. pullulans VOCs; (a) B. cinerea TN080 with A. pullulans A1 VOCs, (b) B. cinerea DAR69764 with A. pullulans A2 VOCs and (c) A. alternata with A. pullulans A2 VOCs, in comparison with non-fumigated controls; (d) B. cinerea TN080, (e) B. cinerea DAR69764 and (f) A. alternata. Arrow heads indicate disrupted germ tubes and conidia.
PCA discriminated between A. pullulans inoculated and non-inoculated headspace
Based on the results of the two antifungal assays described above, A. pullulans isolates were selected for VOCs analysis, A. pullulans isolate A1 which inhibited B. cinerea TN080, isolate A2 which inhibited B. cinerea DAR69764 and A. alternata the greatest and A. pullulans isolate A3 which had the least inhibitory activity towards all fungal pathogens examined in this study.
MCR-ALS analysis resulted in peak areas of forty three compounds from culture headspace comprising volatile substances produced by the antagonist (three A. pullulans isolates), pathogen (B. cinerea TN080, B. cinerea DAR 69764 and A. alternata), culture medium and potential fibre and column contaminants. The normalised peak table was used for PCA. The first latent variable of the scores plot shows the separation along PC1 (Fig. 4a), describing blank and negative pathogen samples separation from negative antagonist and interaction samples. This explains 48% of data variance along PC1. As PCA is an unsupervised method, the clear separation between the two groups instinctively confirms differences in the volatile composition of headspace between antagonist inoculated and non-inoculated samples. The corresponding loadings plot (Fig. 4b) shows variables located on the right side from the origin in PC1 influence the discrimination of A. pullulans inoculated samples where compounds 14, 29 (Table 1) and 37 seem to have a greater influence on this discrimination. There was no clear discrimination between negative antagonist and interaction samples (Fig. 4a).
(a) PCA scores plot of GC-MS normalised peaks obtained from culture vial headspace of 52 samples, showing PC1 vs. PC2. Four categories of samples on PCA scores plot include blank (non-inoculated media), interaction (inoculated with both A. pullulans and respective pathogen), negative antagonist (either A. pullulans A1 or A. pullulans A2 or A. pullulans A3 only) and negative pathogen (either B. cinerea TN080 or B. cinerea DAR 69764 or A. alternata only) each with four replicates. (b) PCA loadings plot shows PC1 vs. PC2 with forty one variables identified from culture headspace. Variables include volatile substances produced by the antagonist, pathogen, culture medium and potential fiber and column contaminants.
Identification of fourteen VOCs from A. pullulans headspace
MCR-ALS extracted spectra for forty four variables (including IS) were imported to NIST mass spectral library and were tentatively identified by comparing with those reference spectra from the library. Fifteen compounds were filtered out with greater than 80% of similarity to those of reference compounds and appear to be produced by A. pullulans (Table 1). Seven alcohols, three ketones and four esters were among the fifteen compounds. One compound was reported as unknown (Table 1). The identity of these compounds were further confirmed by comparing their retention times with authentic standards and with LRI derived from the alkane series run in an identical GC oven ramp. These fifteen compounds were produced by all three isolates of A. pullulans (Supplementary material Fig. S5).
PLS-DA with VIP and SR tests identified four important VOCs for sample discrimination
PLS-DA was performed using peak areas of two sample classes (interaction and blank). According to the VIP and SR scores, four compounds possessed the greatest explanatory power when separating the A. pullulans inoculated interaction samples from blanks along PC1. These compounds were ethanol, 2-methyl-1-propanol, 3-methyl-1-butanol and 2-phenylethanol.
The quality of the fitted model was excellent as shown in Supplementary material Figs. S1,S2. There is a clear distinction between the permuted distribution and the original data based on number of misclassifications, the Q2 values and the AUROC values for both classes (interaction and blank). The AUROC values close to 1 indicate a perfect separation between the classes. The significant p values obtained for Q2 values (P = 0.001 for class interaction, P = 0.024 for class blank) and number of misclassifications (P = 0.001 for class interaction, P = 0.024 for class blank) based on the differences in the permuted and true data, indicate a good predictability of the applied model.
Quantified A. pullulans VOCs in the interaction headspace
Four VOCs identified from the PLS-DA with high discriminatory value were quantified in the interaction headspace (Table 2). As observed in Table 2, ethanol was produced in extremely high concentrations in all three systems when compared to the other three VOCs.
RSM optimised four antifungal VOCs to synergistically minimise B. cinerea and A. alternata colony growth
Concentrations of ethanol, 2-methyl-1-propanol, 3-methyl-1-butanol and 2-phenylethanol were optimised to minimise colony growth of A. alternata and B. cinerea using a Box-Behnken experimental design followed by RSM (Fig. 5, Table 3). The following simplified polynomial response surfaces were fitted on colony diameter of (Y);
Response surface and contour plots showing significant interactive pairs of VOCs. (a) Response surface between 3-methyl-1-butanol and 2-phenylethanol, (c) contour plot between 3-methyl-1-butanol and 2-phenylethanol, show a significant interaction against A. alternata colony growth. (b) Response surface between ethanol and 2-phenylethanol and (d) contour plot between ethanol and 2-phenylethanol show a significant interaction on B. cinerea DAR69764 colony growth reduction. Colony diameter reduces from red to blue on response surfaces and contour plots. Plots have created using the coded levels of concentrations for each VOC.
Model F-values 81.51 (Eq. (2)), 92.07 (Eq. (3)) and 21.08 (Eq. (4)) were significant using p ≤ 0.05. Lack of fit F-values were non-significant (P > 0.05) and residuals were normally distributed for the three models. R2 values 0.9575 (Eq. (2)), 0.9456 (Eq. (3)) and 0.8001 (Eq. (4)) obtained for the three models, indicated that these models were accurate for predicting responses. The main effects of the compounds and second order interactions were determined based on the fitted equations. The sign of the β coefficients of the polynomial surface response indicated a positive (increased growth) or negative (reduced growth) impact on the colony growth. The size of the effect was determined from the level of significance expressed by the p-value (Supplementary material Table S2).
The colony diameter of A. alternata was significantly reduced by ethanol (P < 0.0001, term A) and 2-phenylethanol (P = 0.0170, term D). However, the interaction between these two compounds was not significant (P > 0.05), thus this term was removed from the final model (Eq. (2)). The interaction between 3-methyl-1-butanol and 2-phenylethanol caused a significant (P = 0.0255) positive impact which is an increased growth. The response surface plot (Fig. 5a) and contour plot (Fig. 5c) of 3-methyl-1-butanol and 2-phenylethanol indicated a reduction of the diameter of A. alternata when one of the two compounds of the pair is at its highest concentration and the other compound is at the lowest concentration. According to the fitted linear model (Eq. (3), Supplementary material Table S2), all four compounds individually exert a significant effect (P < 0.05) on the colony growth reduction of B. cinerea TN080. None of the second order interactions between these compounds were significant in terms of the impact on colony growth. B. cinerea DAR69764 colony growth was significantly reduced by ethanol (P < 0.0001, term A) and 2-phenylethanol (P = 0.0002, term D) (Supplementary material Table S2). The interaction between the two compounds was also significant (P = 0.0293). According to the response surface plot (Fig. 5b) and contour plot (Fig. 5d) the lowest colony diameter was observed when both compounds were at their highest concentrations. The three linear models were validated by testing mixtures of three different concentrations of the four compounds.
The minimum colony diameters predicted by RSM were 14.5 mm, 16.84 mm and 10.14 mm for A. alternata, B. cinerea TN080 and B. cinerea DAR69764 respectively. The concentrations of the four compounds were optimised together for the two B. cinerea isolates. The optimised concentrations of the four compounds in the mixture can be observed in Table 3.
Discussion
Fungicides and other chemical-based methods for plant disease management have become less popular in recent years due to public concern about environmental pollution, issues with pesticide resistance and the removal of pesticide registrations32. The utilisation of biocontrol agents such as microbial VOCs represent a more environmentally friendly4,5 alternative for plant disease management. Research on microbial VOCs fumigation has demonstrated successful antifungal effects on a large spectrum of pathogenic fungi and bacteria3,33. Despite these advances, plant diseases caused by B. cinerea and A. alternata remain a challenge14,34.
A. pullulans has been widely studied as a biological control agent7,8. A variety of mechanisms have been postulated to describe its antifungal properties including enhanced natural host defences7, competition for nutrients9,35 and antifungal VOCs production11. The latter has been a recently investigated area and not well characterised to date. In order to better describe the biological control potential of A. pullulans against B. cinerea and A. alternata, its ability to produce antifungal VOCs was studied by incorporating a chemometrics approach.
Knowing the negative effects of microbial VOCs on fungal physiological functions including mycelial growth, spore germination, sporulation and sclerotial development11,33, we studied the antifungal effects of A. pullulans VOCs on two fungi B. cinerea and A. alternata, responsible for disease losses in a wide range of agricultural crop plants. We observed a strong reduction of pathogen mycelial growth and conidia germination of the two pathogens by A. pullulans VOCs. A notable, less dense and weak mycelial mat was evident and a clear disruption of conidial germ tubes was noted in exposed mycelia and conidia (Fig. 3a–c).
Previous studies on volatile analysis have identified a number of VOCs in the A. pullulans culture headspace12. In our study, headspace-SPME-GC-MS analysis incorporated with MCR-ALS revealed fifteen VOCs produced by A. pullulans and fourteen of them were identified. Thus, the observed inhibitory activity is due to the activity of a set of A. pullulans VOCs which include alcohols, ketones and esters. 3-Methyl-1-butanol, 2-methyl-1-propanol and 2-phenylethanol, identified from A. pullulans headspace in our study were previously recorded from A. pullulans headspace and were also tested against B. cinerea, Penicillium and Colletotrichum sp. and observed notable inhibitory activities11. To the best of the authors’ knowledge, 3-methyl-1-hexanol, acetone, 2-heptanone, ethyl butyrate, 3-methylbutyl acetate and 2-methylpropyl acetate were identified for the first time to be produced by A. pullulans isolates. However, these compounds have been previously reported in the culture headspace of other microbial organisms: 3-methyl-1-hexanol from Lysobacter species36, 2-heptanone from Escherichia coli37, 3-methylbutyl acetate from Sporobolomyces roseus12, acetone from Muscodor albus24 and ethyl butyrate and 2-methylpropyl acetate from Saccharomyces cerevisiae38.
Quantitation of VOCs is as crucial as identification, especially in the evaluation of antifungal activity of VOCs in artificial cocktails. Quantification studies of microbial VOCs so far has mainly been based on relative peak areas of compounds on chromatograms17,39 while some of the studies tested random mixtures with various concentrations40. However, a lack of IS makes a comparison of VOCs efficacy challenging mainly due to variations in the SPME performance between injections19. As SPME technique, does not reflect the real situation, previous studies have also mentioned the importance of using standards to quantify real VOCs atmosphere concentration11. We observed an area ratio of 2:1:1.2 between ethanol: 3-methyl-1-butanol and 2-phenylethanol while actual quantification using authentic standards and IS resulted in a ratio of 397:1:2.5 for the three compounds respectively in the A. pullulans (A1) and B. cinerea TN080 interaction headspace. This indicates the necessity of accurate quantification of VOCs. A previous study reported that Muscodor albus VOCs did not mimic the natural atmospheric conditions on some of the test organisms such as Candida albicans, Sclerotinia sclerotiorum and Aspergillus fumigatus24. The authors suspected a key ingredient was missing in the artificial test atmosphere to cause a biocidal activity on those fungal cells. However, artificial volatile formulations based on incorrect proportions could be an alternative possibility for dissimulation of the natural atmospheric conditions.
Our study introduces a novel setup (Fig. 1) to directly extract VOCs from the culture headspace by automated SPME-GC-MS analysis, where both antagonist and pathogen are present simultaneously and actively interacting; unlike previous studies, we also used an IS to facilitate the quantification of VOCs. This is the first report to introduce a direct system for automated extraction of microbial VOCs from a dual culture setup which also facilitates quantification of volatiles. In a previous study41, microbial VOCs quantification was carried out using IS, in which aliquots of the original liquid cultures were transferred to headspace vials and an IS was added followed by SPME-GC-MS analysis. Headspace analysis of the original culture provides a reliable quantitative approach, as it prevents losing a portion of VOCs during transfer, which was actually produced in the culture and partitions into the headspace. This is important, particularly given that volatility varies among different compounds. In this regard, the technique proposed here represents a more reliable and robust approach for the accurate quantification of microbial VOCs. As we observed in the preliminary experiments of the present study, selecting suitable IS which are non-inhibitory to the test organisms and persist in the culture system throughout the growth period without being absorbed by the mycelia is challenging. These reasons may have discouraged the application of IS in previous bio-control studies. Moreover, a promising method to introduce a suitable IS to a culture system without disturbing its headspace upon completion of the growth period is required. Our study has introduced a reliable, robust approach to overcome this issue (Fig. 1).
When combined the untargeted feature extraction process with PLS-DA it is possible to determine which features, or combination of features, from the extracted data table of peak areas enable separation of specific sample groups when these are modelled. By numerically defining specific class membership (−1 or +1) and using a projection to latent structures approach, the PLS algorithm seeks to predict the class membership by maximising the co-variance between the peak area table and the class membership values. In the present investigation we defined class membership for PLS-DA as ‘media blank’ + ‘pathogen’ and ‘interaction samples’ being those samples with growth of the antagonist and pathogen. Effectively peak areas that are derived from the media blank and from growth of the pathogen only, are mathematically subtracted from the peak areas of the remaining samples so that discrimination of the modelled classes is achieved. Thus, PLS-DA successfully discriminated between A. pullulans inoculated interaction samples and the non-inoculated culture headspace (Fig. 4b). Permutation testing metrices of Q2, AUROC and number of misclassifications (Supplementary material Fig. S1,S2) confirmed the robustness of the true model to explain the discrimination between A. pullulans and pathogen inoculated class interaction and the class blank (P < 0.001). Ethanol, 2-methyl-1-propanol, 3-methyl-1-butanol and 2-phenylethanol were identified as the most effective VOCs to discriminate two sample classes (Fig. 4b). Our RSM (Fig. 5, Supplementary material Table S2) with these four VOCs revealed, ethanol and 2-phenylethanol have significant effects on supressing colony growth of both B. cinerea and A. alternata. Significant differences are evident between the different antagonist isolates and controls for inhibition of the pathogen mycelial growth. This is in agreement with the PCA scores plot (Fig. 4a) which shows a clear discrimination of blanks and pathogen samples from A. pullulans and interaction samples along PC1 axis. No clear separation between different A. pullulans isolates or between specific antagonist-pathogen combinations is apparent (Supplementary material Fig. S3).
The efficacy of ethanol as a biocontrol agent has been previously demonstrated in other studies. Previous work on ethanol fumigation on table grapes at low temperature postharvest storage successfully controlled B. cinerea42,43. Also ethanol in combinations with other alternatives such as potassium sorbate44 and calcium chloride42, was used to improve disease control against B. cinerea. Exposure to ethanol solutions at low temperatures reduced spore germination of A. alternata43. However, 1,000 µg/mL of ethanol only had a transitory inhibitory effect on A. alternata45 which agrees with our findings that the optimised concentration of ethanol by RSM is in the order of 106 to control A. alternata in vitro. This means ethanol could be effective in extremely high concentrations. Likewise, 2-phenylethanol has also been demonstrated to have antimicrobial activity against various pathogenic fungi such as Penicillium italicum46, Aspergillus flavus47, B. cinerea and Colletotrichum acutatum11.
Various mechanisms have been hypothesised for microbial VOCs mediated antifungal effects. 2-Phenylethanol mediated inhibition of P. italicum cells identified to be highly associated with interactions with the mitochondria and the nucleus47. The mechanism behind the toxicity of alcohols, including ethanol and larger alkanols is likely due to their role in membrane disruption, leading to the dissipation of the proton gradient of affected cells48 which subsequently interferes and/or inhibits regular cell metabolic functions49.
There are differences in sensitivity to these compounds among fungal species50,51. Thus, optimal treatments for different fungal pathogens will be different. Differential sensitivity could be explained by differences in the composition of membranes (e.g. lipid to protein ratio) and cell walls between fungal species and involvement of other consequences such as oxidative stress and imbalanced cellular redox homeostasis to different degrees upon volatile fumigation52 and impacts on enzymes related to energy-generating pathways and metabolism53. This may explain why we found A. alternata was less sensitive to A. pullulans VOCs than B. cinerea.
The use of RSM in our study revealed interactive effects between A. pullulans VOCs in the inhibition of B. cinerea and A. alternata colony growth. Many studies report a single VOCs is not lethal to a particular pathogen when studied in isolation, and hypothesise a synergistic or additive effect of different compounds24. Microbial VOCs are produced at low concentrations by organisms, which also suggest their effect to be synergistic or additive25. These assumptions were further confirmed in our study (Fig. 5). Interestingly, 3-methyl-1-butanol11,54 and 2-phenylethanol11,46 which are two well-known antifungal compounds, increased the growth of A. alternata when present in an interaction whereas their individual effects were negative. Further experiments are essential to confirm and investigate the mechanisms behind these positive interactive effects.
There are instances where artificial environments did not fully replicate the inhibitory effects of pathogens which were observed in natural antagonist headspace16,24. In addition to relying on relative peak areas of VOCs, the absorption of VOCs to the culture medium also needs to be considered when preparing accurate formulations of artificial VOCs mixtures for antifungal assays. However, no prior studies appear to have considered this effect. According to our observations, it was evident that higher concentrations of VOCs were required to be added to the artificial system (Supplementary material Table S1) compared to original headspace concentrations (Table 2). This indicates antagonists may produce these compounds in very high levels whereas we measure the amounts in headspace which are in an equilibrium after absorption into the growth medium.
In conclusion, a novel extraction setup was designed for automated extraction of VOCs by headspace-SPME-GC-MS for quantification of volatiles, when both antagonist and pathogen are in an interaction. The proposed system allows the introduction of an IS, without losing compounds. A total of fourteen VOCs were identified from A. pullulans by untargeted metabolomics followed by MCR-ALS. These included seven alcohols, three ketones and four esters. PCA and PLS-DA were further employed to discriminate between the volatile profiles of antagonist inoculated and non-inoculated samples. Four VOCs were identified as the most important features in discriminating samples based on the SR and VIP scores. Ethanol and 2-phenylethanol were found as key inhibitory A. pullulans VOCs for both B. cinerea and A. alternata. RSM optimised a cocktail of ethanol, 3-methyl-1-butanol, 2-methyl-1-propanol and 2-phenylethanol for the first time to minimise colony growth of B. cinerea and A. alternata in vitro.
Data availability
All relevant datasets generated during and/or analysed during the current study are promptly available from the corresponding author upon request.
References
Yuan, Y. et al. Potential of endophytic fungi isolated from cotton roots for biological control against Verticillium wilt disease. PLoS One 12, e0170557, https://doi.org/10.1371/journal.pone.0170557 (2017).
Tahir, H. A. et al. Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci. Rep. 7, 40481, https://doi.org/10.1038/srep40481 (2017).
Raza, W., Ling, N., Yang, L., Huang, Q. & Shen, Q. Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci. Rep. 6, 24856, https://doi.org/10.1038/srep24856 (2016).
Qin, X., Xiao, H., Cheng, X., Zhou, H. & Si, L. Hanseniaspora uvarum prolongs shelf life of strawberry via volatile production. Food Microbiol. 63, 205–212, https://doi.org/10.1016/j.fm.2016.11.005 (2017).
Spadaro, D. & Droby, S. Unraveling the mechanisms used by antagonistic yeast to control postharvest pathogens on fruit. Acta Hortic, 63–70, https://doi.org/10.17660/ActaHortic.2016.1144.9 (2016).
Pertot, I. et al. Combining biocontrol agents with different mechanisms of action in a strategy to control Botrytis cinerea on grapevine. Crop. Prot. 97, 85–93, https://doi.org/10.1016/j.cropro.2017.01.010 (2017).
Madhupani, Y. D. S. & Adikaram, N. K. B. Delayed incidence of stem-end rot and enhanced defences in Aureobasidium pullulans-treated avocado (Persea americana Mill.) fruit. J. Plant. Dis. Prot. 124, 227–234, https://doi.org/10.1007/s41348-017-0086-8 (2017).
Rathnayake, R. M. S. P., Savocchia, S., Schmidtke, L. M. & Steel, C. C. Characterisation of Aureobasidium pullulans isolates from Vitis vinifera and potential biocontrol activity for the management of bitter rot of grapes. Eur. J. Plant. Pathol. 151, 593–611, https://doi.org/10.1007/s10658-017-1397-0 (2018).
Australian Pesticides and Veterinary Medicines Authority. Public release summary on the evaluation of the new active Aureobasidium pullulans (strains DSM 14940 and DSM 14941) in the product botector fungicide, https://apvma.gov.au/node/27401 (2017).
Chen, P. H., Chen, R. Y. & Chou, J. Y. Screening and evaluation of yeast antagonists for biological control of Botrytis cinerea on strawberry fruits. Mycobiology 46, 33–46, https://doi.org/10.1080/12298093.2018.1454013 (2018).
Di Francesco, A., Ugolini, L., Lazzeri, L. & Mari, M. Production of volatile organic compounds by Aureobasidium pullulans as a potential mechanism of action against postharvest fruit pathogens. Biol. Control. 81, 8–14, https://doi.org/10.1016/j.biocontrol.2014.10.004 (2015).
Verginer, M., Leitner, E. & Berg, G. Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 58, 8344–8350, https://doi.org/10.1021/jf100393w (2010).
Elad, Y., Williamson, B., Tudzynski, P. & Delen, N. Botrytis: biology, pathology and control. (Springer, 2004).
Estiarte, N., Crespo-Sempere, A., Marin, S., Sanchis, V. & Ramos, A. J. Effect of 1-methylcyclopropene on the development of black mold disease and its potential effect on alternariol and alternariol monomethyl ether biosynthesis on tomatoes infected with Alternaria alternata. Int. J. Food Microbiol. 236, 74–82, https://doi.org/10.1016/j.ijfoodmicro.2016.07.001 (2016).
Steel, C. C., Blackman, J. W. & Schmidtke, L. M. Grapevine bunch rots: impacts on wine composition, quality, and potential procedures for the removal of wine faults. J. Agric. Food Chem. 61, 5189–5206, https://doi.org/10.1021/jf400641r (2013).
Rouissi, W., Ugolini, L., Martini, C., Lazzeri, L. & Mari, M. Control of postharvest fungal pathogens by antifungal compounds from Penicillium expansum. J. Food Prot. 76, 1879–1886, https://doi.org/10.4315/0362-028X.JFP-13-072 (2013).
Zheng, M., Shi, J. Y., Shi, J., Wang, Q. G. & Li, Y. H. Antimicrobial effects of volatiles produced by two antagonistic Bacillus strains on the anthracnose pathogen in postharvest mangos. Biol. Control. 65, 200–206, https://doi.org/10.1016/j.biocontrol.2013.02.004 (2013).
Fialho, M. B., Toffano, L., Pedroso, M. P., Augusto, F. & Pascholati, S. F. Volatile organic compounds produced by Saccharomyces cerevisiae inhibit the in vitro development of Guignardia citricarpa, the causal agent of citrus black spot. World J. Microbiol. Biotechnol. 26, 925–932, https://doi.org/10.1007/s11274-009-0255-4 (2009).
Rebiere, L., Clark, A. C., Schmidtke, L. M., Prenzler, P. D. & Scollary, G. R. A robust method for quantification of volatile compounds within and between vintages using headspace-solid-phase micro-extraction coupled with GC-MS–application on Semillon wines. Anal. Chim. Acta 660, 149–157, https://doi.org/10.1016/j.aca.2009.10.029 (2010).
Fortini, M., Migliorini, M., Cherubini, C., Cecchi, L. & Calamai, L. Multiple internal standard normalization for improving HS-SPME-GC-MS quantitation in virgin olive oil volatile organic compounds (VOO-VOCs) profile. Talanta 165, 641–652, https://doi.org/10.1016/j.talanta.2016.12.082 (2017).
Roussel, S., Preys, S., Chauchard, F. & Lallemand, J. In Process Analytical Technology for the Food Industry Food Engineering Series Ch. Chapter 2, 7–59 (2014).
Khakimov, B., Gürdeniz, G. & Engelsen, S. B. Trends in the application of chemometrics to foodomics studies. Acta Aliment. 44, 4–31, https://doi.org/10.1556/AAlim.44.2015.1.1 (2015).
Schmidtke, L. M., Blackman, J. W., Clark, A. C. & Grant-Preece, P. Wine metabolomics: objective measures of sensory properties of semillon from GC-MS profiles. J. Agric. Food Chem. 61, 11957–11967, https://doi.org/10.1021/jf403504p (2013).
Strobel, G. A., Dirkse, E., Sears, J. & Markworth, C. Volatile antimicrobials from Muscodor albus, a novel endophytic fungus. Microbiology 147, 2943–2950, https://doi.org/10.1099/00221287-147-11-2943 (2001).
Mercier, J. & Jimenez, J. I. Control of fungal decay of apples and peaches by the biofumigant fungus Muscodor albus. Postharvest Biol. Tec. 31, 1–8, https://doi.org/10.1016/j.postharvbio.2003.08.004 (2004).
Gotor-Vila, A. et al. Antifungal effect of volatile organic compounds produced by Bacillus amyloliquefaciens CPA-8 against fruit pathogen decays of cherry. Food Microbiol. 64, 219–225, https://doi.org/10.1016/j.fm.2017.01.006 (2017).
Schueuermann, C. et al. A GC-MS untargeted metabolomics approach for the classification of chemical differences in grape juices based on fungal pathogen. Food Chem. 270, 375–384, https://doi.org/10.1016/j.foodchem.2018.07.057 (2019).
van Den Dool, H. & Dec. Kratz, P. A generalization of the retention index system including linear temperature programmed gas—liquid partition chromatography. J. Chromatogr. A 11, 463–471, https://doi.org/10.1016/s0021-9673(01)80947-x (1963).
Westerhuis, J. A. et al. Assessment of PLSDA cross validation. Metabolomics 4, 81–89, https://doi.org/10.1007/s11306-007-0099-6 (2008).
Szymanska, E., Saccenti, E., Smilde, A. K. & Westerhuis, J. A. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 8, 3–16, https://doi.org/10.1007/s11306-011-0330-3 (2012).
Farrés, M., Platikanov, S., Tsakovski, S. & Tauler, R. Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J. Chemom. 29, 528–536, https://doi.org/10.1002/cem.2736 (2015).
Wisniewski, M., Droby, S., Norelli, J., Liu, J. & Schena, L. Alternative management technologies for postharvest disease control: The journey from simplicity to complexity. Postharvest Biol. Tec. 122, 3–10, https://doi.org/10.1016/j.postharvbio.2016.05.012 (2016).
Massawe, V. C. et al. Volatile organic compounds of endophytic Bacillus spp. have biocontrol activity against Sclerotinia sclerotiorum. Phytopathology, https://doi.org/10.1094/PHYTO-04-18-0118-R (2018).
Saravanakumar, D., Clavorella, A., Spadaro, D., Garibaldi, A. & Gullino, M. L. Metschnikowia pulcherrima strain MACH1 outcompetes Botrytis cinerea, Alternaria alternata and Penicillium expansum in apples through iron depletion. Postharvest Biol. Tec. 49, 121–128, https://doi.org/10.1016/j.postharvbio.2007.11.006 (2008).
Bencheqroun, S. K. et al. In vitro and in situ study of postharvest apple blue mold biocontrol by Aureobasidium pullulans: Evidence for the involvement of competition for nutrients. Postharvest Biol. Tec. 46, 128–135, https://doi.org/10.1016/j.postharvbio.2007.05.005 (2007).
Lazazzara, V. et al. Growth media affect the volatilome and antimicrobial activity against Phytophthora infestans in four Lysobacter type strains. Microbiol. Res. 201, 52–62, https://doi.org/10.1016/j.micres.2017.04.015 (2017).
Chen, J., Tang, J., Shi, H., Tang, C. & Zhang, R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J. Basic. Microbiol. 57, 228–237, https://doi.org/10.1002/jobm.201600505 (2017).
Morales, M. L., Fierro-Risco, J., Callejon, R. M. & Paneque, P. Monitoring volatile compounds production throughout fermentation by Saccharomyces and non-Saccharomyces strains using headspace sorptive extraction. J. Food Sci. Technol. 54, 538–557, https://doi.org/10.1007/s13197-017-2499-6 (2017).
Toffano, L., Fialho, M. B. & Pascholati, S. F. Potential of fumigation of orange fruits with volatile organic compounds produced by Saccharomyces cerevisiae to control citrus black spot disease at postharvest. Biol. Control. 108, 77–82, https://doi.org/10.1016/j.biocontrol.2017.02.009 (2017).
Xie, S., Zang, H., Wu, H., Uddin Rajer, F. & Gao, X. Antibacterial effects of volatiles produced by Bacillus strain D13 against Xanthomonas oryzae pv. oryzae. Mol. Plant. Pathol. 19, 49–58, https://doi.org/10.1111/mpp.12494 (2018).
Gethins, L. et al. Influence of carbon and nitrogen source on production of volatile fragrance and flavour metabolites by the yeast Kluyveromyces marxianus. Yeast 32, 67–76, https://doi.org/10.1002/yea.3047 (2015).
Chervin, C., Westercamp, P. & Monteils, G. Ethanol vapours limit Botrytis development over the postharvest life of table grapes. Postharvest Biol. Tec. 36, 319–322, https://doi.org/10.1016/j.postharvbio.2005.02.001 (2005).
Gabler, F. M., Mansour, M. F., Smilanick, J. L. & Mackey, B. E. Survival of spores of Rhizopus stolonifer, Aspergillus niger, Botrytis cinerea and Alternaria alternata after exposure to ethanol solutions at various temperatures. J. Appl. Microbiol. 96, 1354–1360 (2004).
Guha, P. & Basak, S. Optimization of ethanol and potassium sorbate treatment for minimizing postharvest losses in onion bulbs (Allium cepa L. Cv. Sukhsagar). Int. J. Agriculture Food Sci. Technol. 4, 561–570 (2013).
Prusky, D., Perez, A., Zutkhi, Y. & Ben-Arie, R. Effect of modified atmosphere for control of black spot, caused by Alternaria alternata, on stored persimmon fruits. Postharvest Pathol. Mycotoxins 87, 203–208 (1997).
Liu, P. et al. Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiol. 14, 242, https://doi.org/10.1186/s12866-014-0242-2 (2014).
Hua, S. S., Beck, J. J., Sarreal, S. B. & Gee, W. The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 30, 71–78, https://doi.org/10.1007/s12550-014-0189-z (2014).
Alpha, C. J., Campos, M., Jacobs-Wagner, C. & Strobel, S. A. Mycofumigation by the volatile organic compound-producing fungus Muscodor albus induces bacterial cell death through DNA damage. Appl. Env. Microbiol. 81, 1147–1156, https://doi.org/10.1128/AEM.03294-14 (2015).
Dantigny, P., Tchobanov, I., Bensoussan, M. & Zwietering, M. H. Modeling the effect of ethanol vapor on the germination time of Penicillium chrysogenum. J. Food Prot. 68, 1203–1207, https://doi.org/10.4315/0362-028x-68.6.1203 (2005).
Dao, T. & Dantigny, P. Control of food spoilage fungi by ethanol. Food Control. 22, 360–368, https://doi.org/10.1016/j.foodcont.2010.09.019 (2011).
Gao, H., Li, P., Xu, X., Zeng, Q. & Guan, W. Research on volatile organic compounds from Bacillus subtilis cf-3: Biocontrol effects on fruit fungal pathogens and dynamic changes during fermentation. Front. Microbiol. 9, 456, https://doi.org/10.3389/fmicb.2018.00456 (2018).
Fialho, M. B., Carvalho, G., Martins, P. F., Azevedo, R. A. & Pascholati, S. F. Antioxidative response of the fungal plant pathogen Guignardia citricarpa to antimicrobial volatile organic compounds. Afr. J. Microbiol. Res. 8, 2077–2084, https://doi.org/10.5897/ajmr2014.6719 (2014).
Fialho, M. B. et al. Proteomic response of the phytopathogen Phyllosticta citricarpa to antimicrobial volatile organic compounds from Saccharomyces cerevisiae. Microbiol. Res. 183, 1–7, https://doi.org/10.1016/j.micres.2015.11.002 (2016).
Dalilla, C. R., Mauricio, B. F., Simone, C. B., Silvia, B. & Sergio, F. P. Antimicrobial activity of volatile organic compounds and their effect on lipid peroxidation and electrolyte loss in Colletotrichum gloeosporioides and Colletotrichum acutatum mycelia. Afr. J. Microbiology Res. 9, 1527–1535, https://doi.org/10.5897/ajmr2015.7425 (2015).
Goodner, K. L. Practical retention index models of OV-101, DB-1, DB-5, and DB-Wax for flavor and fragrance compounds. LWT 41, 951–958, https://doi.org/10.1016/j.lwt.2007.07.007 (2008).
Santos, A., Fernandes, A., Wagner, R., Jacob-Lopes, E. & Zepka, L. Biogeneration of volatile organic compounds produced by Phormidium autumnale in heterotrophic bioreactor. J. Appl. Phycol. 28, 1561–1570, https://doi.org/10.1007/s10811-015-0740-0 (2015).
Li, J. et al. Characterization of volatile compounds in grass carp (Ctenopharyngodon idellus) soup cooked using a traditional chinese method by GC-MS. J. Food Process. Preserv 41, https://doi.org/10.1111/jfpp.12995 (2017).
Sonmezdag, A. S., Kelebek, H. & Selli, S. Pistachio oil (Pistacia vera L. cv. Uzun): Characterization of key odorants in a representative aromatic extract by GC-MS-olfactometry and phenolic profile by LC-ESI-MS/MS. Food Chem. 240, 24–31, https://doi.org/10.1016/j.foodchem.2017.07.086 (2018).
Verzera, A., Campisi, S., Zappala, M. & Bonaccorsi, I. SPME-GC-MS analysis of honey volatile components for the characterization of different floral origin. Am. Laboratory 33, 18–21 (2001).
Lieb, V. M., Esquivel, P., Cubero Castillo, E., Carle, R. & Steingass, C. B. GC-MS profiling, descriptive sensory analysis, and consumer acceptance of Costa Rican papaya (Carica papaya L.) fruit purees. Food Chem. 248, 238–246, https://doi.org/10.1016/j.foodchem.2017.12.027 (2018).
Du, X. & Rouseff, R. Aroma active volatiles in four southern highbush blueberry cultivars determined by gas chromatography-olfactometry (GC-O) and gas chromatography-mass spectrometry (GC-MS). J. Agric. Food Chem. 62, 4537–4543, https://doi.org/10.1021/jf500315t (2014).
Liu, J., Arneborg, N., Toldam-Andersen, T. B., Petersen, M. A. & Bredie, W. L. Effect of sequential fermentations and grape cultivars on volatile compounds and sensory profiles of Danish wines. J. Sci. Food Agric. 97, 3594–3602, https://doi.org/10.1002/jsfa.8218 (2017).
Author information
Authors and Affiliations
Contributions
S.M.Y.D., L.M.S. and C.C.S. designed and planned the study. S.M.Y.D. conducted laboratory experiments. S.M.Y.D., L.M.S. and J.M.G. analysed the experimental data. S.M.Y.D. drafted the manuscript and created the drawing of Figure 1. L.M.S. drafted the PLS-DA permutation testing experimental procedure. All authors reviewed and contributed to the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The author(s) declare no competing interests. This work was financially supported by the Australian Research Training Program scholarship by Charles Sturt University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yalage Don, S.M., Schmidtke, L.M., Gambetta, J.M. et al. Aureobasidium pullulans volatilome identified by a novel, quantitative approach employing SPME-GC-MS, suppressed Botrytis cinerea and Alternaria alternata in vitro. Sci Rep 10, 4498 (2020). https://doi.org/10.1038/s41598-020-61471-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61471-8
This article is cited by
-
Inhibition activity of tomato endophyte Bacillus velezensis FQ-G3 against postharvest Botrytis cinerea
Folia Microbiologica (2024)
-
Microbial Volatile Compounds: Prospects for Fungal Phytopathogens Management, Mechanisms and Challenges
Journal of Crop Health (2024)
-
Antagonistic fungal volatiles as potential biocontrol countermeasure for microbial postharvest fruit diseases
Egyptian Journal of Biological Pest Control (2023)
-
Contribution of Leaf-Associated Microorganisms from Native Andean Ericaceae against Botrytis cinerea in Vaccinium corymbosum Cultivars
Journal of Soil Science and Plant Nutrition (2023)
-
Using off-gas for insights through online monitoring of ethanol and baker’s yeast volatilome using SESI-Orbitrap MS
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.