Abstract
We investigated T-cell receptor variable β chains binding to the superantigen staphylococcal enterotoxin C3 (SEC 3) with structure information in different stages of affinity maturation. Metadynamics in combination with molecular dynamics simulations allow to access the micro-to-millisecond timescale and reveal a strong effect of energetically significant mutations on the flexibility of the antigen-binding site. The observed changes in dynamics of the complementarity determining region (CDR) loops, especially the CDR 2, and HV 4 loop on this specific pathway of affinity maturation are reflected in their structural diversity, thermodynamics of conformations and kinetics of structural transitions. In addition, this affinity maturation pathway follows the concept of conformational selection, because even without the presence of the antigen the binding competent state is present in this pre-existing ensemble of conformations. In all stages of this affinity maturation process we observe a link between specificity and reduced flexibility.
Similar content being viewed by others
Introduction
T-cells play an important signaling role in the adaptive immune system and are activated upon antigen recognition by their T-cell receptors (TCRs)1. TCRs are expressed on two types of cells, cytotoxic T-cells and helper T-cells. TCRs consist of an α and ß chain, which can be divided into a constant and a variable domain, homologous to the immunoglobins2. TCR α chains are made from V and J genes, analogous to light chains in antibodies, while TCR ß chains, same as for antibody heavy chains, are assembled from the V, D and J genes3,4,5,6. Sequence and structural diversity are concentrated on six hypervariable loops, also known as complementarity determining regions (CDRs), each three loops on the α and ß variable domains, which are critical for antigen recognition7,8,9. TCR CDR 1 and CDR 2 loops show interactions with the major histocompatibility complexes (MHC I and MHC II), while CDR 3 has contacts with peptide antigens. Similar to antibodies the TCR CDRs can adopt well-characterized canonical conformations which facilitate reliable structure prediction based on sequence information3. Still a clear characterization of structure and dynamics is essential to understand the antigen-binding process, the involved conformational changes and the associated biological implications10. Various studies focused already on understanding the evolution of an antibody-antigen interface during affinity maturation11,12,13,14,15. However, elucidating the physicochemical principles that are involved in protein-protein interactions, would improve the understanding of biological systems16. Structural basis of affinity maturation has been established for an in vitro designed protein-protein interaction system which provides with the help of strong structural experimental data molecular snapshots of the affinity maturation process and the involved remodeling of the protein interface17,18. The energetic contributions of individual amino acid residues to the complex have been investigated to get a better understanding of intramolecular cooperativity and to be able to predict specificity17,18. The correlation between enhanced specificity and rigidification is often discussed in terms of conformational selection19,20,21. Promiscuity might arise from a multitude of weakly populated conformations, each of which is able to bind different binding partners. Rigidification shifts the probability toward a small number of states and thereby reduces the amount of possible binding partners20,22,23,24.
We analyzed an affinity maturation pathway of murine TCR Vß variants with structural information for every stage to elucidate the underlying mechanism of the affinity maturation process and to identify differences in their conformational diversity. We focused on the CDR 2 and the HV 4 loop to investigate changes in dynamics as a consequence of point-mutations, because they directly interact with the superantigen staphylococcal enterotoxin C3 (SEC 3) and are the mutation hotspots in this affinity maturation pathway.
Results
A previous study investigated the cooperative and energetic influence of point-mutations in murine TCR Vß variants17,18. Experimental structure information was available for all stages of affinity maturation. Figure 1 summarizes all available crystal structures, all point-mutations and their positions, color coded respectively, on a representative TCR Vß variant structure.
Figure 2 shows the ionic and hydrogen bond interactions formed in the TCR Vß variants in different stages of affinity maturation. A clear structural stabilization can be observed for the most affinity matured variant compared to the wildtype.
Structural interaction analysis of Vß crystal structures in different stages of affinity maturation. The CDR2 and HV 4 loop are colored yellow and orange, respectively, while in blue the interaction partners in different stages of affinity maturation are illustrated. Strong ionic and hydrogen-bond interactions are present in the X-ray structures with the accession codes 1BEC, 2APF and 2APX.
As described in the methods section, the metadynamics simulations for each available starting structure was clustered using the same distance cut-off with the aim of having representative structures distributed over the entire sampled conformational space. This strategy resulted in 241 clusters of the CDR 2 and the HV 4 loop for the wildtype TCR Vß variant 1BEC and in 43 clusters for the wildtype TCR Vß 1JCK complexed with SEC 3. The conformational diversity of 1BEC upon binding to SEC 3 (1JCK) in the resulting ensemble is substantially decreased and represents a prime example of conformational selection. The TCR variant 2AQ3 is the most similar to the wildtype and displays a comparable conformational diversity reflected in 210 cluster representatives. Significantly reduced conformational diversity was observed in the most matured TCR Vß variant 2APX which leads to 48 cluster representatives. An overview of the resulting CDR 2 and HV 4 conformational ensembles of the whole affinity maturation pathway is illustrated in Fig. 3. A more detailed perspective on the CDR 2 and HV 4 loop ensembles upon affinity maturation is shown in SI Fig. S1. The variant 2AQ2 was simulated with the antigen SEC 3 present to investigate the influence of the antigen-binding on the observed dynamics. The TCR variant 2APF binds with the same binding affinity as the 2AQ2 variant and is crystallized without SEC 3. By comparing the conformational diversity of 2APF with 2AQ2 the number of accessible CDR 2 and HV 4 loop conformations substantially decreases from 88 to 9 clusters.
Overview of all analyzed TCR variants with the resulting conformational ensembles of the CDR 2 and the HV 4 loop colored in blue of each 1 µs metadynamics simulations. The diversity of the conformational ensemble is characterized by the number of clusters for each simulation and decreases with higher stages of maturation. The wildtype 1JCK and the affinity matured variant 2AQ2 were additionally simulated with the antigen present to investigate the role of binding to SEC 3 on the observed conformational ensemble. The resulting ensembles clearly follow the paradigm of conformational selection.
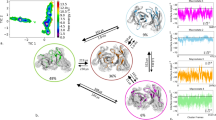
Figure 4 shows the combined tICA analyses using the same coordinate system for five TCR Vß variants in different stages of affinity maturation. The tICA space displays conformational shifts on this affinity maturation pathway and a substantial reduction in conformational space. Below the resulting conformational ensembles of the CDR 2 and HV 4 loop are illustrated with the color coded positions of their point mutations.
TICA analyses for five TCR Vß variants in different stages of affinity maturation clearly show a significant rigidification of CDR 2 and the HV 4 loop. All TCR Vß variants are projected onto the combined tICA space of 24.1 µs trajectories of 1BEC, 9.5 µs trajectories of 2APB, 7 µs trajectories of 2APT, 8.8 µs trajectories of 2APF and 4.8 µs trajectories of 2APX. The X-ray structures crystallized without antigen are colored in orange, while the X-ray structures crystallized with antigen are colored blue. Both of them are projected into the tICA analyses. Additionally, a graphical representation of the loop flexibility is shown as representative ensemble below the tICA analyses.
Based on the observed decrease in conformational diversity depending on the presence of the antigen SEC 3, the 2APF/2AQ2 variants clearly follows the paradigm of conformational selection and the resulting population shift upon binding is visualized in Fig. 5. Figure 5 illustrates the resulting conformational ensembles of the hierarchical clustering of the 8.8 µs trajectories of the 2APF and the 1 µs trajectories of the 2AQ2 of the CDR 2 and HV 4 loops by aligning on the whole TCR ß variable domain. This clustering led to 14 clusters for the 2APF and to 4 clusters for the 2AQ2 bound to SEC 3. Figure 6 provides kinetic characterization of conformational transitions and results in short transition timescales for the wildtype 1BEC TCR Vß variant, while it reveals slightly longer timescales in the higher microsecond timescale for the matured 2APX variant. The Markov-state model of the 2APX TCR Vß variant reveals one dominantly populated state, while the 1BEC has three probable states with fast exchanges between these macrostates.
In the systems studied CDR 2 and HV 4 are the major loops binding the antigen, thus we primarily focused on these two loops. Surprisingly, the CDR 3 loop, even though not directly involved in binding to SEC 3, still is significantly rigidified during affinity maturation. Figure 7 illustrates the resulting conformational ensembles of the CDR 3 loop by applying the hierarchical clustering with a distance cut-off of 2 Å and reveals a significant decrease in conformational diversity in different stages of affinity maturation.
CDR 3 loop ensemble of the hierarchical clustering of the 24.1 µs 1BEC trajectories, the 9.5 µs 2APB trajectories, the 7.0 µs 2APT trajectories, the 8.8 µs 2APF trajectories and the 4.8 µs 2APX trajectories. Even though the CDR 3 loop is not directly involved in binding it is still rigidified during affinity maturation.
Discussion
This present study characterizes the conformational diversity during affinity maturation of the CDR 2 and HV 4 loops, because of their strong involvement in antigen binding. Strong experimental structural information together with molecular dynamics simulations allow the investigation of conformational selection, the consequences of point-mutations on flexibility and changes in kinetics and thermodynamics during this affinity maturation pathway.
The concept of conformational selection suggests that within this pre-existing ensemble of conformations the binding competent state is selected, accompanied by a population shift and redistribution of the conformational states20,24,25. The theory of having an ensemble of pre-existing conformations out of which the functional ones are selected was suggested Pauling25,26 and demonstrated by Milstein and Wedemayr27,28. This view has been supported by the population shift or conformational selection model, originated from the Monod–Wyman–Changeux model29. This new view on proteins, i.e., that one sequence can show high structural diversity, facilitated the understanding and evolution of new functions and structures21. Proper characterization of the CDR loops, especially the loops which are mainly involved in the binding process, is crucial to understand protein-protein interactions and antigen binding30,31. Figure 2 compares the intramolecular interactions of three variants in different stages of affinity maturation. While there is no difference in intramolecular stabilizing hydrogen bond or ionic interactions in the binding site between the wildtype and a further matured variant 2APF the most matured TCR Vß variant 2APX forms a significantly higher number of hydrogen bonds and ionic interactions. This indicates a substantial stabilization on a static structural level. The mutations in the CDR 2 and HV 4 loops are in line with the observed rigidification (Fig. 3) of these loops upon affinity maturation, which are directly involved in binding to SEC 3. Also the CDR 3 loop without showing interactions with the antigen SEC 3 reveals as a consequence of mutations a rigidification upon affinity maturation. Various studies already focused on elucidating the affinity maturation process and the numerous diverse involved mechanisms, including improved shape complementarity in the binding site, an increase in buried surface area upon complex formation, additional interfacial polar or hydrophobic interactions and rigidification of the binding site18,28,32,33,34,35. Figure 3 shows an overview of all simulated variants with their resulting number of cluster representatives by using the same distance cut-off. The number of resulting clusters is one possible way to characterize flexibility and reveals a substantial rigidification especially when comparing the most matured variant with the wildtype. Another possibility to compare the flexibility among the variants is to take the conformational space into account. We used tICA coordinates, because we were interested in retaining the kinetics of exchanges between conformational sub-states occurring on different timescales. Figure 4 shows tICA plots of five exemplary variants in distinct maturation stages using the same coordinate system in combination with a representative ensemble of CDR 2 and HV 4 loop structures, including colored point-mutations corresponding to Fig. 1. The conformational diversity of the representative ensembles in Fig. 4 decreases significantly from wildtype to the most matured variant. Also, the conformational space of the matured 2APX variant is substantially reduced compared to the wildtype 1BEC. Additionally, a clear population shift can be observed throughout all considered variants during this maturation pathway. The available X-ray structures, crystallized with and without antigen, are located in the same local side minimum, which becomes the dominant minimum in solution of the most matured 2APX variant. The resulting conformational selection is illustrated in Fig. 5. We clearly see a strong population shift of the 2APF variant to the bound 2AQ2 and a significantly decreased conformational space of the CDR 2 and HV 4 loops if the antigen SEC 3 is present. Figure 6 characterizes the transition timescales of sub-states of the accessible conformational space and the resulting kinetics reveal fast transition timescales for the wildtype 1BEC, while the matured 2APX displays slower transition kinetics. This indicates that the rigidification shifts the probability toward a small number of states and thereby increases specificity. The probabilities of the most matured 2APX variant are shifted towards the binding competent state, while the wildtype 1BEC has higher probabilities for various other sub-states which are accessible in the nanosecond timescale. In this affinity maturation study the CDR 3 loop is not involved in the antigen-binding process, but it still remains one of the most challenging loops in terms of structure prediction and characterization in solution36. Therefore we used our long-timescale simulations to analyze the conformational diversity of the CDR 3 loop in solution. Figure 7 reveals a decrease in flexibility of the CDR 3 loop during the affinity maturation pathway reflected in the reduced number of clusters.
Methods
A previously published method characterizing the CDR-H3 loop ensemble upon antigen binding in solution12,22 was used to investigate the conformational diversity of CDR 2 and HV 4 loops of TCR Vß variants in different stages of affinity maturation. Experimental structure information was available for all stages of affinity maturation and we used all available crystal structures as starting structures for molecular dynamics simulations. The TCR Vß variant with the PDB accession code 2AQ2 was simulated with the antigen SEC 3 bound. All other available complex structures were simulated without the presence of the antigen. The starting structures for simulations were prepared in MOE (Molecular Operating Environment, Chemical Computing Group, version 2018.01) using the Protonate3D tool37,38. To neutralize the charges we used the uniform background charge39,40,41. Using the tleap tool of the AmberTools1639,40 package, the crystal structures were soaked with cubic water boxes of TIP3P water molecules with a minimum wall distance of 10 Å to the protein42. For all crystal structures parameters of the AMBER force field 14SB were used43. The TCR Vß variants were carefully equilibrated using a multistep equilibration protocol44.
Metadynamics simulations
To enhance the sampling of the conformational space well-tempered metadynamics45,46,47 simulations were performed in GROMACS48,49 with the PLUMED 2 implementation50. As collective variables, we used a linear combination of sine and cosine of the ψ torsion angles of the CDR 2 loop and the HV 4 loop calculated with functions MATHEVAL and COMBINE implemented in PLUMED 250. As discussed previously the ψ torsion angle captures conformational transitions comprehensively37,38. The decision to additionally include the HV 4 loop ψ torsion angles is based on the strong involvement of the HV 4 loop in the binding to SEC 3 as evident from the X-ray structure of the complex. The simulations were performed at 300 K in an NpT ensemble. The height of the Gaussian was determined according to minimal distortion of the TCR systems, resulting in a Gaussian height of 10.0 kcal/mol. Gaussian deposition occurred every 1000 steps and a biasfactor of 10 was used. 1 µs metadynamics simulations were performed for each available TCR Vß variant crystal structure. The resulting trajectories were clustered in cpptraj40,51 by using the average linkage hierarchical clustering algorithm with a distance cut-off criterion of 1.2 Å resulting in a large number of clusters. The cluster representatives for the TCR Vß variants in different stages of affinity maturation with the PDB accession codes 1BEC, 2APB, 2APT, 2APF, 2APX without the antigen present and 2AQ2 with the antigen present were equilibrated and simulated for 100 ns using the AMBER1639 simulation package.
Molecular dynamics simulations
Molecular dynamics simulations were performed in an NpT ensemble using pmemd.cuda52. Bonds involving hydrogen atoms were restrained by applying the SHAKE algorithm53, allowing a time step of 2.0 fs. Atmospheric pressure of the system was preserved by weak coupling to an external bath using the Berendsen algorithm54. The Langevin thermostat55 was used to maintain the temperature during simulations at 300 K.
For the resulting trajectories cluster analyses were performed on the CDR 2 and HV 4 loop using the hierarchical clustering algorithm implemented in cpptraj40 with a distance cut-off criterion of 2 Å to get a representative ensemble of structures. Additionally, a time-lagged independent component analysis (tICA) was performed using the python library PyEMMA 2 employing a lag time of 10 ns56. Thermodynamics and kinetics were calculated with a Markov-state model57 by using PyEMMA 2, which uses the k-means clustering algorithm58 to define microstates and the PCCA+ clustering algorithm59 to coarse grain the microstates to macrostates. PCCA+ is a spectral clustering method, which discretizes the sampled conformational space based on the eigenvectors of the transition matrix. The sampling efficiency and the reliability of the Markov-state model (e.g., defining optimal feature mappings) can be evaluated with the Chapman-Kolmogorov test60,61, by using the variational approach for Markov processes62 and by taking into account the fraction of states used, as the network states must be fully connected to calculate probabilities of transitions and the relative equilibrium probabilities. To build the Markov-state model we used the backbone torsions of the CDR 2 and the HV 4 loops, defined 150 microstates using the k-means clustering algorithm and applied a lag time of 10 ns.
The images presented in this paper were created by using the PyMOL molecular graphics system63.
Conclusion
To elucidate the mechanism of affinity maturation we analyzed a series of TCR Vß variants with strong experimental structural information in different stages of affinity maturation. Besides the structural intramolecular stabilization, we observed a substantial reduction in flexibility and resulting conformational diversity of the CDR 2 and HV 4 loop, which are directly involved in antigen binding. The investigated variants follow the conformational selection paradigm accompanied by a significant population shift towards the binding competent conformation in the further matured 2APF variant. We characterized the transition kinetics between different conformational states and observed a conformational shift during this affinity maturation pathway towards the binding competent state in the matured 2APX variant. Our results clearly reveal a rigidification and a strong population shift during the affinity maturation process.
References
Wilson, I. A. & Christopher Garcia, K. T-cell receptor structure and TCR complexes. Curr. Opin. Struct. Biol. 7, 839–848 (1997).
Alcover, A., Alarcón, B. & Di Bartolo, V. Cell Biology of T Cell Receptor Expression and Regulation. Annu. Rev. Immunol. 36, 103–125 (2018).
Wong, W. K., Leem, J. & Deane, C. M. Comparative analysis of the CDR loops of antigen receptors. bioRxiv 709840, https://doi.org/10.1101/709840 (2019).
Wucherpfennig, K. W., Gagnon, E., Call, M. J., Huseby, E. S. & Call, M. E. Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb. Perspect. Biol. 2, a005140–a005140 (2010).
Market, E. & Papavasiliou, F. N. V(D)J Recombination and the Evolution of the Adaptive Immune System. PLoS Biol. 1, e16 (2003).
Bassing, C. H., Swat, W. & Alt, F. W. The Mechanism and Regulation of Chromosomal V(D)J Recombination. Cell 109, S45–S55 (2002).
Leem, J., de Oliveira, S. H. P., Krawczyk, K. & Deane, C. M. STCRDab: the structural T-cell receptor database. Nucleic Acids Res. 46, D406–D412 (2018).
Chothia, C. & Lesk, A. M. Canonical structures for the hypervariable regions of immunoglobulins. J. Mol. Biol. 196, 901–917 (1987).
Al-Lazikani, B., Lesk, A. M. & Chothia, C. Canonical structures for the hypervariable regions of T cell αβ receptors. J. Mol. Biol. 295, 979–95 (2000).
Stanfield, R. L. & Wilson, I. A. Antigen-induced conformational changes in antibodies: A problem for structural prediction and design. Trends Biotechnol. 12, 275–279 (1994).
Li, Y., Li, H., Yang, F., Smith-Gill, S. J. & Mariuzza, R. A. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat. Struct. Mol. Biol. 10, 482–488 (2003).
Fernández-Quintero, M. L. et al. Characterizing the Diversity of the CDR-H3 Loop Conformational Ensembles in Relationship to Antibody Binding Properties. Front. Immunol. 9, 3065 (2019).
Li, B. et al. The Protein-Protein Interface Evolution Acts in a Similar Way to Antibody Affinity Maturation. J. Biol. Chem. 285, 3865–3871 (2010).
Schmidt, A. G. et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. 110, 264 (2013).
Adhikary, R., Yu, W., Oda, M., Zimmermann, J. & Romesberg, F. E. Protein Dynamics and the Diversity of an Antibody Response. J. Biol. Chem., https://doi.org/10.1074/jbc.M112.372698 (2012).
Glanville, J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017).
Cho, S. et al. Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction. Struct. Lond. Engl. 1993 13, 1775–1787 (2005).
Yang, J. et al. Dissecting Cooperative and Additive Binding Energetics in the Affinity Maturation Pathway of a Protein-Protein Interface. J. Biol. Chem. 278, 50412–50421 (2003).
Ma, B., Kumar, S., Tsai, C.-J. & Nussinov, R. Folding funnels and binding mechanisms. Protein Eng. Des. Sel. 12, 713–720 (1999).
Tsai, C.-J., Kumar, S., Ma, B. & Nussinov, R. Folding funnels, binding funnels, and protein function. Protein Sci. 8, 1181–1190 (1999).
James, L. C. & Tawfik, D. S. Conformational diversity and protein evolution – a 60-year-old hypothesis revisited. Trends Biochem. Sci. 28, 361–368 (2003).
Fernández-Quintero, M. L., Kraml, J., Georges, G. & Liedl, K. R. CDR-H3 Loop Ensemble in Solution – Conformational Selection upon Antibody Binding. mAbs null-null. https://doi.org/10.1080/19420862.2019.1618676 (2019).
Wang, W. et al. Conformational Selection and Induced Fit in Specific Antibody and Antigen Recognition: SPE7 as a Case Study. J. Phys. Chem. B 117, 4912–4923 (2013).
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Pauling, L. A Theory of the Structure and Process of Formation of Antibodies*. J. Am. Chem. Soc. 62, 2643–2657 (1940).
James, L. C., Roversi, P. & Tawfik, D. S. Antibody multispecificity mediated by conformational diversity. Science 299, 1362–1367 (2003).
Foote, J. & Milstein, C. Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. USA 91, 10370–10374 (1994).
Wedemayer, G. J., Patten, P. A., Wang, L. H., Schultz, P. G. & Stevens, R. C. Structural Insights into the Evolution of an Antibody Combining Site. Science 276, 1665 (1997).
Monod, J., Wyman, J. & Changeux, J.-P. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 (1965).
Hare, B. J. et al. Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor. Nat. Struct. Biol. 6, 574–581 (1999).
Willcox, B. E. et al. TCR Binding to Peptide-MHC Stabilizes a Flexible Recognition Interface. Immunity 10, 357–365 (1999).
Mishra, A. K. & Mariuzza, R. A. Insights into the Structural Basis of Antibody Affinity Maturation from Next-Generation Sequencing. Front. Immunol. 9, 117–117 (2018).
Laffy, J. M. J. et al. Promiscuous antibodies characterised by their physico-chemical properties: From sequence to structure and back. Prog. Biophys. Mol. Biol. https://doi.org/10.1016/j.pbiomolbio.2016.09.002 (2016).
Schreiber, G. & Keating, A. E. Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 21, 50–61 (2011).
Kaur, H. & Salunke, D. M. Antibody promiscuity: Understanding the paradigm shift in antigen recognition. IUBMB Life 67, 498–505 (2015).
Hughes, M. M. et al. T cell receptor CDR3 loop length repertoire is determined primarily by features of the V(D)J recombination reaction. Eur. J. Immunol. 33, 1568–1575 (2003).
Labute, P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 75, 187–205 (2009).
1010 Sherbrooke St. West, Suite #910, Montreal, QC: Chemical Computing Group ULC, Canada, H3A 2R7. (2018).
Case, D. A. et al. AMBER 2016, University of California, San Francisco. (2016).
Roe, D. R. & Cheatham, T. E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Hub, J. S., de Groot, B. L., Grubmüller, H. & Groenhof, G. Quantifying Artifacts in Ewald Simulations of Inhomogeneous Systems with a Net Charge. J. Chem. Theory Comput. 10, 381–390 (2014).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Maier, J. A. et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Wallnoefer, H. G., Liedl, K. R. & Fox, T. A challenging system: Free energy prediction for factor Xa. J. Comput. Chem. 32, 1743–1752 (2011).
Barducci, A., Bussi, G. & Parrinello, M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 100, 020603 (2008).
Biswas, M., Lickert, B. & Stock, G. Metadynamics Enhanced Markov Modeling of Protein Dynamics. https://doi.org/10.1021/acs.jpcb.7b11800 (2018).
Barducci, A., Bonomi, M. & Parrinello, M. Metadynamics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 1, 826–843 (2011).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C. & Bussi, G. PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014).
Shao, J., Tanner, S. W., Thompson, N. & Cheatham, T. E. Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theory Comput. 3, 2312–2334 (2007).
Salomon-Ferrer, R., Götz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Miyamoto, S. & Kollman, P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Adelman, S. A. & Doll, J. D. Generalized Langevin equation approach for atom/solid‐surface scattering: General formulation for classical scattering off harmonic solids. J. Chem. Phys. 64, 2375–2388 (1976).
Scherer, M. K. et al. PyEMMA 2: A Software Package for Estimation, Validation, and Analysis of Markov Models. J. Chem. Theory Comput. 11, 5525–5542 (2015).
Chodera, J. D. & Noé, F. Markov state models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol. 25, 135–144 (2014).
Likas, A., Vlassis, N. & J. Verbeek, J. The global k-means clustering algorithm. Biometrics 36, 451–461 (2003).
Röblitz, S. & Weber, M. Fuzzy spectral clustering by PCCA+: application to Markov state models and data classification. Adv. Data Anal. Classif. 7, 147–179 (2013).
Karush, J. On the Chapman-Kolmogorov Equation. Ann Math Stat. 32, 1333–1337 (1961).
Miroshin, R. N. Special solutions of the Chapman–Kolmogorov equation for multidimensional-state Markov processes with continuous time. Vestn. St Petersburg Univ. Math. 49, 122–129 (2016).
Wu, H. & Noé, F. Variational approach for learning Markov processes from time series data. (2017).
Schrodinger. The PyMOL Molecular Graphics System, Version 1.8. (2015).
Acknowledgements
This work was supported by the Austrian Science Fund (FWF) via the grant P30565.
Author information
Authors and Affiliations
Contributions
Monica L. Fernández-Quintero and Klaus R. Liedl conceived the study. The research was performed by Monica L. Fernández-Quintero and Clarissa A. Seidler under the supervision of Klaus R. Liedl. Monica L. Fernández-Quintero and Clarissa A. Seidler curated the data and analyzed it. Monica L. Fernández-Quintero drafted the manuscript. All authors contributed to the paper by critically reviewing it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fernández-Quintero, M.L., Seidler, C.A. & Liedl, K.R. T-Cell Receptor Variable β Domains Rigidify During Affinity Maturation. Sci Rep 10, 4472 (2020). https://doi.org/10.1038/s41598-020-61433-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61433-0
This article is cited by
-
Antibodies exhibit multiple paratope states influencing VH–VL domain orientations
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.