Abstract
Though chickens (Gallus gallus domesticus) are globally ubiquitous today, the timing, location, and manner of their domestication is contentious. Until recently, archaeologists placed the origin of the domestic chicken in northern China, perhaps as early as 8,000 years ago. Such evidence however complicates our understanding of how the chicken was domesticated because its wild progenitor – the red jungle fowl (Gallus gallus) – lives in tropical ecosystems and does not exist in northern China today or in the recent past. Increasingly, multiple lines of evidence suggest that many of the archaeological bird remains underlying this northern origins hypothesis have been misidentified. Here we analyze the mitochondrial DNA of some of the earliest purported chickens from the Dadiwan site in northern China and conclude that they are pheasants (Phasianus colchicus). Curiously, stable isotope values from the same birds reveal that their diet was heavy in agricultural products (namely millet), meaning that they lived adjacent to or among some of the earliest farming communities in East Asia. We suggest that the exploitation of these baited birds was an important adaptation for early farmers in China’s arid north, and that management practices like these likely played a role in the domestication of animals – including the chicken – in similar contexts throughout the region.
Similar content being viewed by others
Introduction
Northern China is one of the few places where agriculture evolved independently ca. 9,000-7,000 calibrated years before the present (cal BP) and with a suite of plant and animal domesticates unique to the region, namely Panicum and Setaria millets, pigs, dogs, and a medium-size bird most typically identified as chicken1,2. The causes of this agricultural revolution are the subject of much debate3,4,5, as is the direct evidence for it6. Among the most contentious of these is the role of the chicken (Gallus gallus domesticus) in the agricultural origins of East Asia7,8,9,10. Indeed, there is no scientific consensus on where, when, or how the domestic chicken evolved, despite the fact that chickens are the most ubiquitous domestic animal on earth today11.
Chinese archaeological contexts that contain other evidence for agricultural life, often contain bones of medium-sized birds identified as “chicken,” the oldest of which have been touted as the epicenter of chicken domestication. This is problematic because some of the oldest locations are in the arid regions of northern China where the wild ancestor of the domestic chicken - the tropically adapted “red jungle fowl” (Gallus gallus) - does not thrive today12. Here we present evidence that the birds exploited by the some of the earliest agricultural peoples of arid northwest China were not chickens at all, but rather pheasants (Phasianus spp.), and closely related to the wild pheasants that today thrive in the arid environments of northern Eurasia. Importantly, as with other domestic and commensal animals throughout China and around the world, these birds were dependent upon an environment of resources that exists only in an agricultural ecosystem managed and perpetuated by humans.
Study Specimens
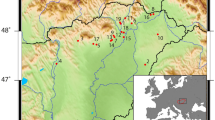
The present study is an extension of a broader effort to understand the origins of agriculture and the co-evolution of domestic plants, animals, and people in arid East Asia2,3,13,14. Specimens analyzed here were first collected during the 1978-1984 excavations15 of the Dadiwan Neolithic site in Gansu Province, PRC (Fig. 1). Though human hunter-gatherers visited the Dadiwan site sporadically over the past 80,000 years16, the earliest evidence for agricultural behavior (i.e., storage features, domestic structures, carbonized remains of cereals, and isotopic evidence that the local dogs had been foddered with those cereals year-round) does not appear until ~7,800 cal BP, during the Laoguantai cultural period2. By 6,300 cal BP (the Yangshao cultural period), Neolithic life at Dadiwan was intensively agricultural and culturally complex, with densely packed communities marked by permanent architecture, storage facilities, craft production zones, lavish burials, two kinds of morphologically domestic millets, and isotopic evidence that both dogs, and pigs (in abundance) were provisioned throughout the year with stored grain2.
Map of North China showing Dadiwan and the Neolithic culture areas referenced in the text. Regional boundaries are approximate. Basemap is a digital elevation model built from 90 m SRTM data (available at http://srtm.csi.cgiar.org/) using ArcMap 10.7.1 (http://www.esri.com) and Adobe Illustrator CS5 15.0.0 (http://www.adobe.com).
Previous research2,6 demonstrates that birds in this Neolithic village farming community were also provisioned with the grains and perhaps vegetation of C4 plants (most likely millets), which only grow during the summer. At Dadiwan, the stable isotopic values on the collagen from medium-sized bird bones fall within a range indicative of a year-round diet of both C4 and C3 vegetation (Fig. 2). This accords well with isotopic differences across the wild-domestic feeding spectrum, for example among modern wild-foraging and pen-raised Phasianus17, as well as between ancient domestic turkeys (Meleagris gallopavo) and contemporary wild turkeys from the American southwest18. As with many of the pigs and some of the dogs from Dadiwan, the stable isotope values from the bird bone found at the same site suggest that those birds exploited a mixed-bag of resources, including those they could forage on their own (as if they did not live in the human biome) and those they could only acquire through a life in proximity to human millet cultivators.
Though the Dadiwan birds were initially identified as “Gallus sp.,” the original analysts did acknowledge that some of the individuals might also be identified as Alectoris, i.e. “chukar” (石鸡) or simply Phasianidae, i.e. “pheasant” (雉)15. To resolve the taxonomic uncertainty, we selected for genetic species identification only those specimens previously identified as Gallus which had already been assayed for stable isotopic proxies of diet. All of these specimens were recovered from buried contexts dating to either the Laoguantai (7,900-7,200 cal BP; n = 3) or Yangshao (6,300-5,900 cal BP; n = 5) cultural periods (Table 1; Supplemental Information Fig. 3). Because the eight specimens come from eight unique depositional contexts and two completely different prehistoric cultural components (separated by ~900 years), initially we assumed them to represent eight different individuals.
Methods
Here we report the methods used for genetic species identification. The methods and rationale behind morphological species identification of zooarchaeological specimens12,15, stable isotope analyses of bone collagen2,6, and radiocarbon dating of associated materials2,16, have all been published elsewhere, and are not described herein. We merely report and evaluate the results of those analyses (Table 1; Fig. 2).
All DNA extraction and pre-polymerase chain reaction (PCR) procedures were conducted in the ancient DNA cleanroom at the Laboratories of Molecular Anthropology and Microbiome Research (LMAMR; lmamr.org) at the University of Oklahoma, Norman, OK. DNA was extracted using ancient DNA methods and various primers were designed to screen the mitochondrial DNA for sections that are informative for discriminating species (Supplemental Information). Once the specimens had been identified as the common ring-necked pheasant (Phasianus colchicus), additional primers were designed to sequence the bulk of the control region. Shotgun DNA libraries were constructed from four of the samples – a subset of the total eight – as an additional means of authenticating our findings (Supplemental Information).
All sequences were aligned to the full Phasianus colchicus mitochondrial genome (NC_015526.1) in Sequencher (version 5.4.6). A comparative data set of 104 control region mtDNA (spanning from np 170 to 950) from three subspecies of ring-necked pheasant (P.colchicus alashanicus, P.colchicus pallasi, P.colchicus strauchi19,20,21,22,23 was compiled from Genbank. These data along with control region sequences from samples LM-091, LM-092, LM-093, and LM-094 were aligned with DNA Alignment (version 1.3.3.2), converted to a Nexus file, and imported into PopART version 1.724. A median-joining network was created using the default parameters in PopART.
These same sequences were aligned using MUSCLE25 in Seqotron26 prior to manual curation. The Bayesian Information Criterion (BIC)27 in jModelTest228 was used to estimate the model of evolution for the control region sequence in Phasianus colchicus. A neighbor-joining tree was constructed using PAUP* version 4.0a 16529 with HKY + G_I as the model of evolution30 at default parameters for an initial evaluation of population structure. A comparative control region sequence from Elliot’s pheasant (Syrmaticus ellioti; Genbank accession AB164624) was defined as the outgroup31. Following the neighbor-joining tree, a Bayesian analysis sans the outgroup was performed in BEAST 2.532 using the same model of evolution under a constant population prior. Three independent runs sampling every 5,000th generation out of 500 million iterations of the Markov chain Monte Carlo were run. The tip dates of the samples were not included in any of the runs. Each run was then resampled using LogCombiner version 1.10.4 to pull every 50,000th generation. All three runs were combined, and 10% of the burnin removed. The final tree was generated based on maximum clade credibility derived from median node heights.
To confirm the Bayesian analyses, a maximum likelihood using PAUP* was run using the same model, HKY + G + I, with 1000 bootstrap replicates. No bootstrap values exceeded 50% for all internal and terminal nodes. Low bootstrap support in the consensus trees may have been related to the high number of shared haplotypes resulting in distortion. A secondary maximum likelihood was run for the unique haplotypes (n = 54) using the same parameters, and the topology was consistent with both the median-joining network and the Bayesian tree.
Consensus sequences produced from high throughput sequencing were aligned with 34 complete mitochondrial genomes belonging to organisms in the Phasianinae sub-family, followed by construction of a Maximum likelihood tree using RAxML33 (GTRGAMMA substitution model, 100 bootstrap replicates).
Results
All specimens analyzed were identified as the common ring-necked pheasant (Phasianus colchicus) based on sequences produced with the numerous primer pairs employed in our study (Supplemental Information). Independent amplifications confirm the species identification of seven specimens. Only a single amplicon from sample LM-088 was sequenced successfully using primer set LophuraF/R and identified as the common ring-necked pheasant, however this result could not be replicated. This may be due to the fact that the LM-088 specimen was burned and/or partially calcined in prehistory and contains little intact, original collagen (Table 1, Supplemental Fig. 1). Contamination was not detected in the extraction negative controls or in any PCR negative controls.
Six sets of control region primers (C149F/C325R, C266F/C438R, C400F/C580R, C539F/C711R, C660F/C833R, and C791F/C971R), were sequenced to further identify at the sub-species level. Four samples (LM-091, LM-092, LM-093, and LM-094) belong to two matrilines, which are shared to published Genbank sequences of three ring-necked pheasant subspecies (P. c. alaschanicus, P. c. pallasi, or P. c. strauchi). The median-joining network (Fig. 3) and Bayesian tree (Supplemental Fig. 2) illustrate haplotype sharing, and further depict the relationship between the Dadiwan specimens and 108 comparative ring-necked pheasant sequences. Samples LM-089, LM-090, and LM-095 were excluded from this analysis due to lack of comparative mitochondrial control region sequences (i.e., spanning from np 170 to 950). As illustrated in the network, one lineage is found in the central clade and is shared by all subspecies. The other lineage is a derived form of the central clade that is shared only with P. c. pallasi. Furthermore, though four of these birds are a 100% match with modern individuals belonging to three regional ring-necked pheasant subspecies: “strauchi”, “pallasi” and “alashanicus” (Fig. 3), they are genetically distinct from the dozens of other subspecies found elsewhere throughout Eurasia. The Bayesian tree is congruent with the network, illustrating two clades with strong support (Supplemental Fig. 2). Both internal and terminal nodes are poorly supported with posterior probabilities thus, the trees are not accurate representations of evolutionary divergence. More genetic information is required to full resolve the phylogenetic placement of both modern and ancient Phasianus colchicus subspecies. The topologies reveal a general pattern that suggests the hypervariable region of the Phasianus mitochondrial lineages are very similar across subspecies with a likelihood of lineage sharing. More genetic information is required to fully resolve the phylogenetic placement of Phasianus colchicus subspecies in this analysis. This would involve generating full mitogenome sequences from both ancient and contemporary specimens to supplement the few Phasianus mitogenomes currently available in genomic databases.
Median-joining network (constructed using 780 bp of the control region that spans from np 170-950) illustrating the relationship between the Dadiwan specimens and 108 comparative control region sequences from Genbank. There are two lineages depicted in the network. One lineage is found in the central clade and is shared by all of the subspecies included. The other lineage is a derived form of the central clade that is shared only with P. c. pallasi.
A total of 18.4 million read pairs were generated across the four sample libraries (4.7 M ± 1.8 M; mean ± s.d.). Post-quality filtering, between 80–91% of the reads were retained for samples LM-091, LM-092, and LM-093 (Supplemental Table 4). Comparison of analysis-ready reads from these three samples to the P. colchicus reference genome (GCA_004143745) resulted in mapping rates of between 79–90%. Sample LM-094 had a large fraction of adapter dimers (~56%), attributed to the low starting DNA quantities used during library preparation. Consequently, read mapping rates for LM-094 when compared to the P. colchicus reference genome was ~9% (Supplemental Table 4). Fragment distributions for mapped reads are consistent with those expected for ancient DNA specimens34,35, ranging between a median fragment size of 47–76 bp for the four libraries (Fig. 4a, Supplemental Table 4). Terminal deamination follows patterns expected for ancient DNA libraries generated with partial-UDG treatment36,37, with 5′ C- > T rates between 3.4%–11.7%, and 3′ G- > A rates between 2.8%–11.6% (Fig. 4b, Supplemental Table 4). Mapping of analysis-ready reads from samples LM-091, LM-092, and LM-093, to the P. colchicus mitochondrial genome (NC_015526) showed coverage with five reads or higher across >99% of sites along the reference sequence. Phylogenetic analysis of consensus mtDNA sequences generated from these alignments showed clustering of these specimens with P. colchicus (100% bootstrap support; Fig. 4c).
Authentication of endogenous ancient DNA. (a) Fragment size distribution of reads mapped to P. colchicus genome, (b) Terminal base damage rates for reads mapped to P. colchicus genome, and (c) Maximum likelihood tree built from whole mitochondrial genomes depicting phylogenetic placement of Dadiwan samples in relation to members of the sub-family Phasianinae. Sample LM-094 was removed prior to phylogenetic analysis due to limited sequence coverage (127 sites with >5x coverage).
Based on the similarities and differences of age, archaeological provenience, skeletal element morphology, diet, and mitogenome, the assemblage of bird bone presented here could come from as few as four but as many as eight different individual birds (Supplemental Information, Supplemental Tables 6 and 7).
Discussion
Because Dadiwan is among the oldest unequivocally agricultural settlements in China, and the earliest in the arid northwest, a fuller understanding of agricultural evolution, and potentially chicken domestication, requires greater confidence in the taxonomic identity of the faunal remains recovered at the site.
Our analyses reveal that the birds living in the human biome surrounding the Dadiwan site, and subsequently exploited by people, were neither domestic chickens, nor their wild ancestors the red jungle fowl. Instead, we identify them as pheasants (Phasianus colchicus). Importantly, the three extant subspecies that match the prehistoric individuals from Dadiwan are all found in northern China today in a mixture of northern latitude environments that include deserts, steppe, and arid highlands38. Moreover, when the Dadiwan sequences were compared to the tropical taxa of Asia they deviated greatly, suggesting that the prehistoric provisioning and exploitation of large meaty birds at Dadiwan was a local (i.e. not imported) adaptation.
Currently we have no evidence that these Dadiwan birds were kept in pens, that they provided eggs for human consumption, or even that they were under direct human control. What we can say is they do not resemble wild-foraging animals, and that they must have spent a sizeable portion of their lives eating things that only humans could provide. In turn, the humans ate the birds fed either on their own grain stores, or on the vegetative waste associated with the harvesting and processing of these domesticated millets.
We refer to this simple symbiosis as “low-level bird production” and suggest that it was an intentional, and low-cost human strategy for enhancing local resource yields. Because the abundance of birds could be tuned up or down simply by modifying the abundance of agricultural waste (either within the village grounds, or in the adjacent agricultural plots), it was an effective means of low-level food production39. Furthermore, the practice could be initiated every time the farming community fissioned or relocated, without having to transport caged or incubated birds. Birds endemic to the local environmental context simply preyed upon the village refuse in pursuit of their own self-interest and in turn the people preyed upon those birds.
The isotopic pattern in bird bone collagen is repeated elsewhere in Neolithic north China, suggesting the practice of low-level bird production was adaptive across a broad swath of ethno-linguistic groups inhabiting temperate environmental contexts, through time and space (Figs. 1 and 2). A spatio-temporal study of archaeological bird remains like these might reveal whether low-level bird production was limited to pheasants, and may provide insight about the degree to which pheasants, or any other large, meaty, ground-dwelling birds – including chickens and their wild ancestors – might have been managed in prehistory.
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files. Complete sequence data have been submitted to NCBI under the accession ID PRJNA578639 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA578639).
References
Larson, G. et al. Current perspectives and the future of domestication studies. Proceedings of the National Academy of Sciences 111, 6139–6146 (2014).
Barton, L. et al. Agricultural origins and the isotopic identity of domestication in northern China. Proceedings of the National Academy of Sciences 106, 5523–5528 (2009).
Bettinger, R. L., Barton, L. & Morgan, C. T. The origins of food production in North China: a different kind of agricultural revolution. Evolutionary Anthropology 19, 9–21 (2010).
Bar-Yosef, O. Climatic fluctuations and early farming in West and East Asia. Current Anthropology 52, S175–S193 (2011).
Yan, W. Nong ye fa sheng yu wen ming qi yuan (The origin of agriculture and civilization). (Kexue Chubanshe (Science Press), 2000).
Barton, L. Early Food Production in China’s Western Loess Plateau, University of California, Davis, (2009).
Xiang, H. et al. Early Holocene chicken domestication in northern China. Proceedings of the National Academy of Sciences 111, 17564–17569 (2014).
Peters, J. et al. Questioning new answers regarding Holocene chicken domestication in China. Proceedings of the National Academy of Sciences 112, E2415 (2015).
Eda, M. et al. Reevaluation of early Holocene chicken domestication in northern China. Journal of Archaeological Science 67, 25–31 (2016).
Peng, M. S., Shi, N. N., Yao, Y. G. & Zhang, Y. P. Caveats about interpretation of ancient chicken mtDNAs from northern China. Proceedings of the National Academy of Sciences 112, E1970–E1971 (2015).
Bennett, C. E. et al. The broiler chicken as a signal of a human reconfigured biosphere. Royal Society Open Science 5, 180325 (2018).
Peters, J., Lebrasseur, O., Deng, H. & Larson, G. Holocene cultural history of Red jungle fowl (Gallus gallus) and its domestic descendant in East Asia. Quaternary Science Reviews 142, 102–119 (2016).
Zhang, D. J. et al. Gansu Dadiwan yizhi jujin liuwannian lai de kaogu jilu yu hanzuo nongye qiyuan (Archaeological records of Dadiwan in the past 60 ka and the origin of millet agriculture). Zhongguo Kexue Yuan (Chinese Science Bulletin) 55, 887–894 (2010).
Bettinger, R. L. et al. In Late Quaternary Climate Change and Human Adaptation in Arid China Developments in Quaternary Science (Eds David B. Madsen, Xing Gao, & Fa Hu Chen) 83–101 (Elsevier, 2007).
GPICRA. (Gansu Provincial Institute of Cultural Relics and Archaeology) Qin’an Dadiwan xinshiqi shi dai yizhi fa jue baogao (Dadiwan in Qin’an: report on excavations at a Neolithic site). (Cultural Relics Publishing House, 2006).
Bettinger, R. L. et al. The transition to agriculture at Dadiwan, People’s Republic of China. Current Anthropology 51, 703–714 (2010).
Jensen, P. M., Madsen, P., Jensen, L. S. & Pipper, C. B. Differences in carbon and nitrogen stable isotope signatures amongst wild and released pheasant populations. European Journal of Wildlife Research 58, 755–760 (2012).
Lipe, W. D. et al. Cultural and genetic contexts for early turkey domestication in the northern Southwest. American Antiquity 81, 97–113 (2016).
Kozyrenko, M. M., Fisenko, P. V. & Zhuravlev, L. Genetic variation of Manchurian pheasant (Phasianus colchicus pallasi Rotshild, 1903) inferred from mitochondrial DNA control region sequences. Genetika 45 (2009).
Liu, Y., Zhan, X., Wang, N., Chang, J. & Zhang, Z. Effect of geological vicariance on mitochondrial DNA differentiation in Common Pheasant populations of the Loess Plateau and eastern China. Molecular Phylogenetics and Evolution 55, 409–417 (2010).
Qu, J., Zhang, J. & Liu, N. Ecological genetics of Strauch’s Pheasant (Phasianus colchicus strauchi): correlation between environmental factors and population genetic variability. Genbank unpublished (2008).
Xu, X. L., Fu, Y., Wang, Y. & Bai, S. Y. Genetic divergence in northeast subspecies of the ring-necked pheasant. Genbank unpublished (2013).
Zhao, C., Liu, Z., Sun, Y. & Gao, H. Phasianus colchicus alaschanicus isolate HLSHJZ16688 mitochondrion, complete genome. Genbank unpublished (2015).
Leigh, J. W. & Bryant, D. PopART: full-feature softwared for haplotype network construction. Methods in Ecology and Evolution 6, 1110–1116 (2015).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004).
Fourment, M. & Holmes, E. C. Segotron: a user-friendly sequence editor for Mac OS X. BMC Research Notes 9, 106 (2016).
Schwarz, G. Estimating the dimensions of a model. Annals of Statistics 6, 461–464 (1978).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. j. ModelTest 2: more models, new heuristics and high performance computing. Nature Methods 9, 772 (2012).
PAUP*. Phylogenetic analysis using parsimony (*and other methods) (Sinauer Associates, Sunderland, MA, 2003).
Hasegawa, M., Kishino, H. & Yano, T. Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. Journal of Molecular Evolution 22, 160–74 (1985).
Kato, S., Nishibori, M. & Yasue, H. Pheasant complete mitochondrial genome. Genbank unpublished (2004).
Bouckaert, R. et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology 15, e1006650 (2019).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Dabney, J., Meyer, M. & Paabo, S. Ancient DNA damage. Cold Spring Harbor Perspectives in Biology 5, a012567 (2013).
Jonsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. & Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013).
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philosophical Transactions of the Royal Society B 3370, 20130624 (2015).
Neparaczki, E. et al. Revising mtDNA haplotypes of the ancient Hungarian conquerors with next generation sequencing. PLoS ONE 12, e0174886 (2017).
Kayvanfar, N., Aliabadian, M., Niu, X., Zhang, Z. & Liu, Y. Phylogeography of the common pheasant Phasianus colchicus. Ibis 159, 430–442 (2017).
Smith, B. D. Low-level food production. Journal of Archaeological Research 9, 1–43 (2001).
Atahan, P. et al. Subsistence and the isotopic signature of herding in the Bronze Age Hexi Corridor, NW Gansu, China. Journal of Archaeological Science 38, 1747–1753 (2011).
Chen, X. L. et al. Raising practices of Neolithic livestock evidenced by stable isotope analysis in the Wei River Valley, North China. International Journal of Osteoarchaeology, https://doi.org/10.1002/oa.2393 (2014).
Dai, L., Kan, X. & Zhang, X. An investigation into the strategy of pig husbandry combining zooarchaeological and stable isotopic approaches at Neolithic Houjiazhai, China. International Journal of Osteoarchaeology, https://doi.org/10.1002/oa.2788 (2019).
Acknowledgements
Wang Hui of the Gansu Province Institute of Archaeology and Cultural Relics graciously provided access to the Dadiwan fauna housed in the Gansu Provincial Museum, Lanzhou, China. Junyang Cao provided translation of Chinese taxonomic nomenclature in the Dadiwan monograph. The original isotopic analyses of these remains were conducted in collaboration with Seth Newsome (then at the Carnegie Institution of Washington) and Chen Fahu (Lanzhou University), with funding from the Wenner-Gren Foundation (awarded to L.B.). The University of Oklahoma provided funding for all molecular analyses (through B.M.K.).
Author information
Authors and Affiliations
Contributions
L.B., B.B. and B.M.K. wrote the main manuscript text; L.B. collected the specimens; B.B. conducted laboratory analyses; K.S. performed shotgun DNA sequencing analysis and produced Fig. 4; C.M. and B.B. produced the median joining network analysis and Fig. 3; A.T. produced the Bayesian analysis and Supplemental Fig. 2; all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barton, L., Bingham, B., Sankaranarayanan, K. et al. The earliest farmers of northwest China exploited grain-fed pheasants not chickens. Sci Rep 10, 2556 (2020). https://doi.org/10.1038/s41598-020-59316-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59316-5
This article is cited by
-
Intensive exploitation of pheasants at the Early Holocene site of Xiaogao in Northern China
Archaeological and Anthropological Sciences (2024)
-
Archaeological and molecular evidence for ancient chickens in Central Asia
Nature Communications (2024)
-
Contextualizing Ancestral Pueblo Turkey (Meleagris gallopavo spp.) Management
Journal of Archaeological Method and Theory (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.