Abstract
Thyroid volume of Hashimoto’s thyroiditis (HT) patients varies in size over the course of disease and it may reflect changes in biological function of thyroid gland. Patients with subclinical hypothyroidism predominantly have increased thyroid volume whereas patients with more pronounced hypothyroidism have smaller thyroid volumes. Suggested mechanism for thyroid atrophy is thyrocyte death due to apoptosis. We performed the first genome-wide association study (GWAS) of thyroid volume in two groups of HT patients, depending on levothyroxine (LT4) therapy, and then meta-analysed across. Study included 345 HT patients in total and 6 007 322 common autosomal genetic variants. Underlying hypothesis was that genetic components that are involved in regulation of thyroid volume display their effect in specific pathophysiologic conditions of thyroid gland of HT patients. We additionally performed immunohistochemical analysis using thyroid tissues and analysed differences in expression levels of identified proteins and apoptotic marker between HT patients and controls. We found genome-wide significant association of two loci, both involved in apoptosis, with thyroid volume of HT patients: rs7212416 inside apoptosis-antagonizing transcription factor AATF (P = 8.95 × 10−9) and rs10738556 near chromatin-remodeling SMARCA2 (P = 2.83 × 10−8). In immunohistochemical analysis we observed that HT patients with homozygous AATF risk genotypes have decreased AATF expression (0.46-fold, P < 0.0001) and increased apoptosis (3.99-fold, P = 0.0001) in comparison to controls. HT patients with heterozygous SMARCA2 genotypes have decreased SMARCA2 expression, albeit without reaching statistical significance (1.07-fold, P = 0.5876), and significantly increased apoptosis (4.11-fold, P < 0.0001). By two lines of evidence we show that two highly plausible genetic loci, AATF and SMARCA2, may be involved in determining the thyroid volume of HT patients. The results of our study significantly add to the current knowledge of disturbed biological mechanisms in thyroid gland of HT patients.
Similar content being viewed by others
Introduction
The main functional unit of thyroid gland is group of cells called follicle. Follicle consists of epithelial follicular cells, known as thyrocytes, which surround colloid filled with thyroid hormone precursor thyroglobulin. The size of follicles, number of thyrocytes and the size of colloid are all dependent on thyroid gland biological activity and can affect thyroid gland size1,2.
Thyroid volume can be enlarged in number of thyroid diseases1 whereas its size can also be decreased, as seen in progressed autoimmune hypothyroidism3,4,5. Moreover, Hashimoto’s thyroiditis (HT), also known as autoimmune thyroiditis, can be presented as goiter on one side of the spectrum or as atrophic thyroid gland on the other4,6. A study demonstrated that patients with autoimmune hypothyroidism have larger dispersion of thyroid volume size in comparison to controls indicating that some patients have increased and some decreased thyroid volumes, i.e. goiter and thyroid atrophy just represent the extremes in the distribution3. Further studies linked thyroid volume size with thyroid function, as it was found that patients with smallest thyroid volumes have more pronounced hypothyroidism3,5, whereas patients with subclinical hypothyroidism predominantly have increased thyroid volume4. It was also suggested that progression of autoimmune thyroiditis to overt hypothyroidism is accompanied by thyroid atrophy4. In other words, as thyroid volume changes over the course of disease5, it reflects the change in biological function of thyroid gland, as well as stroma changes, including lymphocitic infiltration, local edema, and vascularity. The underlying mechanisms of progressing atrophy are not well known, but the most suspected mechanism is thyrocyte death due to apoptosis4,7. In summary, thyroid mass is affected by the number of thyrocytes that are maintained through the balance between their proliferation and apoptosis8. However, predominant process in HT is increased apoptosis9,10,11,12. Although the knowledge on mechanisms and regulation of apoptosis in thyroid gland is limited, it is suggested that the most important apoptotic pathway includes the Fas death receptor10.
There are several other determinants of thyroid follicular cells volume and thyroid volume, including serum TSH and iodine13,14. In iodine sufficient areas, or in cases of iodine supplementation, thyroid volume of healthy individuals ranges within reference values15,16. Other factors that influence thyroid volume are age17, anthropometric features, such as body surface area (BSA)18,19, gender20,21, smoking22 and alcohol consumption23. Beside these, genetic factors are important regulators of thyroid size13. It has been estimated that genetic factors contribute to about 71% of the total variation in thyroid volume of healthy individuals13. Nevertheless, genetic determinants of healthy thyroid volume are not well understood and to date, only one genome-wide association study (GWAS) of thyroid volume in thyroid disease free individuals has been performed24. It identified four independent loci associated with thyroid volume located inside or near CAPZB, FGF7 and LOC44038924. On the other hand, genetic factors that determine thyroid volume in thyroid diseases are completely unexplored.
This study, for the first time, analyzed genome-wide genetic variants underlying thyroid volume of HT patients. We hypothesize that in specific pathological conditions of thyroid gland, such as in HT, genetic components involved in the regulation of thyroid volume could actually be the same ones regulating the number of thyrocytes and thus, biological activity of thyroid gland. The main aim of our study was to identify genetic determinants underlying these processes and to verify findings by protein expression analysis in thyroid tissues that can further deepen the understanding of biology behind pathologic processes in HT.
Methods
Subjects
A total of 370 HT patients were involved in this study. Patients were recruited after complete examination by nuclear medicine specialist at the Outpatient clinic for thyroid disorders at the Department for Nuclear Medicine at the University Hospital Split (Croatia). All patients were of white European ancestry, adult and living in the southern Croatia, a region considered iodine sufficient since 200315. Patients were diagnosed with HT on the basis of clinical examination and diffuse thyroid autoimmune disease established by characteristic thyroid ultrasound (US) imaging (unhomogenic thyroid tissue with diffusely reduced echo levels). Diagnosis was further complemented with biochemical measurements of thyroid hormones and antibodies (increased TSH, decreased T3, T4 and/or increased TPOAb and/or TgAb) according to ETA recommendations and guidelines for Management of Subclinical Hypothyroidism25. During recruitment period, from 2013 to 2016, we were sequentially recruiting newly diagnosed patients as they were referring to Outpatient clinic for thyroid disorders, mostly due to positive thyroid antibodies or abnormal TSH levels established during routine screenings, or because of suspected clinical manifestations of thyroid disorders. We additionally involved patients with previously diagnosed HT that came to physicians on follow-up. All HT patients that were recruited through our study are part of “Croatian biobank of patients with Hashimoto’s thyroiditis” (CROHT) that was established with principal aim of identifying genetic variants associated with HT26 and HT-related traits, such as thyroid antibodies27 and thyroid volume.
Dimensions of thyroid gland in HT patients were determined by thyroid ultrasonography performed using Medison Accuvix V10 (Samsung Medison Co., Ltd, Seoul 135–280, Korea) high frequency linear probe (8–12 MHz). Thyroid volume was calculated as a sum of volumes of both lobes of thyroid gland, where the volume of each lobe was calculated as length × width × depth × 0.47928. The body surface area (BSA) in m2 was estimated as W0.425 × H0.725 × 0.007184, where W is the weight in kg and H is the height in cm29. Thyroid hormones and antibodies levels in plasma of HT patients were determined by immunoassay reaction conducted in a fully automated instrument “Liaison” Biomedica Chemiluminescence Analyzer in the Laboratory of Biochemistry, University Hospital Split. Genomic DNA of HT patients was extracted from peripheral blood leukocytes, using Nucleon Genomic DNA Extraction Kit BACC3, according to the manufacturer’s instructions (GE Healthcare, Little Chalfont, Buckinghamshire, UK). DNA concentration was determined by the Nanodrop ND-1000 Spectrophotometer (ND-1000, Thermo Fisher Scientific, USA). The final concentration of DNA was uniformed across all samples and brought to 100 µg/µl prior genotyping.
Written informed consent was obtained from all study participants, and the study was approved by two separate Ethics Committees: the University of Split, School of Medicine (Classification no. 003-08/14-03/0001; Registry no. 2181-198-03-04-14-0028) and University Hospital Split (Classification no. 530-02/13-01/11; Registry no. 2181-147-01/06/J.B.-14-2). Both Ethics Committees declared that study is in accordance with the provisions of the Code of Ethics and the Helsinki Declaration.
Clinical characteristics of HT patients
Descriptive statistics was used to summarize clinical characteristics of patients. None of continuous variables showed normal distribution (tested by Kolmogorov-Smirnov test), thus were presented as median (first quartile - third quartile), while nominal variables (gender) were presented as number of individuals (%) per category. To compare clinical characteristics between two groups of HT patients, depending on levothyroxine (LT4) therapy, Mann–Whitney-U test was used for continuous variables and χ2-test was used for nominal variables.
Genotyping, quality control (QC) and imputation
All participants were genotyped using Illumina Infinium HumanCoreExome genotyping platform that contains 551 839 markers. We performed quality control (QC) of genotype data, using PLINK and R software, following the standard procedure.
In the sample QC, we excluded all samples with call rate less than 95% and all samples with heterozygosity rate deviating more than three standard deviations from the mean. We cross-checked reported sex with the sex inferred from the genotypes – no exclusions were made based on this criterion. We checked for ethnicity by performing multidimensional scaling analysis (MDS) and visually inspecting the MDS plot (Supplementary Fig. 1). No individuals were excluded based on this criterion. We also checked pair-wise identity by descent (IBD) and did not detect duplicate samples. Twenty-five individuals did not pass QC, thus leaving a total of 345 individuals for imputation and GWAS. After removal of individuals failing QC, we excluded all genetic variants with call rate less than 98% and all variants deviating from Hardy-Weinberg equilibrium (HWE) (P < 10−4).
Following QC, we performed genotype imputation on clean dataset. All SNPSs were aligned to the forward strand prior to the imputation. Genotypes were firstly pre-phased using SHAPEIT2 software30 and then imputed with IMPUTE2 software31 using 1000 Genomes (Phase3, all populations) as reference panel. We checked concordance tables to assess an overall imputation quality, mean genotype concordance in our dataset was 95,51%. We also compared the concordance rate and INFO (type 0) metric of directly typed SNPs to detect poorly genotyped SNPs or possible strand flips. Performed analysis showed that all directly genotyped SNPs used in imputation were well genotyped and correctly aligned to the forward strand. After imputation, we excluded all genetic variants with MAF less than 5%, variants with INFO metric less than 0.4 and variants with HWE P < 10−4. A total of 6 007 542 autosomal genetic variants passed post-imputation QC and were selected for GWAS.
Genome-wide association analyses
We performed GWAS of thyroid volume in two groups of HT patients separately, depending on levothyroxine (LT4) therapy. There were 130 patients that were on LT4 therapy and 215 patients without LT4 therapy. Thyroid volume was firstly adjusted for covariates under standard linear regression model in R: age, gender, TSH and BSA were used as covariates in both groups of HT patients, as they are known to influence thyroid volume. We used LT4 dosage as an additional covariate in the group of HT patients that were on LT4 therapy, as LT4 therapy reduces thyroid volume32,33. Residuals derived from linear regression model were quantile transformed in R and then used as a new phenotype in association analyses, instead of thyroid volume. Association analyses between 6 007 542 common autosomal genetic variants and adjusted thyroid volume were performed under the linear mixed model implemented in GEMMA software which accounts for population stratification and relatedness34.
GWAS meta-analysis
Individual GWAS summary results were meta-analyzed in METAL using fixed-effects inverse variance method which weights genetic variant’s effect sizes (β-coefficients) by their standard errors (SE)35. Manhattan plot of GWAS meta-analysis results was generated using R package qqman. Regional association plots were generated using LocusZoom software36. We checked cluster plots of all directly genotyped variants with P-value less than 0.1 located +/− 400 kb from top-associated variants, to eliminate possible spurious associations. Cluster genotyping plots were generated using Ilummina GenomeStudio software. For genetic variants associated with thyroid volume on the genome-wide level of significance (P < 5 × 10−8), we assessed the distribution of thyroid volume per genotype and generated box plots using R statistical software.
Verification of results using immunohistochemical analysis
Thyroid tissue paraffin blocks were obtained from HT patients who are part of our CROHT biobank and control participants from “10001 Dalmatians” biobank37, with available genome-wide data, who underwent thyroidectomy at University Hospital of Split. Tissues were reviewed by expert pathologist for signs of HT and for confirmation of healthy (control) tissue. We analyzed expression of AATF, SMARCA2 and apoptotic marker cytochrome C (CytC) in thyroid follicles. Immunohistochemical analysis was used to test AATF/SMARCA2 and apoptotic marker expression between HT cases and controls of the same genotype i.e. if HT patients with one/two risk alleles have decreased AATF/SMARCA2 expression and increased apoptosis in comparison to controls with the same genotype. As the risk allele T of rs7212416 (AATF) has allele frequency of 85%, we were able to acquire tissues from 3 HT patients and 7 controls with TT genotypes (individuals with two risk alleles). Due to allele frequency of 50% of the risk allele T of rs10738556 (SMARCA2), there were no thyroid specimens from individuals with homozygous TT genotypes in our Biobanks, therefore, we focused on analysis of heterozygous individuals. We were able to acquire tissues from 5 HT patients and 7 controls with heterozygous CT genotypes (i.e. individuals with one risk allele).
Formalin fixed and paraffin embedded tissue sections were deparaffinized and run through process of antigen retrieval in citrate buffer. Nonspecific binding was blocked by Protein Block (Abcam, Cambridge, UK). Tissue sections were incubated with primary antibodies to AATF (Abcam, Cambridge, UK), SMARCA2 (Abcam, Cambridge, UK) and CytC (Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4 °C. Staining was visualized by incubation with secondary antibodies labeled with red (donkey anti rabbit Cy3 polyclonal, Dako, Glostrup, Denmark) and green (donkey anti mouse labeled with AF488, Invitrogen, Carlsbad, CA, USA) fluorochromes. Finally, samples were counterstained with DAPI (4′,6-diamidino-2-phenylindole).
Photo-micrographs were shot by SPOT Insight digital camera (Diagnostic Instruments, USA), mounted on Olympus BX61 fluorescence microscope (Olympus, Tokyo, Japan). Camera settings were set using image acquisition software CellA® at 1360 × 1024 resolution, exposition of 1/333.3 s with noise reduction filter. Ten micro-photographs of thyroid follicles, under the magnification of 200×, were shot per slide in all of 3 fluorescent channels. Furthermore, florescence intensity histograms were acquired for red and green fluorescence channels in ImageJ software (NIH, Bethesda, MD, USA). Region of positive signal was determined by using the slides stained with secondary antibodies only, thus quantifying the autofluorescence and fluorescence due to unspecific binding of secondary antibodies. Region of positive signal was defined as the one that excluded 99.99% of the signal obtained from florescence intensity histograms of slides stained with secondary antibodies only. Expression of AATF and SMARCA2 was quantified as the area under the curve (AUC) of florescence intensity histograms. On the other hand, since CytC release from mitochondria into cytoplasm is a marker of apoptosis, unlike its expression, we quantified it as a percentage of micro-photograph area covered by positive signal. AUCs and their interval estimates were calculated by using AUC analysis routine in GraphPad Prism 8.0 software (Graph Pad, La Jolla, CA, USA). Statistical significance and effect sizes as well as respective 95%CI were calculated by t-test with Welch correction (Graph Pad, La Jolla, CA, USA). For comparisons of percentage of area covered by positive signal t-test was used.
Results
Clinical characteristics of HT patients
Clinical characteristics of HT patients divided in two sets, depending on LT4 therapy, are summarized in Table 1. HT patients without LT4 therapy have mean TSH levels just slightly above the upper reference values for our population, while their median T3 and T4 levels are in normal reference ranges. Expectedly, hormone values of patients without therapy reflect the fact that these patients are newly diagnosed or in follow-up with preserved thyroid function. HT patients that are on LT4 therapy have significantly lower TSH levels (P = 0.0001), and significantly higher T4 (P < 0.0001) and fT4 (P < 0.0001) levels, however their T3 and T4 levels are in reference ranges for our population, suggesting that their LT4 supplementation is appropriate. Male to female ratio is in accordance to expected distribution of HT according to the gender, with higher incidence in females38. Additionally, HT patients that are on LT4 therapy have significantly lower thyroid volume (P = 0.0024). There are no significant differences between two groups of HT patients in age, gender, BSA, and T3 levels. We also do not observe differences in median TgAb/TPOAb levels between two groups of patients.
GWAS meta-analysis
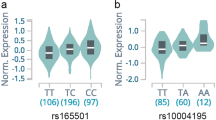
We identified two genome-wide significant hits in this first GWAS meta-analysis of thyroid volume in HT patients (Fig. 1). The most associated SNPs representing each signal are rs7212416 inside apoptosis-antagonizing transcription factor AATF (P = 8.95 × 10−9, β = −0.589, SE = 0.102 for allele T) and rs10738556 located 7 kb from chromatin-remodeling SMARCA2 (P = 2.83 × 10−8, β = −0.44, SE = 0.079 for allele T) (Table 2). Regional association plots of these genetic variants are shown in Supplementary Fig. 2. Box plots of distribution of thyroid volume per genotype for both genetic variants are shown in Fig. 2.
We identified additional six suggestively associated loci with thyroid volume (P < 10−6) (Fig. 1): rs201666766 inside DNAH5 (P = 1.21 × 10−7, β = −1.051, SE = 0.199 for allele A), rs170884 inside FAM129A (P = 1.22 × 10−7, β = 0.452, SE = 0.085 for allele C), rs4747268 close to PTER (P = 1.26 × 10−7, β = −0.539, SE = 0.102 for allele A), rs193145729 close to MIR8054 (P = 5.25 × 10−7, β = 0.538, SE = 0.107 for allele G), rs11247367 inside IGF1R (P = 6.59 × 10−7, β = 0.786, SE = 0.158 for allele G) and rs10767013 between MIR8054 and CCDC179 (P = 7.44 × 10−7, β = 0.362, SE = 0.073 for allele G) (Table 2). Regional association plots of these genetic variants are shown in Supplementary Fig. 3.
Immunohistochemical analysis of AATF and SMARCA2
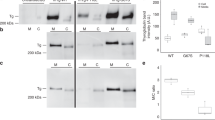
In order to examine expression patterns on protein level, we measured expression of AATF along with apoptosis in thyroid tissues of samples with rs7212416 TT genotypes. Patients with HT had significant 0.46-fold decrease in AATF levels when compared to control patients (95% CI: −0.69 to −0.24, p < 0.0001). This was accompanied by significant increase in apoptosis by 3.99-fold in comparison to control samples (95% CI: 2.74 to 4.57, p = 0.0001), as shown in Fig. 3A and Supplementary Fig. 4.
Differences in expression of (A) AATF and CytC between HT patients (n = 3) and controls (n = 5) with rs7212416 TT genotypes (B) SMARCA2 and CytC in HT patients (n = 5) and controls (n = 7) with rs10738556 CT genotypes. Data is represented as mean and 95% CI of fold change in relative difference between groups, normalized to control group. ****p < 0.0001.
We also analyzed expression of SMARCA2 and apoptosis in thyroid tissue of samples with rs10738556 CT genotypes. SMARCA2 levels were decreased by 1.07-fold in HT group in comparison to control group, however without reaching statistical significance (95% CI: −19 to 0.67, p = 0.5876), whereas we observe significant increase in apoptosis by 4.11-fold in HT group (95% CI: 2.9 to 4.32, p < 0.0001) (Fig. 3B, Supplementary Fig. 4).
Discussion
We have identified two genome-wide significant loci with thyroid volume of HT patients, one in apoptosis-antagonizing transcription factor AATF (also known as Che-1) and other near chromatin remodeling SMARCA2, associated with transcriptional regulation. Additionally, we have identified six loci suggestively associated with thyroid volume, of which two, IGF1R and FAM129A, play a role in apoptosis, thyroid morphogenesis and thyroid carcinomas, respectively. Identification of mentioned genes with clear connection to apoptosis is particularly interesting as apoptosis of thyrocytes affects the mass of thyroid gland and is considered to be the landmark of HT8,9,10,11,12. We additionally performed immunohistochemical analysis of expression levels of AATF, SMARCA2 and apoptotic marker in thyroid tissues of HT patients and controls to further explore involvement of identified proteins in pathophysiological processes of thyroid gland of HT patients.
Here, we observe a negative effect of the major allele T (allele frequency 85%) of the most associated AATF genetic variant, rs7212416, on thyroid volume. Patients with major allele on this SNP have decreased thyroid volume (Table 2, Fig. 2A). Our hypothesis is that in the microenvironment of thyroid gland of HT patients, underlying genetic variants may affect AATF activity and possibly regulate the magnitude of apoptosis. As AATF has anti-apoptotic role, increased apoptotic activity in thyroid gland of HT patients might be modulated by decreased activity of AATF, due to its lower expression or ubiquitin-dependent degradation39. We additionally performed immunohistochemical analysis in which we were able to demonstrate that HT patients (homozygous for AATF rs7212416 risk allele T), have significantly decreased AATF expression and significantly increased apoptosis in comparison to controls with the same genotype (Fig. 3A, Supplementary Fig. 4). These results provide further support of involvement of AATF in HT pathophysiology. However, additional experiments need to be performed to drive conclusion about association of identified SNP with AATF expression levels and apoptosis.
We also observe in GWAS analysis that the effect is stronger (absolute β value) in the group of HT patients that are in progressed stage of disease (overt hypothyroidism and on LT4 therapy) in comparison to newly diagnosed HT patients without LT4 therapy (Table 2), which is in line with the hypothesis that thyroid atrophy is a proxy for underlying apoptosis and is more pronounced in later stages of HT.
Beside an anti-apoptotic role, AATF has several other versatile, but correlative roles in transcriptional regulation, induction of cell proliferation, cell cycle checkpoint control and response to DNA damage through activation of p5340,41,42,43,44. Bruno et al. suggested interesting role of AATF as a novel effector in the regulation of p53 pathway41. In this model, DNA damage-induced phosphorylation and stabilization of AATF, set off cellular arrest by increasing transcription of p53 and p2141. An anti-apoptotic role of AATF is further suggested because activation of DNA damage checkpoints by AATF promotes DNA repair and cell survival41, whereas in the case of extensive DNA damage, a cell is destined for apoptosis and AATF is targeted for degradation39. Similarly, experimental silencing of AATF resulted with decreased cell proliferation and promotion of apoptosis42,45. Additionally, upon AATF silencing, pro-apoptotic genes, including Fas and FasL, were up regulated, while multiple anti-apoptotic genes were down-regulated45. Since the apoptosis via Fas death receptor is the hallmark of HT10, involvement of AATF in Fas/FasL signaling is compelling.
As we have previously performed GWAS study of 405 HT cases and 430 controls26, we were able to check allele frequencies of the most associated AATF genetic variant (rs7212416) in both groups. We do not observe difference in allele frequency distribution of this SNP between HT patients and controls implying that an effect of this SNP on thyroid volume may be triggered by pathologic conditions in thyroid of HT patients. A further indication of suggested mechanism is demonstrated by our immunohistochemical analysis where we found significantly decreased AATF levels and increased apoptosis in thyroid follicles of HT patients in comparison to controls with the same rs7212416 TT genotype. We therefore propose that SNP effect may be secondary to inflammatory events in the thyroid of HT patients where inflammatory cytokines initiate autoimmunity and play regulative roles in expression of apoptotic signaling46,47. Studies have shown that thyrocytes, alongside immune cells, contribute to secretion of proinflammatory cytokines of Th1 type that stimulate autocrine/paracrine apoptosis induction of thyrocytes48. However, as already marked, true association between identified genetic variant and apoptosis needs to be analyzed in additional tests.
Interestingly, a genomic region encompassing AATF has already been strongly associated with chemokine macrophage inflammatory protein 1b levels (MIP-1-beta)49, which is involved in cytokine signaling in immune system50. This chemokine has already been associated with autoimmune pathology of adrenal gland51. Furthermore, MIP-1-beta has also been linked to Fas-FasL mediated apoptosis52. As chemokines, together with Th1 cytokines, are considered the main regulators of autoimmune process in HT, a finding of association of the same genomic region with MIP-1-beta and regulation of thyroid volume is intriguing and provides multiple lines of evidence of involvement of this genomic region with apoptosis.
This study presents yet another important link between our two major hits, AATF and SMARCA2. A transcriptional regulator SMARCA2, also known as hBRM, is a component of SWI/SNF family of proteins that changes chromatin structure through altering DNA-histone contacts (www.genecards.org). hSWI/SNF chromatin remodelers have already been linked to apoptosis53,54. Overexpression of SMARCA2 has been associated with decrease in apoptosis of ovarian cancer cells55 and pancreatic cancer cells56. On the other hand, SMARCA2 is found to be cleaved during apoptosis57 and its truncated levels correlate with cell damage and apoptosis after virus infection58. It has been shown that SMARCA2 also interacts with HDAC159, the deacetylase known for its role in regulation of the cell cycle progression through Rb binding that is inhibited by AATF60. In brief, Bruno et al. have proven that AATF directly inactivates HDAC1/Rb/E2F1 transcriptional inhibition by competition with HDAC1 for the Rb binding site, thus inducing cellular growth and proliferation. On the other hand, a glucocorticoid receptor-induced apoptosis is found to be dependent on Rb and SMARCA261. From our results, we can speculate that both AATF and SMARCA2, through common interaction with HDAC1 and Rb, could be involved in the regulation of transcription that blocks cell proliferation and subsequently drives thyrocytes to apoptosis in thyroid gland of HT patients.
We also demonstrate that HT patients (heterozygous for SMARCA2 rs10738556) have decreased SMARCA2 expression, albeit without reaching statistical significance, and significantly increased apoptosis in comparison to controls (Fig. 3B, Supplementary Fig. 4). We assume that due to analysis of heterozygous individuals i.e. individuals with only one risk allele, we observed a weaker decrease in SMARCA2 expression. Although these results suggest involvement of SMARCA2 in molecular processes specific to HT, more tests need to be performed to give definite conclusion on association of identified genetic variant with differential SMARCA2 expression and induction of apoptosis.
Furthermore, SMARCA2 is involved in the same pathway as our next suggestively associated locus, IGF1R. They both belong to hepatocellular carcinoma (HCC) pathway that has in turn been associated with abnormal thyroid hormone signaling62 further linking our genetic variants with thyroid dysfunction. IGF1R has pleiotropic roles in humans, among which two roles are relevant for our study. Firstly, it is an anti-apoptotic marker and, secondly, it has a direct role in functionality of thyroid gland. In in vitro conditions, IGF1R works together with TSH in stimulating thyrocyte growth and is actually found to be essential for regulation of thyroid function and TSH-stimulated goitrogenesis63. Important role in maintaining homeostasis and normal thyroid morphogenesis was further demonstrated in animal model64. These data suggest an important role of IGF1R with thyroid growth morphogenesis thus providing a concrete support for our finding of association of this gene with thyroid volume. We believe that this is a true candidate gene for maintaining thyroid volume albeit our study was not powerful enough to detect its association on a genome-wide level.
Finally, we bold out our second suggestively associated locus, FAM129A, also known as C1orf24 and NIBAN, as it was also found to act in anti-apoptotic manner, in regulation of p53-mediated apoptosis65. FAM129A is highly expressed in carcinomas including thyroid carcinomas66,67 and is used as one of several markers that may differentiate benign from malignant thyroid lesions, therefore, it is considered to be the best predictor for thyroid cancer68,69,70,71. FAM129A silencing in thyroid carcinoma cell lines directly leads to apoptosis, cell migration inhibition and cell cycle progression70. Finally, immunohistochemical staining showed that FAM129A, beside in thyroid tumors, is weekly expressed in subsets of scattered cells with oxyphilic cell metaplasia in HT and not in normal thyroid tissue71. Taken together, the current knowledge on this gene and our results, imply the role of FAM129A in the apoptosis regulation in thyroid tissue of HT patients.
Beside the role of FAM129A in thyroid cancer, the loss of SMARCA2 was also found to be associated with thyroid cancer, particularly with anaplastic thyroid cancer72. Given the fact that some studies report an increased incidence of well-differentiated thyroid cancer in chronic autoimmune thyroiditis73,74, it is possible that there is a genetic link between HT and thyroid cancer, i.e. that changes in expression of SMARCA2 and FAM129A in HT can predispose to development of some forms of thyroid cancer. If this holds true, then SMARCA2/FAM129A could be potential biomarkers for stratifying HT patients who maybe of higher risk of developing thyroid cancer. However, more research using prospective studies with full genetic profiling is needed to test this hypothesis, as our study was not designed to give answer on genetic correlation between HT and thyroid carcinomas.
Further, we have identified another four suggestively associated variants located inside or near DNAH5, PTER and MIR8054. Genetic variants inside these genomic regions were previously associated with number of traits, such as bacterial meningitis (DNAH5 region)75, hepatitis B (PTER region)75, cancer (MIR8054 region)76, allergic rhinitis (MIR8054 region)77.
Finally, we reflect on limits of our study, the main one being the sample size. Although the sample size of 345 individuals, as used in our study, is considered to be small dataset for GWAS studies, we have carefully selected our phenotype and study population to increase genetic effect sizes and thus the power of our study. It is known that a thoughtful selection of investigated phenotypes, for example for the disease severity, may enrich for genetic causes78. Thyroid volume in HT patients is considered to change over the course of disease and it is smaller in patients with prolonged disease duration, representing tissue degradation5. This is exactly what we observe in our cohort, individuals with prolonged disease and in overt hypothyreosis (those that are taking LT4 therapy), have smaller thyroid volumes than newly diagnosed individuals (Table 1). We have used this information to increase the power of our study and analyzed thyroid volume separately in two subsets of HT patients, depending on LT4 therapy, as these two subsets reflect the severity of disease, and then meta-analyzed across. Finally, our initial hypothesis was that a change in size of thyroid tissue may be seen as a proxy for change in biological activity of thyroid but also as manifestation of underlying apoptotic processes. The results of our study validate the design of our GWAS analysis. However, as we do not have replication cohort, and to our knowledge, there is currently no other cohort of HT patients with available GWAS data and thyroid dimensions, it is very important to replicate and confirm our findings in other genetic and functional studies. Furthermore, we found significantly decreased AATF and indication of decreasing SMARCA2 expression levels accompanied by significantly increased apoptosis in HT patients with two or one risk allele, respectively. Our study is not designed to provide direct evidence that identified genetic variants are associated with decrease in AATF/SMARCA2 expression and increase in apoptosis, therefore, for confirmation of our hypothesis, a further study on causality between association of genetic variants with expression of AATF/SMARCA2 and apoptosis in thyroid cell lines is needed. Additionally, due to lack of available thyroid tissue, we could not evaluate expression levels of AATF/SMARCA2 and apoptosis in individuals among three different genotypes.
In summary: (a) our GWAS study of thyroid volume in HT patients identified association of two genetic variants in AATF/SMARCA2, thus implying that regulation of thyroid volume of patients with HT has a genetic component; (b) further analysis of expression patterns showed differential expression levels of AATF and SMARCA2 between HT cases and controls of the same genotype accompanied by significantly increased apoptosis in HT patients, further suggesting their role in pathophysiological processes of thyroid gland of HT patients. Additional clarification of mechanisms through which identified genetic variants exhibit their effect is needed. Based on results of our study and function of associated proteins, we speculate that these genetic variants may display their effect under specific pathophysiologic conditions of thyroid gland of HT patients, possibly through modulation of AATF/SMARCA2 expression levels and induction of apoptosis. Our study provides suggestive evidence of involvement of six more loci in regulation of thyroid volume, of which IGF1R and FAM129A have also been involved in apoptosis and thyroid gland functionality. If manipulation of apoptosis would be possible in HT patients, as suggested to be one of therapeutic strategies for autoimmune diseases79, then these proteins, especially AATF, could be good therapeutic target candidates for testing the prevention of further thyroid damage in HT patients. In conclusion, we identified two highly plausible genetic loci, AATF and SMARCA2, which seem to be involved in determining the thyroid volume in HT. The results of our study significantly add to the current knowledge of disturbed biological mechanisms in thyroid gland of HT patients and pathogenesis of HT on molecular level.
Data availability
Genotype data are deposited in Croatian Digital Academic Archives and Repositories (DABAR) and are available following this link: https://urn.nsk.hr/urn:nbn:hr:171:066445.
References
Jamjoom, A. The influence of concomitant intradural pathology on the presentation and outcome of patients with acute traumatic extradural haematoma. Acta Neurochir. 115, 86–89 (1992).
Albi, E. et al. The thyroid lobes: the different twins. Arch. Biochem. Biophys. 518, 16–22, https://doi.org/10.1016/j.abb.2011.12.001 (2012).
Carle, A. et al. Thyroid volume in hypothyroidism due to autoimmune disease follows a unimodal distribution: evidence against primary thyroid atrophy and autoimmune thyroiditis being distinct diseases. J. Clin. Endocrinol. Metab. 94, 833–839, https://doi.org/10.1210/jc.2008-1370 (2009).
Bulow Pedersen, I. et al. A population study of the association between thyroid autoantibodies in serum and abnormalities in thyroid function and structure. Clin. Endocrinol. 62, 713–720, https://doi.org/10.1111/j.1365-2265.2005.02284.x (2005).
Nordmeyer, J. P., Shafeh, T. A. & Heckmann, C. Thyroid sonography in autoimmune thyroiditis. A prospective study on 123 patients. Acta Endocrinol. 122, 391–395 (1990).
Raber, W., Gessl, A., Nowotny, P. & Vierhapper, H. Thyroid ultrasound versus antithyroid peroxidase antibody determination: a cohort study of four hundred fifty-one subjects. Thyroid. 12, 725–731, https://doi.org/10.1089/105072502760258712 (2002).
Phelps, E., Wu, P., Bretz, J. & Baker, J. R. Jr. Thyroid cell apoptosis. A new understanding of thyroid autoimmunity. Endocrinol. Metab. Clin. North. Am. 29, 375–388, viii (2000).
Tamura, M. et al. Role of apoptosis of thyrocytes in a rat model of goiter. A possible involvement of Fas system. Endocrinol. 139, 3646–3653, https://doi.org/10.1210/endo.139.8.6140 (1998).
Xu, C. et al. Excess iodine promotes apoptosis of thyroid follicular epithelial cells by inducing autophagy suppression and is associated with Hashimoto thyroiditis disease. J. Autoimmun. 75, 50–57, https://doi.org/10.1016/j.jaut.2016.07.008 (2016).
Kaczmarek, E., Lacka, K., Jarmolowska-Jurczyszyn, D., Sidor, A. & Majewski, P. Changes of B and T lymphocytes and selected apopotosis markers in Hashimoto’s thyroiditis. J. Clin. Pathol. 64, 626–630, https://doi.org/10.1136/jcp.2010.086553 (2011).
Wang, S. H. & Baker, J. R. The role of apoptosis in thyroid autoimmunity. Thyroid. 17, 975–979, https://doi.org/10.1089/thy.2007.0208 (2007).
Wang, S. H. et al. A unique combination of inflammatory cytokines enhances apoptosis of thyroid follicular cells and transforms nondestructive to destructive thyroiditis in experimental autoimmune thyroiditis. J. Immunol. 168, 2470–2474 (2002).
Hansen, P. S. et al. Genetic and environmental causes of individual differences in thyroid size: a study of healthy Danish twins. J. Clin. Endocrinol. Metab. 89, 2071–2077, https://doi.org/10.1210/jc.2003-031999 (2004).
Rognoni, J. B., Penel, C., Bastiani, P., Roccabianca, M. & Lemarchand-Beraud, T. Down regulation of hypertrophied follicular cell volume in thyroid hyperplastic gland. Histol. Histopathol. 4, 193–200 (1989).
Kusic, Z. et al. Croatia has reached iodine sufficiency. J. Endocrinol. Invest. 26, 738–742 (2003).
Bonofiglio, D. et al. Beneficial effects of iodized salt prophylaxis on thyroid volume in an iodine deficient area of southern Italy. Clin. Endocrinol. 71, 124–129, https://doi.org/10.1111/j.1365-2265.2008.03432.x (2009).
Krejbjerg, A. et al. Iodine fortification may influence the age-related change in thyroid volume: a longitudinal population-based study (DanThyr). Eur. J. Endocrinol. 170, 507–517, https://doi.org/10.1530/EJE-13-0918 (2014).
Boas, M. et al. Association of thyroid gland volume, serum insulin-like growth factor-I, and anthropometric variables in euthyroid prepubertal children. J. Clin. Endocrinol. Metab. 94, 4031–4035, https://doi.org/10.1210/jc.2009-0939 (2009).
Fuse, Y., Saito, N., Tsuchiya, T., Shishiba, Y. & Irie, M. Smaller thyroid gland volume with high urinary iodine excretion in Japanese schoolchildren: normative reference values in an iodine-sufficient area and comparison with the WHO/ICCIDD reference. Thyroid. 17, 145–155, https://doi.org/10.1089/thy.2006.0209 (2007).
Tarnoki, A. D. et al. Genetic and environmental influence on thyroid gland volume and thickness of thyroid isthmus: a twin study. Arch. Endocrinol. Metab. 59, 487–494, https://doi.org/10.1590/2359-3997000000110 (2015).
Gomez, J. M., Maravall, F. J., Gomez, N., Guma, A. & Soler, J. Determinants of thyroid volume as measured by ultrasonography in healthy adults randomly selected. Clin. Endocrinol. 53, 629–634 (2000).
Knudsen, N. et al. Association of tobacco smoking with goiter in a low-iodine-intake area. Arch. Intern. Med. 162, 439–443 (2002).
Knudsen, N. et al. Alcohol consumption is associated with reduced prevalence of goitre and solitary thyroid nodules. Clin. Endocrinol. 55, 41–46 (2001).
Teumer, A. et al. Genome-wide association study identifies four genetic loci associated with thyroid volume and goiter risk. Am. J. Hum. Genet. 88, 664–673, https://doi.org/10.1016/j.ajhg.2011.04.015 (2011).
Pearce, S. H. et al. ETA Guideline: Management of Subclinical Hypothyroidism. Eur. Thyroid. J. 2, 215–228, https://doi.org/10.1159/000356507 (2013).
Brcic, L. et al. Genome-wide association analysis suggests novel loci for Hashimoto’s thyroiditis. J Endocrinol Invest, https://doi.org/10.1007/s40618-018-0955-4 (2018).
Brcic, L. et al. Genome-wide association analysis suggests novel loci underlying thyroid antibodies in Hashimoto’s thyroiditis. Sci. Rep. 9, 5360, https://doi.org/10.1038/s41598-019-41850-6 (2019).
Brunn, J. et al. [Volumetric analysis of thyroid lobes by real-time ultrasound (author’s transl)]. Dtsch. Med. Wochenschr. 106, 1338–1340, https://doi.org/10.1055/s-2008-1070506 (1981).
Du Bois, D. & Du Bois, E. F. A formula to estimate the approximate surface area if height and weight be known. 1916. Nutrition 5, 303–311; discussion 312–303 (1989).
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6, https://doi.org/10.1038/nmeth.2307 (2013).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529, https://doi.org/10.1371/journal.pgen.1000529 (2009).
Scarpa, V. et al. Treatment with thyroxine reduces thyroid volume in euthyroid children and adolescents with chronic autoimmune thyroiditis. Horm. Res. Paediatr. 73, 61–67, https://doi.org/10.1159/000271917 (2010).
Dorr, H. G. et al. Levothyroxine Treatment of Euthyroid Children with Autoimmune Hashimoto Thyroiditis: Results of a Multicenter, Randomized, Controlled Trial. Horm. Res. Paediatr. 84, 266–274, https://doi.org/10.1159/000437140 (2015).
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824, https://doi.org/10.1038/ng.2310 (2012).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. 26, 2190–2191, https://doi.org/10.1093/bioinformatics/btq340 (2010).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinforma. 26, 2336–2337, https://doi.org/10.1093/bioinformatics/btq419 (2010).
Rudan, I. et al. 10001 Dalmatians:” Croatia launches its national biobank. Croat. Med. J. 50, 4–6 (2009).
Mincer, D. L. & Jialal, I. In StatPearls (2019).
De Nicola, F. et al. The prolyl isomerase Pin1 affects Che-1 stability in response to apoptotic DNA damage. J. Biol. Chem. 282, 19685–19691, https://doi.org/10.1074/jbc.M610282200 (2007).
Passananti, C., Floridi, A. & Fanciulli, M. Che-1/AATF, a multivalent adaptor connecting transcriptional regulation, checkpoint control, and apoptosis. Biochem. Cell Biol. 85, 477–483, https://doi.org/10.1139/O07-062 (2007).
Bruno, T. et al. Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell 10, 473–486, https://doi.org/10.1016/j.ccr.2006.10.012 (2006).
Bruno, T. et al. Che-1 promotes tumor cell survival by sustaining mutant p53 transcription and inhibiting DNA damage response activation. Cancer Cell 18, 122–134, https://doi.org/10.1016/j.ccr.2010.05.027 (2010).
Hopker, K. et al. AATF/Che-1 acts as a phosphorylation-dependent molecular modulator to repress p53-driven apoptosis. EMBO J. 31, 3961–3975, https://doi.org/10.1038/emboj.2012.236 (2012).
Bruno, T., Iezzi, S. & Fanciulli, M. Che-1/AATF: A Critical Cofactor for Both Wild-Type- and Mutant-p53 Proteins. Front. Oncol. 6, 34, https://doi.org/10.3389/fonc.2016.00034 (2016).
Sharma, M. Apoptosis-antagonizing transcription factor (AATF) gene silencing: role in induction of apoptosis and down-regulation of estrogen receptor in breast cancer cells. Biotechnol. Lett. 35, 1561–1570, https://doi.org/10.1007/s10529-013-1257-8 (2013).
Khan, F. A., Al-Jameil, N., Khan, M. F., Al-Rashid, M. & Tabassum, H. Thyroid dysfunction: an autoimmune aspect. Int. J. Clin. Exp. Med. 8, 6677–6681 (2015).
Wang, S. H. et al. Microarray analysis of cytokine activation of apoptosis pathways in the thyroid. Endocrinol. 148, 4844–4852, https://doi.org/10.1210/en.2007-0126 (2007).
Qin, Q. et al. The increased but non-predominant expression of Th17- and Th1-specific cytokines in Hashimoto’s thyroiditis but not in Graves’ disease. Braz. J. Med. Biol. Res. 45, 1202–1208 (2012).
Ahola-Olli, A. V. et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am. J. Hum. Genet. 100, 40–50, https://doi.org/10.1016/j.ajhg.2016.11.007 (2017).
Chaisavaneeyakorn, S. et al. Levels of macrophage inflammatory protein 1 alpha (MIP-1 alpha) and MIP-1 beta in intervillous blood plasma samples from women with placental malaria and human immunodeficiency virus infection. Clin. Diagn. Lab. Immunol. 10, 631–636 (2003).
Bellastella, G. et al. Simultaneous evaluation of the circulating levels of both Th1 and Th2 chemokines in patients with autoimmune Addison’s disease. J. Endocrinol. Invest. 34, 831–834, https://doi.org/10.3275/7414 (2011).
Mita, A. et al. Expression of Fas ligand by hepatic macrophages in patients with fulminant hepatic failure. Am. J. Gastroenterol. 100, 2551–2559, https://doi.org/10.1111/j.1572-0241.2005.00265.x (2005).
Wang, L. et al. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol. Cell Biol. 25, 7953–7965, https://doi.org/10.1128/MCB.25.18.7953-7965.2005 (2005).
Naidu, S. R., Love, I. M., Imbalzano, A. N., Grossman, S. R. & Androphy, E. J. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 28, 2492–2501, https://doi.org/10.1038/onc.2009.121 (2009).
Xu, X. et al. Overexpression of SMARCA2 or CAMK2D is associated with cisplatin resistance in human epithelial ovarian cancer. Oncol. Lett. 16, 3796–3804, https://doi.org/10.3892/ol.2018.9109 (2018).
Zhang, Z. et al. BRM/SMARCA2 promotes the proliferation and chemoresistance of pancreatic cancer cells by targeting JAK2/STAT3 signaling. Cancer Lett. 402, 213–224, https://doi.org/10.1016/j.canlet.2017.05.006 (2017).
Biggs, J. R. et al. The human brm protein is cleaved during apoptosis: the role of cathepsin G. Proc. Natl Acad. Sci. USA 98, 3814–3819, https://doi.org/10.1073/pnas.071057398 (2001).
Dudek, A. H., Pfaff, F., Bolte, H., Kontchou, C. W. & Schwemmle, M. Partial inactivation of the chromatin remodelers SMARCA2 and SMARCA4 in virus-infected cells by caspase-mediated cleavage. J. Virol. https://doi.org/10.1128/JVI.00343-18 (2018).
Kotlyar, M., Pastrello, C., Sheahan, N. & Jurisica, I. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 44, D536–541, https://doi.org/10.1093/nar/gkv1115 (2016).
Bruno, T. et al. Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell 2, 387–399 (2002).
Singh, P., Chan, S. W. & Hong, W. Retinoblastoma protein is functionally distinct from its homologues in affecting glucocorticoid receptor-mediated transcription and apoptosis. J. Biol. Chem. 276, 13762–13770, https://doi.org/10.1074/jbc.M100137200 (2001).
Manka, P. et al. Thyroid hormone in the regulation of hepatocellular carcinoma and its microenvironment. Cancer Lett. 419, 175–186, https://doi.org/10.1016/j.canlet.2018.01.055 (2018).
Ock, S. et al. IGF-1 receptor deficiency in thyrocytes impairs thyroid hormone secretion and completely inhibits TSH-stimulated goiter. FASEB J. 27, 4899–4908, https://doi.org/10.1096/fj.13-231381 (2013).
Muller, K. et al. TSH compensates thyroid-specific IGF-I receptor knockout and causes papillary thyroid hyperplasia. Mol. Endocrinol. 25, 1867–1879, https://doi.org/10.1210/me.2011-0065 (2011).
Ji, H. et al. AKT-dependent phosphorylation of Niban regulates nucleophosmin- and MDM2-mediated p53 stability and cell apoptosis. EMBO Rep. 13, 554–560, https://doi.org/10.1038/embor.2012.53 (2012).
Majima, S., Kajino, K., Fukuda, T., Otsuka, F. & Hino, O. A novel gene “Niban” upregulated in renal carcinogenesis: cloning by the cDNA-amplified fragment length polymorphism approach. Jpn. J. Cancer Res. 91, 869–874 (2000).
Ito, S. et al. Frequent expression of Niban in head and neck squamous cell carcinoma and squamous dysplasia. Head. Neck 32, 96–103, https://doi.org/10.1002/hed.21153 (2010).
Cerutti, J. M. et al. A preoperative diagnostic test that distinguishes benign from malignant thyroid carcinoma based on gene expression. J. Clin. Invest. 113, 1234–1242, https://doi.org/10.1172/JCI19617 (2004).
Cerutti, J. M. et al. Diagnosis of suspicious thyroid nodules using four protein biomarkers. Clin. Cancer Res. 12, 3311–3318, https://doi.org/10.1158/1078-0432.CCR-05-2226 (2006).
Carvalheira, G., Nozima, B. H. & Cerutti, J. M. microRNA-106b-mediated down-regulation of C1orf24 expression induces apoptosis and suppresses invasion of thyroid cancer. Oncotarget 6, 28357–28370, https://doi.org/10.18632/oncotarget.4947 (2015).
Matsumoto, F. et al. A novel tumor marker, Niban, is expressed in subsets of thyroid tumors and Hashimoto’s thyroiditis. Hum. Pathol. 37, 1592–1600, https://doi.org/10.1016/j.humpath.2006.06.022 (2006).
Kasaian, K. et al. The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer 15, 984, https://doi.org/10.1186/s12885-015-1955-9 (2015).
Zeng, R. et al. Relationship between Hashimoto’s thyroiditis and papillary thyroid carcinoma in children and adolescents. Eur. Rev. Med. Pharmacol. Sci. 22, 7778–7787, https://doi.org/10.26355/eurrev_201811_16401 (2018).
Uhliarova, B. & Hajtman, A. Hashimoto’s thyroiditis - an independent risk factor for papillary carcinoma. Braz. J. Otorhinolaryngol. 84, 729–735, https://doi.org/10.1016/j.bjorl.2017.08.012 (2018).
Tian, C. et al. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 8, 599, https://doi.org/10.1038/s41467-017-00257-5 (2017).
Liu, W., Jiao, X., Thutkawkorapin, J., Mahdessian, H. & Lindblom, A. Cancer risk susceptibility loci in a Swedish population. Oncotarget 8, 110300–110310, https://doi.org/10.18632/oncotarget.22687 (2017).
Bunyavanich, S. et al. Integrated genome-wide association, coexpression network, and expression single nucleotide polymorphism analysis identifies novel pathway in allergic rhinitis. BMC Med. Genomics 7, 48, https://doi.org/10.1186/1755-8794-7-48 (2014).
Schaid, D. J., Chen, W. & Larson, N. B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19, 491–504, https://doi.org/10.1038/s41576-018-0016-z (2018).
Kawakami, A. & Eguchi, K. Involvement of apoptotic cell death in autoimmune diseases. Med. Electron. Microsc. 35, 1–8, https://doi.org/10.1007/s007950200000 (2002).
Acknowledgements
This work was supported by Croatian Science Foundation under the project “Genome-wide association analysis of Hashimoto’s thyroiditis” (Grant No. 4950).
Author information
Authors and Affiliations
Contributions
V.B.P. conceived the idea and designed the project, L.B. analyzed the data, A.B., M.Br., S.G., V.Š. and A.P. were involved in patients’ recruitment, V.P.P. reviewed thyroid tissues for signs of HT and for confirmation of healthy tissue, B.B. performed immunohistochemical analysis, L.B. and V.B.P. interpreted and discussed the results and wrote the manuscript, D.K., T.Z., M.Ba., I.N. and A.P. interpreted the results and edited the manuscript. All authors reviewed the results and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brčić, L., Barić, A., Benzon, B. et al. AATF and SMARCA2 are associated with thyroid volume in Hashimoto’s thyroiditis patients. Sci Rep 10, 1754 (2020). https://doi.org/10.1038/s41598-020-58457-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-58457-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.