Abstract
The common toad Rhinella arenarum is widely distributed in Argentina, where it is utilised as an autochthonous model in ecotoxicological research and environmental toxicology. However, the lack of a reference genome makes molecular assays and gene expression studies difficult to carry out on this non-model species. To address this issue, we performed a genome-wide transcriptome analysis on R. arenarum larvae through massive RNA sequencing, followed by de novo assembly, annotation, and gene prediction. We obtained 57,407 well-annotated transcripts representing 99.4% of transcriptome completeness (available at http://rhinella.uncoma.edu.ar). We also defined a set of 52,800 high-confidence lncRNA transcripts and demonstrated the reliability of the transcriptome data to perform phylogenetic analysis. Our comprehensive transcriptome analysis of R. arenarum represents a valuable resource to perform functional genomic studies and to identify potential molecular biomarkers in ecotoxicological research.
Similar content being viewed by others
Introduction
Amphibians are poikilothermic vertebrates with morphological and ecological adaptations that allow them to occupy diverse terrestrial environments associated with humid ecosystems1,2. They are the only terrestrial vertebrates that preserve free-living larvae and produce large oocytes with a transparent vitelline membrane that allows for the direct observation of the different stages of embryonic development. These characteristics have been exploited in various research areas such as toxicology, physiology, ecology, and evolution3,4,5,6,7. The South American common toad Rhinella arenarum [ex. Bufo arenarum (Hensel, 1867)] is amply distributed in Argentina and breeds in shallow-water areas such as ponds and ditches5,8,9.
Amphibian research models can be easily and inexpensively established. However, only six anuran genomes are available to date: Pyxicephalus adspersus10, Nanorana parkeri11, Rana catesbeiana6, Rhinella marina (Bioproject: PRJEB24695, ID: 445546), Xenopus laevis12, and Xenopus tropicalis13. Furthermore, several conserved morphological characteristics shared by anurans make both taxonomic classification and phylogenetic analysis difficult to perform14. This stresses the need for combining novel genomic information with morphological and karyological data, as well as mitochondrial DNA sequencing, in order to improve accuracy in phylogenetic studies.
Next Generation Sequencing (NGS) provides a cost-effective and rapid method to sequence and analyse complete genomes. However, amphibians have a very high DNA content and a large proportion of repetitive and non-coding sequences15; thus, whole-genome assembly is still expensive and bioinformatically challenging. In contrast, high-throughput RNA-sequencing (RNA-Seq) is an affordable NGS technique that provides a convenient platform for transcript profiling and transcriptome sequencing in non-model amphibian species like R. arenarum16,17.
Here, we report for the first time the de novo assembly of R. arenarum transcriptome using massive RNA-Seq, followed by gene annotation and phylogenetic analysis.
Results and Discussion
Quality control, de novo assembly and transcriptome optimisation
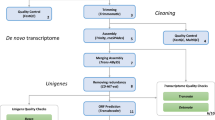
As there is no reference genome for R. arenarum, we performed a de novo transcriptome assembly for the initial tadpole stage following the pipeline showed in Fig. 1a. Larvae at complete operculum stage (Stage 25, according to Del Conte and Sirlin18) were obtained from ten independent samples (see Material and Methods) and processed for bulk transcriptome sequencing. First, we evaluated the RNA-Seq quality profiles using the FASTQC tool (https://github.com/s-andrews/FastQC). Good quality reads of the different profiles are essential for obtaining a consistent transcriptome to be used in further analyses. Thus, a quality trimming step using Trimmomatic 0.3819 was performed to remove low quality bases and adapter sequences. Then, the profiles from the ten independent samples were pooled (Fig. 1b), yielding 382,933,480 raw sequences which assured excellent transcripts coverage and variety to generate a high-quality and complete assembly20,21. Low-quality bases with a Phred quality score below 30 were trimmed from both readings ends. Reads shorter than 45 bases were also discarded, yielding 352,818,318 filtered sequences. Next, the R. arenarum larval transcriptome was de novo assembled using Trinity software, version 2.8 (http://trinityrnaseq.sourceforge.net/) with default parameters22. De novo assemblies are more challenging to achieve in the absence of a reference genome, and entail an additional effort in the case of a polyploid organism due to the presence of subgenomes23. Although polyploidy is common in amphibians, R. arenarum is not a polyploid organism14. Nevertheless, Trinity software version 2.8 improves the handling of non-strand-specific RNA-Seq and high polymorphism containing transcriptomes. Hence, the number of filtered sequences obtained from our procedure assures a high-quality assembly of the R. arenarum transcriptome. A total of 176,409,159 paired reads were used in the de novo assembly, generating a total of 249,729 transcripts and 156,941 Trinity ‘genes’ from 244,861,075 assembled bases, with median and mean transcript lengths of 411 and 980 bp, respectively, 44.6% of GC content, and an N50 of 2,151 bp (Table 1). The annotation of Trinity ‘genes’ comes from the methodology used by this software to generate contigs, cluster them, and finally assign identifiers, i.e. ‘gene’ and ‘isoform’ to the constructed transcripts. These terms are valid under the definition of a gene as “the part of the genome that is active and transcribed”. Afterwards, it is necessary to continue with any annotation pipeline providing transcript identification such as coding sequences, rRNA and lncRNA.
De novo transcriptome assembly of Rhinella arenarum. (a) Flow diagram of the assembly, from raw data to annotated transcripts. (b) For each of the ten samples, representation of total paired reads (blue), total paired reads after adapter removal and quality trimming (orange) and trimmed paired reads mapped-back against the de novo assembled transcriptome (yellow) are shown. (c) The number of transcripts annotated for the species present in reference databases (a: annotation using SwissProt DB; b: annotation using UniRef DB). (d) The number of lncRNAs defined by Annocript pipeline and confirmed by FEELnc tool.
A large proportion of reads mapping back to the assembly (i.e. above 60–70%) indicates proper quality sequence reconstruction and a representative transcriptome22. Using botwie2 v2.3.4.324, we found that more than 80% of the reads were mapped back, as proper pairs in each profile, to the R. arenarum de novo assembled transcriptome (Fig. 1b). Next, we assessed the quality of the assembled transcriptome using TRANSRATE v1.0.325, BUSCO v3.0.226, and DETONATE v1.927. These tools generate several metrics that serve as a guide to understand and evaluate error sources in the assembly process and provide evidence about the quality of the assembled transcriptome. We also assayed the assembled transcriptome through the hierarchical clustering tool CD-HIT, to address the possible generation of chimaeras, redundant transcripts and fragmented assemblies common to the process of de novo assembly28,29. Table 2 contains the principal scores obtained for the three analysis tools before and after CD-HIT processing (complete tables are provided in Supplementary Information as Table 1).
To increase confidence in the quality and completeness of the optimised R. arenarum larval transcriptome, we performed a comparative interpretation through tentative orthologue assignments. BUSCO assessment, which estimates assembly quality based on evolutionarily-informed expectations of gene content from orthologues selected from OrthoDB v930 (https://www.orthodb.org/), showed that the number of “Complete and single-copy” loci increased from 51.8% to 75.7% after CD-HIT processing. The assembled transcriptome included a total of 198,592 transcripts and 155,511 Trinity ‘genes’ with median and mean transcript lengths of 367 and 801 bp, respectively, 44.5% of GC content, and an N50 of 1626 bp (Table 1), representing 99.4% of completeness. Thus, CD-HIT sensibly improved transcriptome assembly, as evidenced by the reduction in the number of transcripts generated by Trinity (Table 1) and the improvement in quality scores (Table 2).
Gene annotation
After quality evaluation and filtration, the assembled transcriptome was interrogated to obtain useful annotations for further analysis. This step was carried out using the pipeline Annocript31. With this tool, the de novo transcriptome was annotated by several BLAST analyses against UniProt and NCBI’s Conserved Domain Database and Nucleotide divisions. Besides, the pipeline added functional annotations for Gene Ontology terms, the Enzyme Commission classification, and Pathway databases. We also used Annocript to identify putative lncRNAs following four criteria: (i) non-annotation, lack of similarity with any protein, domain, or another ncRNA (any RNA species annotated in Rfam, including rRNAs); (ii) transcript length ≥ 200 nucleotides; (iii) an ORF < 100 amino acids; and (iv) non-coding potential score ≥ 0.95.
The statistics for the annotations obtained for the R. arenarum transcriptome are presented in Table 3. Out of 198,592 transcripts obtained after CD-HIT clustering, 18,216 could not be annotated and remain unidentified in available databases up to date (April 2019). Of the remaining transcripts, 57,407 (32.0%) could be annotated based on available information. Of these, 41,336 (72.0%) had hits for Swiss-Prot, 54,851 (95.5%) for UniRef, and 8,340 (14.5%) for non-coding RNAs consisting mainly of tRNAs and rRNAs (i.e. not lncRNAs). From this annotation, 36,478 transcripts (63.5%) shared annotations for SwissProt and UniRef, 4 for Swiss-Prot and ncRNA, 1,360 for UniRef and ncRNA, and 4,639 annotations were shared by the three databases. Besides, there were 215 unique matches for Swiss-Prot, 12,374 for UniRef, and 2,337 hits annotated as other ncRNAs (Fig. 1a and Table 3). Based on strand alignment analysis for Swiss-Prot annotations, we found that 19,546 and 21,790 transcripts were aligned to the positive and negative strands, respectively. For UniRef annotations, in turn, 26,166 and 28,685 transcripts were aligned to the positive and negative strands, respectively. Meanwhile, of 57,407 annotated transcripts, 44,760 (77.9%) were in agreement with the longest ORF. Finally, from the 44,760 longest ORF annotated transcripts, 17,423 (39.9%) were unique transcripts, and 27,337 (60.1%) were isoform transcripts.
We also analysed which were the species closest to our annotated de novo transcriptome (Fig. 1c). On SwissProt, the top five species were Homo sapiens, Mus musculus, Xenopus laevis, Rattus norvegicus, and Xenopus tropicalis, which is not surprising because a high proportion of annotations in this database correspond to H. sapiens and M. musculus (Fig. 1c). However, on the curated multi-species database UniRef90, the top five closest species were Nanorana parkeri (TaxID 125878), Xenopus laevis (TaxID 8355), Lithobates catesbeiana (TaxID 8400), Xenopus tropicalis (TaxID 8364), and Xenopus (TaxID 262014), all belonging to the taxonomic class Amphibia (Fig. 1c).
The Annocript algorithm interpreted 122,969 transcripts as putative lncRNAs (Table 3), which were next tested using the FEELnc tool32. The latter uses a machine-learning method trained with coding transcripts to compute the coding potential score (CPS) for each transcript. The CPS maximises classification performances and infers whether a transcript is coding or non-coding in order to identify high-confidence sets of lncRNAs. From the 122,969 putative lncRNAs defined by Annocript, 52,800 transcripts (42.9%) were confirmed by FEELnc as a set of high-confidence lncRNA transcripts for R. arenarum (Fig. 1d and Supplementary File SF1). The 18,216 transcripts that could not be annotated, as well as the 70,169 putative lncRNAs not confirmed by FEELnc, may be artefacts or misassemblies inherent to the de novo assembly method33,34. Still, another reason why some lncRNAs could not be identified/annotated may be simply the lack of information in current databases. In this regard, it is worth noting that the identification/annotation of lncRNA is still complicated even for well-annotated species with fully sequenced reference genomes such as H. sapiens and M. musculus35,36,37.
The number of R. arenarum transcripts annotated as lncRNAs is higher than those reported for the amphibians Xenopus tropicalis, Xenopus laevis and Lithobates catesbeiana6,38,39,40. In the case of the transcriptome studies in Xenopus, the number of lncRNAs defined depends on the temporal and spatial expression profiling of the samples. In this sense, Necsulea et al.41 studied the evolutionary history of lncRNAs from polyadenylated transcriptomes of 8 organs and 11 species (human, chimpanzee, bonobo, gorilla, orangutan, macaque, mouse, opossum, platypus, chicken and frog) showing that lncRNAs are actively regulated and may function predominantly during the embryonic development. On the other hand, it is essential to note that the annotations obtained from genomes as predicted transcripts are always lower than the plethora and repertoire of transcripts identified in a transcriptome. In our case, lncRNA transcript assessment followed a strict depuration process starting with the four-criteria Annocript pipeline annotation, followed by FEELnc confidence maximisation. Thus, our lncRNA dataset provides the starting point for future lncRNA studies in R. arenarum, which would allow to verify them and to assess their regulation and function. Undoubtedly, the future availability of R. arenarum genome will help curate the lncRNA set using strategies like the identification of promotor regions followed by non-coding regions.
At the moment, no consensus pipeline can be defined as the best for transcriptome assembly42. The state-of-the-art of the de novo assembly of transcripts can be summarised in the selection of raw sequences with good quality, de novo assembly using an appropriate tool, quality validation of the assembly through different software tools and their subsequent availability to the scientific community. Once a set of sequences is released, different research groups making use of them perform its curation. In this sense, we selected some genes from the transcriptome that are of our research interest, designed primers and then sequenced the PCR products by Sanger method in order to validate them. Until now, we have evaluated nine genes using independent biological samples (in Supplementary Information as Table 2), and sequences have been confirmed by alignment against the transcriptome and by annotation comparison with RefSeq and UniProtKB databases. Future sequencing of the R. arenarum genome would represent a unique opportunity since RNA-Seq data could be used to curate the genome assembly and vice versa.
All transcript contigs of this Transcriptome Shotgun Assembly project have been deposited at DDBJ/EMBL/GenBank under the accession number GHCG00000000. The version described in this paper is the first version, GHCG01000000. To facilitate the obtention of gene sequences, we designed a webpage (available at http://rhinella.uncoma.edu.ar) in which the annotated transcriptome for this non-model species can be readily accessed. It allows searching by “gene name” or “name description”, to retrieve data related to the R. arenarum transcriptome. The information contains sequence annotation, nucleotide and peptide sequence data, orthologous information and annotated pathways.
Phylogenetic analysis
To conduct phylogenetic studies with R. arenarum transcriptome data, we downloaded all the protein sequences annotated for the order Anura from UniProtKB taxonomy and constructed a matrix of 5423 anurans × 7376 proteins. Then, we filled the matrix for absent sequences (0; no protein sequence) or present sequences (1; protein sequence) for each anuran. We calculated the Jaccard distance43 using the Philentropy R package44 and selected a final cluster of 55 anurans and 28 protein sequences for each Anura (Supplementary File SF2). Next, we included in the set the corresponding 28 protein sequences derived from the R. arenarum transcriptome and performed multiple alignments for each protein using Muscle algorithm45, available in the MegaX software46. Finally, we concatenated the 28 alignments and applied the Maximum Parsimony method (1000 bootstraps) to construct the evolutionary history47.
Besides, we interrogated the TimeTree database48 with the 56 anurans (55 + R. arenarum) to obtain a consensus taxonomic tree. When we compared our experimental tree against the consensus tree, there were two main differences: first, Engystomops pustulosus clustered together with Allobates femoralis in our analysis, while in TimeTree E. pustulosus clustered in another clade (Supplementary Information – Fig. S1a,b); second, the species Amietia lubrica, Atelognathus reverberii, Breviceps macrops, Callulops wilhelmanus, Cophixalus cheesmanae, Cornufer pelewensis, Craugastor fitzingeri, and Hyperolius bolifambae are not present in the TimeTree database. Nevertheless, R. arenarum was included in the same clade in both trees (Fig. 2a).
Next, we retrieved biodiversity data from the 56 anurans using gbif R package49 and mapped the coordinates for each species on a world map using maps R package50 (Supplementary Information – Fig. S2). Figure 2b shows the anuran species present in the clades closer to R. arenarum. All the species are spread throughout the American continent, except for Bufo gargarizans, which is mainly found in eastern Asia. Since the gene sequence-based phylogenetic analysis of R. arenarum is consistent with the consensus taxonomic tree for this species, we conclude that the R. arenarum transcriptome is highly reliable to perform evolutionary studies, identify new subspecies, and further characterise subspecies like Rhinella arenarum arenarum and Rhinella arenarum mendocinus.
Conclusions
We report for the first time a transcriptome for the non-model organism Rhinella arenarum at the first larvae (complete operculum) stage. The reconstructed transcriptome reached 99.4% of completeness and yielded a set of 57,407 well-annotated transcripts available for downstream analyses (http://rhinella.uncoma.edu.ar). Besides, a high confidence set of 52,800 putative lncRNAs was defined, and the feasibility of phylogenetic analyses was confirmed. The genomic tool delivered here will support biomarker assessment and discovery in ecology and toxicogenomics and facilitate evolutionary and global comparative genomic diversity analyses.
Materials and Methods
Experimental procedures
R. arenarum embryo development
Adult R. arenarum females and males were collected in a pristine environment at Los Barreales Lake (S38.45344 W68.72918) during the breeding (spring) season and maintained in an outdoor terrarium. All efforts were made to minimise the stress and suffering of animals according to the recommended standards of the American Society of Ichthyologists and Herpetologists (ASIH) in Guidelines for the Use of Live Amphibians and Reptiles in Field Research (http://www.asih.org/pubs/). An ad hoc Committee of the Centre for Research in Environmental Toxicology and Agrobiotechnology of Comahue (CITAAC, http://citaac.uncoma.edu.ar), which currently reviews and approves the projects that require the use of laboratory and field animals, approved the Rhinella arenarum project. Also, the collection guide of field specimens and their use in our projects were presented and approved by the Environment Bureau of the Province of Neuquén through the Applied Ecology Center of Neuquén, Argentina. Embryos were obtained by in vitro fertilisation and developed until complete operculum stage (Stage 25, according to Del Conte and Sirlin18) as described before5.
RNA extraction, cDNA library generation and massively parallel sequencing
For deep transcriptome sequencing, R. arenarum larvae were grown in 10 different glass receptacles. From each receptacle, fifteen random larvae were randomly collected and pooled in a tube containing RNAlater® solution (Thermo Fisher Scientific Inc.) (15 larvae/1 tube/1 sample). Then, total RNA of each sample was extracted using the Thermo Scientific GeneJET RNA Purification Kit (Thermo Fisher Scientific Inc.). RNA quantity and quality were evaluated using Agilent RNA 6000 Pico Kit in a 2100 Bioanalyzer (Agilent Technologies). All samples presented an optimal condition [RNA Integrity Number (RIN) > 8]51. The cDNA library for transcriptome analysis was prepared using the TruSeq® RNA v2 kit (Illumina, San Diego, CA, USA). Briefly, mRNA enrichment was carried out from 1 µg of total RNA using oligo dT magnetic beads, followed by chemical fragmentation of the purified mRNA into small pieces, double-stranded cDNA synthesis, end repair, and adenylation processes. Finally, adaptor ligation and enrichment were carried out by PCR. The ten library samples were normalised to 10 nM cDNA and were sequenced in a line of Hiseq. 1500 Illumina, generating non-strand specific “paired-ends” (PE) 2 × 100 bp readings.
Data availability
Raw sequencing data have been deposited in the Bioproject PRJNA485066 in the NCBI repository.
References
Salice, C. J., Rowe, C. L., Pechmann, J. H. K. K. & Hopkins, W. A. Multiple stressors and complex life cycles: Insights from a population-level assessment of breeding site contamination and terrestrial habitat loss in an amphibian. Environ. Toxicol. Chem. 30, 2874–2882 (2011).
Unrine, J. M., Hopkins, W. A., Romanek, C. S. & Jackson, B. P. Bioaccumulation of trace elements in omnivorous amphibian larvae: Implications for amphibian health and contaminant transport. Environ. Pollut. 149, 182–192 (2007).
Mann, R. M., Hyne, R. V., Choung, C. B. & Wilson, S. P. Amphibians and agricultural chemicals: Review of the risks in a complex environment. Environ. Pollut. 157, 2903–2927 (2009).
Rosenbaum, E. A. et al. Response of biomarkers in amphibian larvae to in situ exposures in a fruit-producing region in North Patagonia, Argentina. Environ. Toxicol. Chem. 31, 2311–2317 (2012).
Mardirosian, M. N., Ceschin, D. G., Lascano, C. I. & Venturino, A. Molecular effectors in the chronic exposure to arsenic as early and sensitive biomarkers in developing Rhinella arenarum toads. Aquat. Toxicol. 186, 19–27 (2017).
Hammond, S. A. et al. The North American bullfrog draft genome provides insight into hormonal regulation of long noncoding RNA. Nat. Commun. 8, 1–8 (2017).
Veldhoen, N., Ikonomou, M. G. & Helbing, C. C. Molecular profiling of marine fauna: Integration of omics with environmental assessment of the world’s oceans. Ecotoxicol. Environ. Saf. 76, 23–38 (2012).
Pechen de D’Angelo, A. M. & Venturino, A. Biochemical targets of xenobiotics: Biomarkers in amphibian ecotoxicology. Appl. Herpetol. 2, 335–353 (2005).
Liendro, N., Ferrari, A., Mardirosian, M., Lascano, C. I. & Venturino, A. Toxicity of the insecticide chlorpyrifos to the South American toad Rhinella arenarum at larval developmental stage. Environ. Toxicol. Pharmacol. 39, 525–535 (2015).
Denton, R., Kudra, R., Malcom, J., Du Preez, L. & Malone, J. The African Bullfrog (Pyxicephalus adspersus) genome unites the two ancestral ingredients for making vertebrate sex chromosomes. bioRxiv 1–25 (2018).
Sun, Y.-B. et al. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc. Natl. Acad. Sci. 112, E1257–E1262 (2015).
Session, A. M. et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 (2016).
Hellsten, U. et al. The genome of the western clawed frog xenopus tropicalis. Science (80-.). 328, 633–636 (2010).
Baraquet, M., Valetti, J. A., Salas, N. E. & Martino, A. L. Redescription of the karyotype of five species of the family Bufonidae (Amphibia: Anura) from central area of Argentina. Biologia (Bratisl). 66, 543–547 (2011).
Liedtke, H. C., Gower, D. J., Wilkinson, M. & Gomez-Mestre, I. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nat. Ecol. Evol. 2, 1792–1799 (2018).
Kerksick, C. M., Tsatsakis, A. M., Hayes, A. W., Kafantaris, I. & Kouretas, D. How can bioinformatics and toxicogenomics assist the next generation of research on physical exercise and athletic performance. J. Strength Cond. Res. 29, 270–278 (2015).
Ceschin, D. G. Toxicogenomics: new strategies for ecotoxicology studies in autochthonous species II. The ‘omic’ era in non-model species. Transcriptome analysis for biomarker screening. Int. J. Environ. Heal. 8, 213 (2017).
Del Conte, E. & Sirlin, J. L. Pattern series of the first embryonary stages in Bufo arenarum. Anat. Rec. 112, 125–135 (1952).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Singhal, S. De novo transcriptomic analyses for non-model organisms: An evaluation of methods across a multi-species data set. Mol. Ecol. Resour. 13, 403–416 (2013).
Fang, Z. & Cui, X. Design and validation issues in RNA-seq experiments. Brief. Bioinform. 12, 280–287 (2011).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–512 (2013).
Schatz, M. C. et al. Assembly of large genomes using second-generation sequencing Assembly of large genomes using second-generation sequencing. Genome Res., 0–9, https://doi.org/10.1101/gr.101360.109.20 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–9 (2012).
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M. & Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144 (2016).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Li, B. et al. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 15, 1–21 (2014).
Head, S. R. et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 56, 61–4, 66, 68, passim (2014).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Zdobnov, E. M. et al. OrthoDB v9.1: Cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45, D744–D749 (2017).
Musacchia, F., Basu, S., Petrosino, G., Salvemini, M. & Sanges, R. Annocript: A flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics 31, 2199–2201 (2015).
Wucher, V. et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 45, 1–12 (2017).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 1–19 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Cao, H., Wahlestedt, C. & Kapranov, P. Strategies to Annotate and Characterize Long Noncoding RNAs: Advantages and Pitfalls. Trends Genet. 34, 704–721 (2018).
Li, J. & Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 10, 1–10 (2019).
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R. & Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 19, 535–548 (2018).
Forouzmand, E. et al. Developmentally regulated long non-coding RNAs in Xenopus tropicalis. Dev. Biol. 426, 401–408 (2017).
Paranjpe, S. S., Jacobi, U. G., van Heeringen, S. J. & Veenstra, G. J. C. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics 14 (2013).
Sai, L. et al. Analysis of long non-coding RNA involved in atrazine-induced testicular degeneration of Xenopus laevis. Environ. Toxicol. 34, 505–512 (2019).
Necsulea, A. et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014).
Hölzer, M. & Marz, M. De novo transcriptome assembly: A comprehensive cross-species comparison of short-read RNA-Seq assemblers. Gigascience 8, 1–16 (2019).
Cha, S. Comprehensive Survey on Distance/Similarity Measures between Probability Density Functions. Int. J. Math. Model. Methods Appl. Sci. 1, 300–307 (2007).
Drost, H.-G. Philentropy: Information Theory and Distance Quantification with R. J. Open Source Softw. 3, 765 (2018).
Edgar, R. C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 1–19 (2004).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N. Y). 39, 783–791 (1985).
Hedges, S. B., Marin, J., Suleski, M., Paymer, M. & Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015).
Mcglinn, D., Oldoni, D., Geffert, L., Ram, K. & Chamberlain, M. S. Package ‘ rgbif’ (2018).
Draw, T. & Maps, G. Package ‘maps’ Title Draw Geographical Maps. (2018).
Pereira, M. A., Imada, E. L. & Guedes, R. L. M. RNA‐seq: Applications and Best Practices. Appl. RNA-Seq Omi. Strateg. - From Microorg. to Hum. Heal., https://doi.org/10.5772/intechopen.69250 (2017).
Acknowledgements
Funding for this research was provided by a Servicios Petroleros Mirasal/National University of Comahue partnership and the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) PICT-2017-1529. N.P. has a doctoral fellowship from ANPCyT. D.C., M.M., C.L. and A.V. are staff researchers of CONICET. Part of the bioinformatics analysis was performed in the Cluster Navira, CIDIE - CONICET - UCC (Plataforma Nacional de Bioinformática). We thank Dr. Victor Blanco for writing assistance.
Author information
Authors and Affiliations
Contributions
The R. arenarum transcriptomics project was entirely designed and conducted at the Centre for Research in Environmental Toxicology and Agrobiotechnology of Comahue (CITAAC), National Council of Scientific and Technical Research-National University of Comahue. D.C. and A.V. conceived the study and provided financial support for the project. D.C. performed the bioinformatics analyses and drafted the manuscript. N.P., M.M. and C.L. participated in the obtaining of biological material. N.P., M.M. and C.L. performed laboratory experiments. All the authors read, corrected and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ceschin, D.G., Pires, N.S., Mardirosian, M.N. et al. The Rhinella arenarum transcriptome: de novo assembly, annotation and gene prediction. Sci Rep 10, 1053 (2020). https://doi.org/10.1038/s41598-020-57961-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-57961-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.