Abstract
Most bacteria divide by binary fission using an FtsZ-based mechanism that relies on a multi-protein complex, the divisome. In the majority of non-spherical bacteria another multi-protein complex, the elongasome, is also required for the maintenance of cell shape. Components of these multi-protein assemblies are conserved and essential in most bacteria. Here, we provide evidence that at least three proteins of these two complexes are not essential in the FtsZ-less ovoid planctomycete bacterium Planctopirus limnophila which divides by budding. We attempted to construct P. limnophila knock-out mutants of the genes coding for the divisome proteins FtsI, FtsK, FtsW and the elongasome protein MreB. Surprisingly, ftsI, ftsW and mreB could be deleted without affecting the growth rate. On the other hand, the conserved ftsK appeared to be essential in this bacterium. In conclusion, the canonical bacterial cell division machinery is not essential in P. limnophila and this bacterium divides via budding using an unknown mechanism.
Similar content being viewed by others
Introduction
In all life forms, cell division is a highly conserved and regulated process. Current knowledge about bacterial cell division is largely based on a few model organisms, such as Escherichia coli, Bacillus subtilis and Caulobacter crescentus. Most bacteria divide by binary fission, a widely conserved mechanism based on the interaction between the FtsZ protein and the peptidoglycan (PG) biosynthesis machinery (PG synthesis reviewed in1). FtsZ is an almost universally conserved bacterial cytoskeletal element, homologous to the eukaryotic tubulin2,3,4. This bacterial cytoskeleton is the central member of the cell division machinery, the divisome, a dynamic protein complex composed of more than 20 conserved proteins that assembles at mid-cell5,6. FtsZ filaments were previously believed to form a continuous ring (the Z-ring), closing concentrically during division and splitting both cells. However, the continuity of this ring is currently questioned7 and recent data suggest that FtsZ forms a single-layered structure at a distance of several nanometers from the inner membrane8. The FtsZ filaments treadmill on the inner face of the cytoplasmic membrane and interact with the PG synthases9,10. However, the precise nature of the interaction between the Z-ring protofilaments and their physiological significance remains a mystery11. Whatever its precise organization, this structure determines the future division site and serves as a scaffold to hierarchically recruit the remaining division players8. Additionally, the ring participates in chromosome segregation12.
The Z-ring is a quasi-universal element of cytokinesis in bacteria that possess PG and divide by binary fission. There are however a few prokaryotic exceptions to this FtsZ-dominated division mode. First of all, FtsZ is not the dominant division machinery in Archaea where at least three distinct division systems are found based on FtsZ, ESCRT-III homologs, or actin-related proteins13,14.
Secondly, the gene coding for FtsZ is also absent from the genomes of a limited number of bacteria. These include pathogenic strains from the Tenericutes, the Chlamydiae, and some symbionts such as the gammaproteobacteria “Candidatus Ruthia magnifica”, “Candidatus Vesicomyosocius okutanii HA” and “Candidatus Carsonella ruddii”, the alphaproteobacterium “Candidatus Hodgkinia cicadicola”, and the bacteroidete “Candidatus Sulcia muelleri”15. In most of these cases, the loss of ftsZ might be related to the extreme genome reduction associated with their parasitic or endosymbiotic lifestyle. At least in the case of Wolbachia, a genus of Alphaproteobacteria, it has been shown that the ftsZ gene has been transferred to the Drosophila host genome16. In addition, FtsZ is not found in the Planctomycetes phylum, which is composed of free-living bacteria17,18.
Among the proteins forming the divisome, there are PG synthases and hydrolases that remodel the PG, DNA translocases which establish communication between chromosome replication-segregation and cell division machineries, and proteins that coordinate inner and outer membrane constriction19,20. Twelve of the proteins that compose the divisome are described to be essential and conserved across bacteria: FtsZ, FtsA, ZipA, FtsE, FtsX, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI and FtsN21,22.
Whereas during division the PG is synthesized at the new cell poles23, during cell elongation, the PG precursor, lipid II, is incorporated into the previously existing cylindrical part of the cell wall24. This lateral cell wall synthesis is performed by the elongasome, a protein complex formed by MreBCD, RodA, RodZ, PBP1A, PBP2 as well as MurF, MurG and MraY25,26. The complex is guided by the actin-like protein MreB27,28,29, which is essential for cell elongation and maintenance of the cell shape. MreB is extremely conserved, being found in almost all non-spherical bacteria except the ones exhibiting polar growth. Some examples of MreB-lacking rods are members of the Actinobacteria and Alphaproteobacteria (Rhizobium and Agrobacterium)30,31. In contrast, other Gram-positive rod-shaped bacteria, such as B. subtilis, have multiple mreB copies32.
Apart from binary fission, other cell division mechanisms, such as multiple intracellular offspring, multiple offspring by binary fission, multiple fission or budding, have been reported in some members of Cyanobacteria, Firmicutes, Planctomycetes and the prosthecate proteobacteria33,34. Recent studies also revealed asymmetric division in Chlamydia trachomatis35,36. Bacteria belonging to the PVC superphylum (Planctomycetes-Verrucomicrobia-Chlamydiae)37 are exceptional in many aspects, including cell division38,39,40. The ftsZ gene is not found in any of the genomes of the Planctomycetes and Chlamydiae. These bacteria display a variety of division modes, suggesting that divergent mechanisms have evolved within this superphylum. Despite the lack of FtsZ, some planctomycetes, such as the members of the taxa “Candidatus Brocadiales” and Phycisphaerae, divide by a mechanism similar to binary fission41. Others, like the spherical Gemmata obscuriglobus and the ovoid Planctopirus limnophila, divide by budding17. Although the budding mechanism has been studied in depth in yeast cells, the molecular mechanism of bacterial budding remains entirely unknown.
During canonical cell division, FtsZ interacts directly or indirectly with many PG synthesis enzymes. In contrast to other FtsZ-less bacteria, such as the Tenericutes, members of the Planctomycetes do possess PG42,43,44,45. Although this polymer was long thought to be absent from Chlamydiae and Planctomycetes, it was recently detected in some members of these phyla. This experimental detection combined with phylogenomic analyses, suggested that PG is likely to be present in all members of these phyla. It is now clear that Planctomycetes are not exceptions to but deviations from the Gram-negative cell type46,47.
Cell division and PG synthesis related genes have been previously detected in members of the Planctomycetes17,18,48,49. Genomic analyses have revealed that most classically conserved and essential cell division genes show a punctuated pattern of presence in the genomes of Planctomycetes. On this basis, we and others have suggested that the divisome and elongasome machineries might be divergent, or even lost, in members of this phylum. Due to their essential role in other bacteria, it was however unclear if these complexes were absent or if they were present but consisted of components that had diverged beyond the point of detection.
Here, we investigate the essentiality of the canonical cell division genes ftsI, ftsW, ftsK and mreB coding for homologs of proteins that belong to the divisome and elongasome in model bacteria by performing gene knock-outs in the planctomycete P. limnophila. We show that the ftsI, ftsW and mreB genes are not essential in this species.
Results
We focused here on the P. limnophila homologs of the canonical cell division genes ftsI, ftsW, ftsK and mreB, detected by us and others17,48,49, supporting the identification of the homologs. P. limnophila FtsI (Uniprot ID: D5SXQ2) is homologous to the E. coli protein with 34% identity and similar length: 561 vs 572 amino acids (aa). P. limnophila FtsW (ID: D5STY0) shares 34% identity with the protein from E. coli with similar length, 406 vs 414 aa. P. limnophila contains only one FtsK homolog (ID: D5SYV8) with 46% identity to E. coli protein and size similar to most FtsK homologs (750–900 aa). The MreB homolog (D5SQI2) shares 47% identity to E. coli with similar size, 348 vs 347 aa, and the corresponding gene seems to be in the same transcriptional unit as mreC and mreD, as they are separated by 74 and 3 nucleotides, respectively.
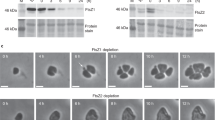
The chromosome and PG are two important cell components inherited during cell division. FtsK is an ATP-dependent DNA pump, most likely responsible for the transfer of the genetic material from mother to daughter cell. It is highly conserved and is an essential component of the divisome in model bacteria50. Despite the punctuated pattern of presence of most of the canonical cell division genes in the planctomycetal genomes, ftsK is conserved across the phylum17, and semi-quantitative RT-PCR assays showed that it is expressed at the same level throughout the cell cycle, as compared to the control gyrA gene, constitutively expressed in most bacteria (Fig. 1). We were unable to generate insertion or deletion mutants of this gene in P. limnophila, suggesting that FtsK plays an essential role in this bacterium (identical results were obtained for G. obscuriglobus ftsK, supporting the proposed essentiality of the gene; ERM unpublished).
Semi-quantitative RT-PCR of the ftsI, ftsW, ftsK, mreB and gyrA genes in Planctopirus limnophila. Decreasing cDNA template concentration from left to right (125, 25, 5, 1 and 0,2 ng respectively). gyrA has been used as housekeeping gene. Positive control: P. limnophila genomic DNA, negative control: no DNA. Uncropped image is available in Supplementary Fig. 2.
Bacterial cell division is tightly linked to PG synthesis. In model organisms, PG incorporation into the growing PG layer is performed by the divisome at the septum and by the elongasome elsewhere. In the periplasm, the PG precursor is incorporated into the cell wall by various penicillin-binding proteins (PBP). FtsI (PBP3) is one of the PG transpeptidases involved in septal PG synthesis51,52. FtsI and FtsW have been shown to interact and to be essential in model bacteria53. The genes coding for FtsI and FtsW are conserved in roughly half of the planctomycetal proteomes17. In order to decipher if these genes are expressed in P. limnophila, we first performed semi-quantitative PCR, demonstrating expression of ftsI and ftsW in the wild type strain at the same level in exponential and stationary phase (Fig. 1). As suggested by their patchy conservation in Planctomycetes17, and in contrast to the situation observed in other bacteria, we were able to generate single deletion mutants for both ftsW and ftsI. Growth curves of the deleted strains did not differ from the wild type (Fig. 2A). Similarly, budding division could not be differentiated from the wild type strain when examined by bright field microscopy (Fig. 2C).
ftsI, ftsW and mreB are non-essential in Planctopirus limnophila. (A) Growth curves (B) morphological measurements (area, circularity, length and width) and (C) bright field imaging of the wild type and the ftsI, ftsW and mreB mutants. Scale bar corresponds to 1 μm. Additional cells for each of the mutants are displayed in Supplementary Fig. 3.
By measuring various cell morphology indexes, the most significant changes were detected for the ftsW deletion mutant. The length of the cells of the ftsW mutant decreases, while their width increase, resulting in a change of circularity (all with pairwise Wilcox test P value < 10−15; Fig. 2B). These changes resulting in slight modifications of the area of the cells (Wilcox test P value > 10−3). Nevertheless, as stated above, no differences could be detected in the growth curves or the division modes and thus, the ftsW gene is not essential for cell division.
The actin homolog MreB is the main component of the elongasome. It is expected to play a scaffolding role during lateral growth to guide cell-wall elongation in rod-shaped bacteria30. Similar to ftsI and ftsW, mreB showed a patchy distribution across the phylum17 and we could generate a deletion mutant. Growth curves, morphology and budding phenotype did not differ from the wild type (Fig. 2).
The lack of a division phenotype for the ΔmreB mutant additionally presented the opportunity to test the effect of the A22 drug in an mreB-negative background (Fig. 3). This compound is classically used as an MreB-inhibitor despite the fact that the possible existence of off-target effects has been raised54. Before fission, the P. limnophila wild type cells increase in cell volume. At the apex of size, the daughter cell bud is formed, and the mother cell decreases in total cell volume. When exposed to A22, wild type and mreB deletion mutants, still increased in size (Supp. Fig. S1A) but no longer initiated cell division (Supp. Fig. S1B). These results confirm that, at least in P. limnophila, the drug has effects on targets other than MreB, corroborating the proposed unspecific effect of A22.
A22 phenotype for the P. limnophila wild type and ΔmreB mutant. Phase contrast images of still frames of time lapse assays of wild type and ΔmreB P. limnophila cells (A) without treatment and (B) treated with MreB inhibitor A22. Asterisks indicate dividing cells without treatment and arrowheads indicate halted division process by A22 drug. Scale bar corresponds to 1 μm.
Discussion
We report the first evidence of non-essentiality for the divisiome and elongasome components FtsI, FtsW and MreB. This raises the possibility that the divisome and elongasome complexes, essential in the vast majority of bacteria, are absent in some members of the Planctomycetes phylum.
Current knowledge about bacterial cell division is predominantly based on model organisms. However, recent studies on various non-model organisms have begun to reveal that some of the statements derived from model organisms do not apply to bacteria universally. A wide variety of different cell division mechanisms is bound to broaden our perspective about bacterial cell division34,55.
New evidence has questioned the central role of FtsZ as the predominant force generator during cell division4,56,57. An emerging model proposes that assembled FtsZ is a platform to recruit PG synthesis enzymes, and that it is PG synthesis per se that directly contributes the forces for septum formation58. As all planctomycetes are devoid of FtsZ, this protein cannot be the force generator during cytokinesis in this phylum. However, P. limnophila divides by budding, not by binary fission. On the other hand, the planctomycetal class Phycisphaerae and order “Candidatus Brocadiales” (i.e. anammox bacteria) divide by a mechanism similar to binary fission without FtsZ. This raises the question of whether there is a different main protein driving cell division in these organisms.
In P. limnophila, ftsI and mreB deletion do not produce phenotypes. However, we observed that ftsW deletion leads to cells that are more spherical compared to the ovoid wild type, without any detectable modification of growth pattern or division mode. FtsW is essential in most organisms containing a single copy of this gene. Depletion of FtsW in rod-shaped bacteria leads to a block in cell division and formation of elongated cells59,60,61. In the polyploid spherical cyanobacterium Synechocystis sp. strain PCC 6803, a ΔftsW heteroploid mutant (still retaining wt copies despite deletion of some ftsW genes) grows slower and generates giant cells62. No such phenotype is observed for the mutants generated in P. limnophila which is ovoid and divides by budding.
MreB is essential in model bacteria as well as in bacteria with a divergent division mode, like the longitudinally dividing bacteria of the genus Thiosymbion55. However, some exceptions exist, such as the ovoid Streptococcus pneumoniae and the rod-shaped Corynebacterium glutamicum, both devoid of MreB63. Many rod-shaped alphaproteobacteria and actinobacteria also lack MreB64. On the other hand, some coccoid cyanobacteria contain the mreB gene.
Planctomycetes belongs to the PVC superphylum that also comprises the Chlamydiae, Verrucomicrobia and Lentisphaerae. The latter two contain FtsZ and divide by binary fission. As for Planctomycetes, the chlamydial genomes do not encode for FtsZ and it has recently been reported that a few members of the Chlamydiae also exhibit asymmetric, budding-like cell division35. In these species, MreB localizes at the cell division site65. In most species lacking FtsZ, it is proposed that MreB might substitute for its function66. This possibility is revoked here as cells lacking MreB do no display any phenotype.
The non-essentiality of divisome and elongasome genes raises the question of the role of PG itself in Planctomycetes and its essentiality to cell integrity and division, despite the importance of these functions in other bacteria. Non-natural forms of PG-deprived cells can be induced in osmoprotective conditions by inhibition of PG synthesis in diverse bacteria67. These so-called L-forms divide and can be maintained indefinitely. It has been shown that ftsZ becomes non-essential in L-form bacteria, at least in B. subtilis68. It has been proposed that, in these cell wall-deficient, propagation occurs by an extrusion-resolution mechanism. In addition, the involvement of “some kind of cytoskeletal system” has been suggested and some MreB homologs were directly pinpointed68. Interestingly, L-form bacteria appear to be resistant to A22 when grown in the presence of a beta-lactam antibiotic, suggesting that MreB is not required for the growth of spherical L-form-like cells69. This is related to our report of A22 susceptibility of the ΔmreB mutant which confirms previous suspicion of side-effects of this widely used drug70.
Finally, loss of PG synthesis can in some cases lead to non-dividing cells that may enlarge considerably. This has been observed in various species, including E. coli and Vibrio cholera. Recently it has been shown that giant bacteria could be formed by eliminating essential functions needed for, or treatment with antibiotics targeting PG synthesis in Acinetobacter baylyi, a diderm gammaproteobacteria. However, these giant cells did not proliferate71. The relevance of these artificially induced cells and division modes and how they compare to Planctomycetes division is unclear.
The non-essentiality of the divisome genes for planctomycetal division might appear to be consistent with the previously suggested unusual status of phylum Planctomycetes and related PVC superphylum bacteria within the bacterial domain. It seems probable that PVC bacteria are derived from diderm bacteria and that their unknown division modes may have evolved from FtsZ-based binary fission. The deciphering of these alternative division modes and their evolution is important for our sampling of the biodiversity and our understanding of evolutionary cell biology.
Altogether, our results demonstrate that Planctomycetes use divergent division modes, urging for the characterization of the molecular mechanisms of division in P. limnophila and in other species of this phylum.
Material and Methods
Bacterial strains and culture conditions
Escherichia coli DH5α strains were grown in Lysogeny broth medium (LB) at 37 °C and P. limnophila DSM 3776T in a modified PYGV medium (DSMZ medium 621 [http://www.dsmz.de]: 0.1% yeast extract, 0.1% peptone, 0.1% glucose, 10 mM HEPES (pH 7,5), vitamin solution and Hutners basal salt solution from DSMZ 590 medium). Planctomycetes were grown at 28 °C. For solid medium, 1.5% bacto-agar was added. To avoid contamination of the planctomycetes when growing for a long time on agar plates, cycloheximide 50 μg ml−1 was added. Cultures were grown aerobically in a shaker (180 rpm). When required, antibiotics were used at the following concentrations (μg ml−1): kanamycin (Km) 25 for E. coli and 50 for P. limnophila.
Plasmid description and genetic modification
Plasmids used for gene deletion in a double event of homologous recombination were derived from the pEX18Tc vector72. To construct knockout plasmids for ftsI, ftsW, ftsK and mreB genes, 1000–1200 bp upstream and downstream fragments of the target gene were amplified by PCR from genomic DNA using the primer pairs listed in Supplementary Table 1. The upstream and downstream fragments were digested with the appropriate enzymes and then cloned into pEX18Tc by three-way ligation. Finally, the kanamycin resistance gene amplified from the pUTminiTn5km plasmid73 was subsequently cloned as a BamHI fragment between the two flanking regions. These plasmids enable the deletion of each of the complete genes.
Genetic transformation of P. limnophila was performed by electroporation as described before74. The cells were then plated onto modified PYGV plates supplemented with kanamycin 50 μg ml−1 and were incubated at 28 °C until colony formation (7–9 days). Colonies were transferred to fresh selection plates and genotyped by Southern Blotting and sequencing.
Mutants sequencing
Deletion mutants were verified by paired-end sequencing on an Illumina MiSeq machine upon library preparation with the Nextera XT DNA Library Prep Kit (Illumina, San Diego, USA). Pre-assembly processing of the reads was done employing Trimmomatic v0.3575, FastQ Screen v0.4.476, PRINSEQ lite v 0.20.477 and FLASH v1.2.178. The processed reads were then assembled using SPAdes v3.7.079. Additionally, all reads were mapped to the original sequence of P. limnophila with Bowtie 280. The raw sequencing data have been deposited at NCBI SRA under acc. no. PRJNA577131.
Semi-quantitative RT-PCR
Total RNA extraction was carried out as previously described81 from 20 ml of wild type cultures at exponential (OD600 ~0,4) and stationary phase (OD600 ~1,5). DNase I treatment was performed with a DNA-free kit (Ambion). The samples were purified using RNAeasy columns (Quiagen) and RNA quality was confirmed by non-denaturing agarose gel electrophoresis. The absence of contaminating DNA was then confirmed by PCR amplification.
Reverse transcription of the RNA samples was performed using the High-Capacity cDNA Archive Kit (Applied Biosystems), with random hexamers as primers to generate cDNAs. The resulting cDNA samples were amplified by semi-quantitative RT-PCR using 1 mM of each primer (Supplementary Table 2). Genomic DNA was used as a positive control of the PCR. Samples were visualized in 10% acrylamide gels.
Microscopy
Bacteria from 2 ml of exponentially growing culture (OD600 ~0.4) were harvested (12000 g, 3 min) and resuspended in 100 μl of fresh medium. A sample of 2 μl was spotted on a glass-bottom dish (MatTek) and covered with a 1% agarose in M3 medium cushion, as described by82. Bright field images were acquired using a 100×/1.46 objective through an 1.6X amplification lens and an EMCCD Andor iXon camera mounted on a Zeiss microscope, resulting in a pixel size of 0.1 × 0.1 µm.
Image analysis
An automatic analysis workflow was design using FIJI until a satisfactory segmentation of the cells was achieved. The image analysis workflow runs as follows under FIJI software: Image Acquisition → Subtract Background → Gaussian Blur → Invert → Enhance Contrast → Unsharp Mask → Watershed Thresholding → Convert to mask → Binary Watershed → Analyze Particles (with a size upper and lower limit of 21 ∙ 10−3 to 3.5 ∙ 10−3 µm. Any particle out of this limit was not considered as a cell. Afterwards, the area of the cells was measured and fitted to an ellipse. In order to extract cell’s width and length, we identified them as the ellipse’s minor and major axis, calculating subsequently the circularity. The values of each single cell were exported for further statistical analysis with R.
A22 inhibition assay
Exponentially growing cultures (OD600 ~0.4) were loaded on a CellASIC ONIX plate for bacteria (EMD Millipore). The plate was then flushed with fresh medium without A22 for at least 12 hours to get the cells accustomed to their new environment. The plate was then flushed with fresh medium constituted with A22 (20 μg ml−1). Images were acquired every five minutes using a Leica DMi8 microscope with a 1000x phase contrast objective using an 8 micron Z-stack.
Statistical methods
All statistical analyses were done with R with >250 observations for each features. Normality was tested with the Bartlett test. The identity of the distributions was evaluated with the kruskal test, and the groups compared pairwise with the Wilcox test for non-parametric statistical tests.
References
Vollmer, W., Blanot, D. & De Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008).
Löwe, J. & Amos, L. A. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 (1998).
Haeusser, D. P. & Margolin, W. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 14, 305–319 (2016).
Erickson, H. P., Anderson, D. E. & Osawa, M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol. Mol. Biol. Rev. 74, 504–528 (2010).
Errington, J., Daniel, R. A. & Scheffers, D.-J. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. MMBR 67, 52–65 (2003).
Rothfield, L., Taghbalout, A. & Shih, Y.-L. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3, 959–968 (2005).
Holden, S. J. et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl. Acad. Sci. 111, 4566 (2014).
Szwedziak, P., Wang, Q., Bharat, T. A. M., Tsim, M. & Löwe, J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3, e04601 (2014).
Ramirez-Diaz, D. A. et al. Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLOS Biol. 16, e2004845 (2018).
Monteiro, J. M. et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018).
Guan, F. et al. Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division. eLife 7, e35578 (2018).
Sánchez-Gorostiaga, A. et al. Life without Division: Physiology of Escherichia coli FtsZ-Deprived Filaments. mBio 7, e01620–16 (2016).
Makarova, K. S. & Koonin, E. V. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol. Direct 5, 33 (2010).
Makarova, K. S., Yutin, N., Bell, S. D. & Koonin, E. V. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8, 731 (2010).
Bernander, R. & Ettema, T. J. FtsZ-less cell division in archaea and bacteria. Curr. Opin. Microbiol. 13, 747–752 (2010).
Kondo, N., Nikoh, N., Ijichi, N., Shimada, M. & Fukatsu, T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc. Natl. Acad. Sci. 99, 14280 (2002).
Rivas-Marín, E., Canosa, I. & Devos, D. P. Evolutionary Cell Biology of Division Mode in the Bacterial Planctomycetes-Verrucomicrobia- Chlamydiae Superphylum. Front. Microbiol. 7, 1964 (2016).
Wiegand, S. et al. Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nat. Microbiol. (2019).
Adams, D. W. & Errington, J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642 (2009).
Lutkenhaus, J., Pichoff, S. & Du, S. Bacterial cytokinesis: From Z ring to divisome. Cytoskelet. Hoboken NJ 69, 778–790 (2012).
Margolin, W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871 (2005).
Du, S. & Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 105, 177–187 (2017).
Den Blaauwen, T., De Pedro, M. A., Nguyen-Distèche, M. & Ayala, J. A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32, 321–344 (2008).
de Pedro, M. A., Quintela, J. C., Höltje, J. V. & Schwarz, H. Murein segregation in Escherichia coli. J. Bacteriol. 179, 2823–2834 (1997).
Egan, A. J. F., Cleverley, R. M., Peters, K., Lewis, R. J. & Vollmer, W. Regulation of bacterial cell wall growth. FEBS J. 284, 851–867 (2017).
Szwedziak, P. & Löwe, J. Do the divisome and elongasome share a common evolutionary past? Curr. Opin. Microbiol. 16, 745–751 (2013).
Carballido-López, R. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. MMBR 70, 888–909 (2006).
Typas, A., Banzhaf, M., Gross, C. A. & Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123 (2011).
Ursell, T. S. et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl. Acad. Sci. USA 111, E1025–E1034 (2014).
Daniel, R. A. & Errington, J. Control of Cell Morphogenesis in Bacteria: Two Distinct Ways to Make a Rod-Shaped Cell. Cell 113, 767–776 (2003).
Brown, P. J. B. et al. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci. 109, 1697 (2012).
Abhayawardhane, Y. & Stewart, G. C. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J. Bacteriol. 177, 765–773 (1995).
Angert, E. R. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3, 214–224 (2005).
Eswara, P. J. & Ramamurthi, K. S. Bacterial Cell Division: Nonmodels Poised to Take the Spotlight. Annu. Rev. Microbiol. 71, 393–411 (2017).
Abdelrahman, Y., Ouellette, S. P., Belland, R. J. & Cox, J. V. Polarized Cell Division of Chlamydia trachomatis. PLOS Pathog. 12, e1005822 (2016).
Liechti, G. et al. Pathogenic Chlamydia Lack a Classical Sacculus but Synthesize a Narrow, Mid-cell Peptidoglycan Ring, Regulated by MreB, for Cell Division. PLoS Pathog. 12, e1005590–e1005590 (2016).
Wagner, M. & Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Environ. Biotechnol. Biotechnol. 17, 241–249 (2006).
Devos, D. P. Gemmata obscuriglobus. Curr. Biol. 23, R705–R707 (2013).
Devos, D. P. & Ward, N. L. Mind the PVCs. Environ. Microbiol. 16, 1217–1221 (2014).
Rivas-Marín, E. & Devos, D. P. The Paradigms They Are a-Changin’: past, present and future of PVC bacteria research. Antonie Van Leeuwenhoek 111, 785–799 (2018).
van Niftrik, L. et al. Linking Ultrastructure and Function in Four Genera of Anaerobic Ammonium-Oxidizing Bacteria: Cell Plan, Glycogen Storage, and Localization of Cytochrome c Proteins. J. Bacteriol. 190, 708 (2008).
Jeske, O. et al. Planctomycetes do possess a peptidoglycan cell wall. Nat. Commun. 6, 7116 (2015).
Liechti, G. W. et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510 (2014).
Pilhofer, M. et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 4, 2856 (2013).
van Teeseling, M. C. F. et al. Anammox Planctomycetes have a peptidoglycan cell wall. Nat. Commun. 6, 6878 (2015).
Devos, D. P. PVC bacteria: variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol. 22, 14–20 (2014).
Boedeker, C. et al. Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 8, 14853 (2017).
Jogler, C. et al. Identification of Proteins Likely To Be Involved in Morphogenesis, Cell Division, and Signal Transduction in Planctomycetes by Comparative Genomics. J. Bacteriol. 194, 6419–6430 (2012).
Pilhofer, M. et al. Characterization and Evolution of Cell Division and Cell Wall Synthesis Genes in the Bacterial Phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and Phylogenetic Comparison with rRNA Genes. J. Bacteriol. 190, 3192–3202 (2008).
Demarre, G., Galli, E. & Barre, F.-X. The FtsK Family of DNA Pumps. in DNA Helicases and DNA Motor Proteins (ed. Spies, M.) 245–262, https://doi.org/10.1007/978-1-4614-5037-5_12 (Springer New York, 2013).
Meeske, A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).
Cho, H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172–16172 (2016).
Fraipont, C. et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 (2011).
Takacs, C. N. et al. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J. Bacteriol. 192, 1671–1684 (2010).
Pende, N. et al. Host-Polarized Cell Growth in Animal Symbionts. Curr. Biol. 28, 1039–1051.e5 (2018).
Meier, E. L., Razavi, S., Inoue, T. & Goley, E. D. A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol. Microbiol. 101, 265–280 (2016).
Xiao, J. & Goley, E. D. Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr. Opin. Microbiol. 34, 90–96 (2016).
England, K., Crew, R. & Slayden, R. A. Mycobacterium tuberculosis septum site determining protein, Ssd encoded by rv3660c, promotes filamentation and elicits an alternative metabolic and dormancy stress response. BMC Microbiol. 11, 79 (2011).
Wang, L., Khattar, M. K., Donachie, W. D. & Lutkenhaus, J. FtsI and FtsW Are Localized to the Septum in Escherichia coli. J. Bacteriol. 180, 2810 (1998).
Reichmann, N. T. et al. SEDS–bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat. Microbiol. 4, 1368–1377 (2019).
Rismondo, J., Halbedel, S. & Gründling, A. Cell Shape and Antibiotic Resistance Are Maintained by the Activity of Multiple FtsW and RodA Enzymes in Listeria monocytogenes. mBio 10, e01448–19 (2019).
Marbouty, M., Mazouni, K., Saguez, C., Cassier-Chauvat, C. & Chauvat, F. Characterization of the Synechocystis Strain PCC 6803 Penicillin-Binding Proteins and Cytokinetic Proteins FtsQ and FtsW and Their Network of Interactions with ZipN. J. Bacteriol. 191, 5123 (2009).
Margolin, W. Sculpting the bacterial cell. Curr. Biol. CB 19, R812–R822 (2009).
Brown, P. J. B., Kysela, D. T. & Brun, Y. V. Polarity and the diversity of growth mechanisms in bacteria. Semin. Cell Dev. Biol. 22, 790–798 (2011).
Jacquier, N., Frandi, A., Pillonel, T., Viollier, P. H. & Greub, G. Cell wall precursors are required to organize the chlamydial division septum. Nat. Commun. 5, 3578 (2014).
Ouellette, S. P., Karimova, G., Subtil, A. & Ladant, D. Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol. Microbiol. 85, 164–178 (2012).
Mercier, R., Kawai, Y. & Errington, J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife 3 (2014).
Leaver, M., Domínguez-Cuevas, P., Coxhead, J. M., Daniel, R. A. & Errington, J. Life without a wall or division machine in Bacillus subtilis. Nature 457, 849 (2009).
Joseleau-Petit, D., Liébart, J.-C., Ayala, J. A. & D’Ari, R. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J. Bacteriol. 189, 6512–6520 (2007).
Karczmarek, A. et al. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol. Microbiol. 65, 51–63 (2007).
Bailey, J. et al. Essential gene deletions producing gigantic bacteria. PLOS Genet. 15, e1008195 (2019).
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J. & Schweizer, H. P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998).
Herrero, M., de Lorenzo, V. & Timmis, K. Transposon vectors containing non-antibiotic selection markers for cloning and stable chromosomal insertion of foreign DNA in Gram-negative bacteria. vol. 172 (1990).
Rivas-Marín, E., Canosa, I., Santero, E. & Devos, D. P. Development of Genetic Tools for the Manipulation of the Planctomycetes. Front. Microbiol. 7 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Wingett, S. FastQ Screen - Contamination screening for NGS data sets. (2011).
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Bankevich, A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 19, 455–477 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Yuste, L. et al. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ. Microbiol. 8, 165–177 (2006).
Fiebig, A., Keren, K. & Theriot, J. A. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol. Microbiol. 60, 1164–1178 (2006).
Acknowledgements
This work was supported by BFU2016-78326-P and MDM-2016-0687, Spanish Ministerio de Economía y Competitividad. Also, part of this work was supported by a visiting scientist grant awarded to ERM by the Soehngen Institute of Anaerobic Microbiology (SIAM, project 024002002) of the Netherlands Organisation for Scientific Research (NWO). SHP is supported by the NWO grant ALWOP.308 and LC by the Spinoza prize awarded to Prof. dr. M.S.M. Jetten.
Author information
Authors and Affiliations
Contributions
D.P.D. and E.R.M. designed the study. E.R.M., S.H.P., L.C. and S.W. conducted the experiments. All authors interpreted the results and discussed the analyses. D.P.D. and E.R.M. wrote the manuscript with feedback from S.P., S.W., C.J. and L.v.N. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rivas-Marin, E., Peeters, S.H., Claret Fernández, L. et al. Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci Rep 10, 66 (2020). https://doi.org/10.1038/s41598-019-56978-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-56978-8
This article is cited by
-
Uncovering the biotechnological capacity of marine and brackish water Planctomycetota
Antonie van Leeuwenhoek (2024)
-
Topographic comparison of the retinal microvascular changes between patients with compressive and glaucomatous optic neuropathies
Scientific Reports (2023)
-
Essential gene complement of Planctopirus limnophila from the bacterial phylum Planctomycetes
Nature Communications (2023)
-
Stieleria tagensis sp. nov., a novel member of the phylum Planctomycetota isolated from Tagus River in Portugal
Antonie van Leeuwenhoek (2023)
-
Stratiformator vulcanicus gen. nov., sp. nov., a marine member of the family Planctomycetaceae isolated from a red biofilm in the Tyrrhenian Sea close to the volcanic island Panarea
Antonie van Leeuwenhoek (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.