Abstract
Recent studies have revealed an association between the dysregulation of the gut microbiome and dementia. However, whether this dysregulation is associated with mild cognitive impairment (MCI), an early stage of cognitive decline, in patients without dementia remains unclear. We performed a cross-sectional analysis to determine the association between the gut microbiome and MCI. Data, including patient demographics, risk factors, cognitive function, and brain imaging, were collected. The gut microbiome was assessed through terminal restriction fragment length polymorphism analysis. Multivariable logistic regression models were used to identify factors independently associated with MCI. Graphical modelling was used to illustrate mutual associations between MCI and identified factors. We analysed 82 patients, 61 of whom exhibited MCI. Patients with MCI had a higher prevalence of Bacteroides. Furthermore, patients with more Bacteroides were more likely to present with white matter hyperintensity and high voxel-based specific regional analysis system for Alzheimer’s Disease (VSRAD) scores, indicating cortical and hippocampal atrophy. A multivariable logistic regression analysis revealed that a greater prevalence of Bacteroides was independently associated with MCI. Graphical modelling also showed a close association between Bacteroides and MCI. In conclusion, an increased prevalence of Bacteroides is independently associated with the presence of MCI in patients without dementia.
Similar content being viewed by others
Introduction
Mild cognitive impairment (MCI) refers to a very early stage of cognitive decline in patients not yet exhibiting dementia and is an important predictive risk factor for dementia1. Many people present to health services with MCI, which occurs in up to 20% of people older than 65 years of age1, and 10–15% of patients with MCI develop dementia annually2. Because the number of patients with dementia has been increasing in Japan, a comprehensive strategy for dementia research has been introduced, which includes analyses of the characteristics of patients with MCI3. A multifactorial assessment of MCI can be useful for preventing the progression from MCI to dementia.
Recently, the relationship between the psychophysiological status (such as depression) of a person and their gut microbiome has become an intriguing research focus4. In addition, several researchers have identified novel associations between the gut microbiome and dementia5,6,7, suggesting that the gut microbiome may modulate host brain function via a microbiome–gut–brain axis8. However, whether the gut microbiome affects cognitive functions has not yet been clarified. A previous study demonstrated the presence of bacterial lipopolysaccharide (LPS) (a component of the outer leaflet of the outer membrane of bacteria) in brain lysates derived from the hippocampus and superior temporal lobe of the neocortex of brains from Alzheimer’s disease (AD) patients9. More specifically, Zhao et al., reported that bacterial LPS, which is a pro-inflammatory neurotoxin, may be able to cross physiological barriers to access the hippocampus, leading to cognitive impairments9, such as memory disturbances. Other studies suggested that the disruption of the neuro-inflammatory system10, vascular inflammation11, or remote relationships driven by various metabolites12 may be involved in the potential mechanisms for cognitive declines caused by the gut microbiome. Specifically, enterobacterial infections may exacerbate AD progression by promoting immune haemocyte recruitment to the brain10. Tang et al., reported that the presence of bacterial products in the systemic circulation may heighten the inflammatory state11. Furthermore, they also reported that microbiota metabolites, such as short-chain fatty acids, trimethylamine/trimethylamine N-oxide, and bile acids, may contribute to life-threatening diseases11. Therefore, analysing the gut microbiome in MCI patients without dementia may reveal previously unidentified risk factors for MCI.
We are conducting a clinical study that was designed to investigate the association between the composition of the gut microbiome and a patient’s clinical condition, as assessed using activities of daily living (ADL) scales and cognitive function measures. This study has been named the Gerontological Investigation of Microbiome: a longitudinal estimation study (Gimlet study)5 and is being performed at the National Center for Geriatrics and Gerontology (NCGG) (Aichi, Japan). During our baseline analysis, we found that the dysregulation of the gut microbiome, as assessed by terminal restriction fragment length polymorphism (T-RFLP) analysis, which is one of the most well-established and reliable 16S ribosomal RNA-based methods, is cross-sectionally and strongly associated with dementia, independent of traditional dementia biomarkers5. Moreover, we found that the presence of dementia, in addition to the presence of cardiovascular risk factors, indicates the advanced dysregulation of the gut microbiome13. In the present sub-study of the Gimlet study, we examined the relationship between the gut microbiome and MCI among patients without dementia who had enrolled in the Gimlet study. We hypothesized that there would be differences in the compositions of the gut microbiome between patients with MCI and those with normal cognitive function (NC).
Results
Patient characteristics
We previously analysed 128 patients in the Gimlet study. Of these, 46 were excluded: 34 had dementia, and 12 provided an insufficient quantity of faecal samples. Therefore, we analysed the remaining 82 eligible patients without dementia for this sub-analysis (female: 52.4%; mean age: 73.9 ± 8.1 years; mean Mini-Mental State Examination [MMSE] score: 27). Patients were stratified according to their levels of cognitive function and their enterotypes: 61 (74.4%) were classified as MCI, and 21 were classified as NC (Tables 1–3); 38 were enterotype I (46.3%), 5 were enterotype II (6.1%), and 39 were enterotype III (47.6%).
MCI vs. NC
Compared with NC patients, those with MCI were older (MCI vs. NC: median years, 77 vs. 69, P < 0.001), had a higher prevalence of hypertension (63.9% vs. 38.1%, P = 0.045) and were more likely to present with indications of cerebral small vessel disease (SVD), such as white matter hypersensitivity (WMH) (34.4% vs. 4.8%, P = 0.009), and impaired instrumental ADL scores (44.3% vs. 9.5%, P = 0.004). Additionally, patients with MCI had higher scores on the Dementia Behaviour Disturbance Scale (DBDS), Zarit Caregiver Burden Interview (ZBI), and voxel-based specific regional analysis system for Alzheimer’s Disease (VSRAD) (median scores: 10 vs. 2, P < 0.001; 11 vs. 6, P = 0.043; and 0.96 vs. 0.52, P = 0.010, respectively), as well as lower Geriatric Depression Scale (GDS) scores (median score: 2 vs. 4, P = 0.032) and reduced cognitive function, as demonstrated by lower scores on the MMSE, Raven’s Coloured Progressive Matrices (RCPM), Frontal Assessment Battery (FAB), and Logical Memory subtests I and II of the Wechsler Memory Scale-Revised (LM-WMSR) (median scores: 25 vs. 29, P < 0.001; 28 vs. 32, P = 0.003; 11 vs. 14, P < 0.001; 8 vs. 16, P = 0.001; and 3 vs. 10, P = 0.001, respectively) and higher scores on the Clinical Dementia Rating Scale-Sum of Boxes (CDR-SB) and Alzheimer’s Disease Assessment Scale-Cognitive Subscale (ADAS-cog) (median scores: 2 vs. 0, P < 0.001; and 8 vs. 5.7, P = 0.003, respectively). Patients with MCI had more enterotype I microbes and fewer enterotype III microbes than patients with NC (55.7% vs. 19.1%; and 37.7% vs. 76.2%, respectively, P = 0.009), suggesting a higher prevalence of Bacteroides and a lower prevalence of ‘other’ bacteria (Tables 1–3).
Comparison by enterotype
As described, we stratified the enrolled patients according to the enterotypes of their microbes and performed the following comparisons: enterotype I vs. non-enterotype I and and enterotype III vs. non-enterotype III (Tables S1 and S2, respectively). When compared with non-enterotype I patients, patients with increased levels of enterotype I microbes were more likely to exhibit a higher prevalence of cerebral SVD components (enterotype I vs. non-enterotype I: silent lacunar infarcts [SLI], 5.3% vs. 2.3%, P = 0.594; WMH, 39.5% vs. 15.9%, P = 0.024; cerebral microbleeds [CMB], 23.7% vs. 11.4%, P = 0.155; and cortical superficial siderosis [CSS], 7.9% vs. 2.3%, P = 0.332), higher VSRAD scores (median score, 0.96 vs. 0.80, P = 0.112), lower global cognitive function (median score, MMSE, 25 vs. 27, P = 0.046; CDR-SB, 2 vs. 0.5, P = 0.002), and impaired memory function (LM-WMSR I, 8 vs. 11, P = 0.134; and LM-WMSR II, 2 vs. 5, P = 0.041). No significant differences were observed for demographics or risk factors (e.g., mean age, 77 vs. 75, P = 0.145) between enterotype I patients and non-enterotype I patients (Table S1). In contrast, compared with non-enterotype III patients, patients with more enterotype III microbes had a lower prevalence of cerebral SVD components (enterotype III vs. non-enterotype III, SLI, 2.6% vs. 4.7%, P = 1.000; WMH, 15.4% vs. 37.2%, P = 0.045; CMB, 10.3% vs. 23.3%, P = 0.148; and CSS, 2.6% vs. 7.0%, P = 0.617) but were more likely to have low VSRAD scores (median score, 0.69 vs. 1.00, P = 0.048) and increased global cognitive function (median score, MMSE, 27 vs. 25, P = 0.071; and CDR-SB, 0.5 vs. 2, P = 0.007) (Table S2). Although there appeared to be significant differences regarding the prevalence of hypertension and alcohol consumption between enterotype III and non-enterotype III patients, no significant differences were observed for the Firmicutes/Bacteroidetes (F/B) ratio (Tables S3 and S4). No significant differences were observed for enterotypes between males and females (Table S5).
Multivariable analysis
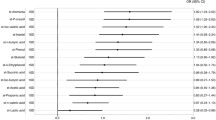
Multivariable logistic regression analyses were performed to identify factors that are independently associated with MCI. Due to the small number of patients, these analyses were performed in step-by-step increments of the number of independent variables: model 1 (adjusted for age, sex, education year, apolipoprotein E [ApoE] ε4 carrier, enterotype, and F/B ratio), model 2 (stepwise adjusted for model 1 and prevalence of risk factors), and model 3 (stepwise adjusted for model 2, magnetic resonance imaging [MRI] findings, and single photon emission computed tomography [SPECT] findings). Multivariable logistic regression analyses revealed that increased levels of enterotype I microbes was associated with the presence of MCI, independent of age, sex, education years, ApoE ε4, traditional risk factors, and brain imaging (model 1: odds ratio [OR 10.2], 95% confidence interval [95% CI], 2.23–62.7, P = 0.002; model 2: OR 5.95, 95% CI, 1.61–28.2, P = 0.006; and model 3: OR 5.36, 95% CI, 1.30–28.7, P = 0.019; Table 4). Conversely, enterotype III was independently associated with the presence of MCI (model 1: OR 0.09, 95% CI, 0.02–0.42, P = 0.002; model 2: OR 0.19, 95% CI, 0.05–0.65, P = 0.008; and model 3: OR 0.19, 95% CI, 0.05–0.65, P = 0.008) (Table 5).
Graphical modelling
Graphical modelling was performed to visualize the mutual associations among factors used in the multivariable logistic regression analyses. Accordingly, modelling showed that the presence of MCI was more likely to be associated with age and was equally associated with ApoE ε4 and enterotype I. Enterotype I was closely associated with the presence of MCI when compared with the other factors. Furthermore, a close association between CMB and WMH was shown by this modelling (Fig. 1).
Graphical modelling of factors used for the multivariable analyses. Line thickness is proportional to the number of patients that contributed to the comparison. Green lines indicate a positive relationship, and red lines indicate a negative relationship. Abbreviations: mci, mild cognitive impairment; apo, apolipoprotein E ε4 carrier; wmh, white matter hyperintensity; en1, enterotype I; age, patient’s age; smk, smoking habit; cmb, cerebral microbleeds.
Discussion
The primary finding of our present study was that the increased prevalence of Bacteroides, defined as enterotype I, was independently associated with the presence of MCI in patients without dementia. Specifically, patients with an increased prevalence of Bacteroides were more likely to have lower global cognitive function (indicated by MMSE and CDR scores) and impaired memory dysfunction (indicated by logical memory subtests) compared with non-enterotype I patients, although there were no significant differences in demographics and risk factors, such as age, sex, and hypertension.
Recent studies have reported controversial findings regarding an association between the gut microbiome and dementia. For example, previous reports have shown both decreased5,6 and increased7 proportions of Bacteroides in patients with dementia. Vogt et al.7, speculated that LPS is the component of Bacteroidetes that potentiates systemic inflammation and amyloid fibrillogenesis, ultimately resulting in amyloid deposition. Furthermore, Zhao and Lukiw et al., also reported that microbiome-derived LPS was enriched in the perinuclear region of the AD brain14, whereas LPS was detected in the hippocampus of patients with AD9. Our findings are consistent with these studies because our patients with more Bacteroidetes had lower logical memory subtest scores, indicating impaired memory function.
The mechanism through which the gut microbiome affects human cognitive functions remains unknown, although animal studies strongly implicate the gut microbiome as a key regulator of the brain and behaviour15. The functional pathways through which the gut microbiome communicates with the brain, also known as gut–microbiome–brain cross-talk, is a bidirectional, functional communication network between microbes and the brain that comprises neuroendocrine, neural, and neuroimmune signalling pathways16. Explicitly, three hypothesis have been proposed: (1) a circulation pathway, indicating the transportation of microbial metabolites, toxins, and pro-inflammatory factors; (2) a neuroendocrine pathway, indicating the activation of neuroendocrine cells that can be attributed to microbial metabolites; and (3) a neural pathway, indicating an interaction between the gut microbiome and the autonomic nervous system. This previous report also showed that an increase in the abundance of a pro-inflammatory gut microbiome taxon and a reduction in the abundance of an anti-inflammatory taxon may be associated with a peripheral inflammatory state in patients with cognitive impairments and brain amyloidosis17. Another report describing the gut–brain module analysis of faecal metagenomes identified the microbial synthesis of metabolites and suggested a potential role for these metabolites in the onset of depression18.
At present, we do not have sufficient data to identify the underlying mechanism that mediates the association between the gut microbiome and cognitive impairment. However, potential differences regarding the presence of WMH may represent supporting evidence that cerebral SVD acts as an intermediate between cognitive impairment and the gut microbiome. Likewise, Ong et al., reported that alterations in brain structure, as assessed by diffusion tensor imaging of MRI scans, were associated with the gut microbiome19, suggesting there may be a definite and direct relationship between the gut microbiome and the brain parenchyma. Further studies examining the association between cerebral SVD and gut microbiome metabolites in the Gimlet study cohort will shed light on this issue.
Here, we found other interesting findings. There was an independent association between WMH and the gut microbiome, although other components of cerebral SVD, such as SLI and CMB, did not show such significant differences. This finding suggests that a potential mechanism regarding the development of WMH may be attributable to the gut microbiome, either directly or indirectly. Further study regarding the association between cerebral SVD and the gut microbiome is warranted.
Our study has several strengths. First, we systematically assessed the cognitive functions of patients, using a comprehensive geriatric assessment in the setting of the memory clinic. These steps have provided detailed analyses of cognitive function. Second, we found a close and strong association between the gut microbiome and MCI, identified by multivariable analyses and graphical modelling, compared with the relationships between the other risk factors and MCI. Applying graphical modelling has strengthened our findings. Third, our comprehensive assessment regarding the gut microbiome and MCI may fill the knowledge gap regarding the mechanisms that connect cognitive decline with the gut microbiome. Finally, because we have established a new relationship and have widened the knowledge of such associations with regard to MCI, our findings may contribute to a better understanding and increased attention being paid to the associations between the gut microbiome and cognitive functions.
Nonetheless, this study has several limitations. A causal relationship between the gut microbiome and MCI could not be established because of the cross-sectional study design. The small number of patients renders our study at risk of being statistically underpowered. The unequal numbers of patients with MCI and those with NC may also be a potential limitation. Selection bias may exist because this was a single hospital-based cohort. Furthermore, potentially confounding factors may exist because the gut microbiome is affected by diet20,21 and the presence of complications, such as blood pressure variability and cardiovascular disease13. In particular, a recent study21 suggests that the modified Mediterranean-ketogenic diet can modulate the gut microbiome and metabolites in association with improved biomarkers regarding Alzheimer’s disease. Likewise, we plan to assess the nutritional status including the diet pattern in the future study because the Japanese diet pattern can alter the intestinal bacteria and have beneficial effects on humans20.
Assessments of the amyloid-β precursor protein may be useful because this factor suggests inflammatory endothelial dysfunction and the risk of cognitive impairment22. The specific mechanism through which the microbiome affects cognitive decline has not yet been clarified. Rather than identifying the roles of specific bacteria, the functional analysis of the gut microbiome as one integrated organ may be more practically useful. Our data indicate both accelerating (in the present study) and decelerating (in a previously published study5) effects of Bacteroides on cognitive decline. However, this relationship may be attributable to unidentified taxa (indicated by ‘other’) and/or metabolites, which may depend on the intraintestinal environment. Next-generation sequencing technologies could be useful for identifying the specific genera or species of microbes that are collectively categorized as ‘other’ bacteria by the T-RFLP method.
To date, various risk factors for cognitive impairment have been proposed, such as aging, education years, hypertension, diabetes mellitus, and social factors1; however, the human gut microbiome has not previously been mentioned as a risk factor. Our findings add the gut microbiome as a new risk factor for cognitive impairment. More importantly, controlling the gut microbiome may represent a possible method for the prevention and intervention of cognitive impairment.
Although this sub-analysis study contains a small number of patients and our analysis is preliminary, our findings provide supporting evidence for a relationship between the gut microbiome and cognitive impairment. Longitudinal assessments of the gut microbiome using next-generation sequencing technology, assessments of the various metabolites produced by the gut microbiome, and assessments of diet patterns should be investigated in future studies to clarify the underlying mechanism that connects the gut microbiome with cognitive function.
Conclusions
We showed that components of the gut microbiome, in particular Bacteroides, may be associated with the presence of MCI in patients without dementia. We speculate that some gut microbiome metabolites could affect cognitive functions through a microbiome–gut–brain axis. Further studies are warranted to examine such relationships.
Methods
Study design
This study was a sub-analysis of our previously published, single-centre observational study (Gimlet study)5. The aim of this study was to investigate the association between the gut microbiome and MCI in patients without dementia. This study complied with the Declaration of Helsinki and was approved by the Institutional Review Board at the National Center for Geriatrics and Gerontology (No. 1191). Informed consent was obtained from all patients and their families before participation in this study. The Gimlet study is registered with the UMIN Clinical Trials Registry (UMIN000031851). Detailed information has been provided in the supplementary file and in the previous report5.
Subjects
Patients were eligible for the Gimlet study if they met the following criteria: (1) were able to undergo a brain MRI; (2) provided informed consent in writing; (3) provided informed consent for the NCGG Biobank to store their clinical data, blood, and faecal samples; and (4) were accompanied by a study partner who could assess the patient’s condition. Patients were excluded if they met the following criteria: (1) were unable to undergo an MRI examination or the MRI scan could not be evaluated because of movement; (2) had local lesions, such as cerebral infarctions, that were detected by MRI before enrolment and that could significantly affect cognitive function; (3) had a history of a major psychological disorders or current, serious or unstable alcohol or drug abuse; (4) had ≤6 years of education; (5) had a history of cancer of the digestive tract; or (6) were judged by an investigator to be ineligible to participate as a study subject (e.g., recent use of antibiotics, brain tumour, encephalitis/meningitis, normal pressure hydrocephalus, Huntington’s disease). Patients were also excluded from this sub-analysis if they met the following criteria: (1) had dementia; or (2) were unable to provide sufficient faecal samples to facilitate the analysis of metabolites both now and in the future.
Baseline assessment
All participants underwent a comprehensive geriatric assessment23 using the following: (1) demographic characteristics; (2) risk factors, such as hypertension, dyslipidaemia, diabetes mellitus, ischaemic heart disease, chronic kidney disease, smoking habits, or a history of stroke and alcohol consumption; (3) basic and instrumental ADL scales, assessed using the Barthel Index24 and Lawton and Brody scale25; (4) global cognitive function, assessed using the MMSE26 and CDR27; (5) neuropsychological testing, using the ADAS-cog28, RCPM29, FAB30, and LM-WMSR31; (6) laboratory variables, including ApoE ε4 as a risk factor for AD; (7) ankle brachial index and pulse wave velocity, as indicators of arteriosclerosis32 and the ‘impact’ of pulse33; (8) brain imaging, such as MRI and SPECT; (9) an assessment of other factors, such as the presence of frailty34 and subjective hearing loss; and (10) an assessment of social and lifestyle factors, e.g., using the Mini-Nutritional Assessment-Short Form (MNA-SF) to assess nutritional status35. Clinical samples and data were provided by the NCGG Biobank, which collects clinical data for research.
Classification of cognitive function
In the Gimlet study5, patients were divided into two categories, using measures that reliably indicate the presence of dementia: (1) a group without dementia (MMSE ≥ 20 and CDR < 1), and (2) a group with dementia (MMSE < 20 and/or CDR ≥ 1). In the present sub-study, the patients without dementia were further divided into two categories: MCI and NC. MCI was defined as MMSE ≥ 20 and CDR = 0.5, indicating possible, very mild dementia and suggesting that the patient has a higher risk of developing dementia36. NC was defined as MMSE ≥ 20 and CDR = 0.
Brain imaging
Patients underwent a 1.5 T MRI of the brain (Philips Ingenia, Eindhoven, the Netherlands). MRI scans were obtained, including diffusion-weighted imaging, fluid-attenuated inversion recovery imaging, T2-weighted imaging, T2*-weighted gradient echo imaging, 3D T1-weighted sagittal and axial coronal views, and 3D time-of-flight MR angiography scans. The presence and components of cerebral SVD were categorised using standards for reporting neuroimaging vascular changes37, including SLI, WMH, CMB, and CSS. Moreover, VSRAD advance software (Eisai Co., Ltd., Tokyo, Japan) was used to quantify cortical and hippocampal atrophy, using standardised z-scores. Increased VSRAD scores suggest the potential presence of AD, as this score reflects hippocampal atrophy, one of the characteristics of the AD brain38. Patients also underwent N-isopropyl-p-[123I]-iodoamphetamine-SPECT. The presence or absence of reduced blood flow in the area of the posterior cingulate gyrus and/or the precuneus was assessed as a surrogate marker of AD39.
Gut microbiome
Faecal samples were collected at home, just after evacuation, by patients or their family members, using scoop collection tubes, while patients were consuming their usual diets, and samples were frozen and preserved at −81 °C at the NCGG Biobank. After all samples were collected, the gut microbiome was analysed by the TechnoSuruga Laboratory (Shizuoka, Japan), using T-RFLP analysis40. T-RFLP analysis is one of the most well-established and reliable 16S ribosomal RNA-based methods, especially when considering its high throughput and reproducibility. First, T-RFLP was used to classify gut microbes into the following 10 groups: Prevotella, Bacteroides, Lactobacillales, Bifidobacterium, Clostridium cluster IV, Clostridium subcluster XIVa, Clostridium cluster IX, Clostridium cluster XI, Clostridium cluster XVIII, and ‘others’. Second, by referencing the Human Faecal Microbiome T-RFLP profile41,42, the gut microbiome was stratified into three enterotypes: enterotype I included Bacteroides at >30%, enterotype II included Prevotella at >15%, and enterotype III included the remaining bacteria. Third, the F/B ratio was examined, as an increase in the F/B ratio is indicative of dysbiosis43. The phylum Firmicutes included the Lactobacillales and Clostridium clusters, and the phylum Bacteroidetes included Bacteroides and Prevotella.
Statistical analysis
Continuous, ordinal, and categorical variables are expressed as the mean ± standard deviation, median and interquartile range, and frequency or proportion (percentage), respectively, and were compared using an unpaired Student’s t-test, a Wilcoxon rank-sum test, and a χ2 test, respectively. First, patients were divided into two groups, according to the presence or absence of MCI, and their clinical characteristics and the compositions of their gut microbiomes were compared using the Wilcoxon rank-sum test and χ2 test, respectively. Second, patients were also divided into two groups, either enterotype I and non-enterotype I, or enterotype III or non-enterotype III, to compare their clinical characteristics based on Gimlet baseline data. This division indicated that there were a small number of patients with enterotype II. Third, multivariable logistic regression models were performed to identify factors that were independently associated with MCI. Backward stepwise multivariable logistic regression analyses were performed by adjusting for patient demographics, ApoE ε4, gut microbiome (enterotype and the F/B ratio), risk factors, brain MRI findings, and blood flow reductions on SPECT images. Last, graphical modelling was generated to illustrate mutual associations among the factors used in the multivariable logistic regression analyses, including traditional risk factors and gut microbiome, to visualize and compare mutual relationships. Appropriately, ORs are presented as 95% CIs. All comparisons were two-tailed, and P < 0.05 was considered to represent statistical significance. Data were analysed using the JMP 11.0 software package (SAS Institute Inc., Cary, NC, USA), and graphical modelling was generated using R (R Language and Environment for Statistical Computing, Vienna, Austria).
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Livingston, G. et al. Dementia prevention, intervention, and care. Lancet. 390, 2673–2734 (2017).
Ganguli, M. et al. Prevalence of mild cognitive impairment by multiple classifications: The Monongahela-Youghiogheny Healthy Aging Team (MYHAT) project. Am J Geriatr Psychiatry. 18, 674–83 (2010).
Saji, N. et al. ORANGE’s challenge: developing wide-ranging dementia research in japan. Lancet Neurol. 15, 661–662 (2016).
Stower, H. Depression linked to the microbiome. Nat. Med. 25, 358 (2019).
Saji, N. et al. Analysis of the relationship between the gut microbiome and dementia: a cross-sectional study conducted in Japan. Sci Rep. 9, 1008 (2019).
Alkasir, R. et al. Human gut microbiota: the links with dementia development. Protein Cell. 8, 90–102 (2017).
Vogt, N. M. et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep 7, 13537 (2017).
Solas, M., Milagro, F. I., Ramírez, M. J. & Martínez, J. A. Inflammation and gut-brain axis link obesity to cognitive dysfunction: plausible pharmacological interventions. Curr Opin Pharmacol. 37, 87–92 (2017).
Zhao, Y., Jaber, V. & Lukiw, W. J. Secretory Products of the Human GI Tract Microbiome and Their Potential Impact on Alzheimer’s Disease (AD): Detection of Lipopolysaccharide (LPS) in AD Hippocampus. Front Cell Infect Microbiol 7, 318 (2017).
Wu, S. C., Cao, Z. S., Chang, K. M. & Juang, J. L. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nature Communications. 8, 24 (2017).
Tang, W. H., Kitai, T. & Hazen, S. L. Gut Microbiota in Cardiovascular Health and Disease. Circ. Res. 120, 1183–1196 (2017).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 472, 57–63 (2011).
Saji, N. et al. Proportional changes in the gut microbiome: a risk factor for cardiovascular disease and dementia? Hypertens Res. 42, 1090–1091 (2019).
Zhao, Y., Cong, L., Jaber, V. & Lukiw, W. J. Microbiome-derived lipopolysaccharide enriched in the perinuclear region of alzheimer’s disease brain. Front Immunol. 8, 1064 (2017).
Cryan, J. F. & Dinan, T. G. Talking about a microbiome revolution. Nat Microbiol. 4, 552–553 (2019).
Welcome, M. O. Current perspectives and mechanisms of relationship between intestinal microbiota dysfunction and dementia: A review. Dement Geriatr Cogn Disord Extra. 8, 360–381 (2018).
Cattaneo, A. et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiology of Aging. 49, 60–68 (2017).
Valles-Colomer, M. et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 4, 623–632 (2019).
Ong, I. M. et al. Gut microbiome populations are associated with structure-specific changes in white matter architecture. Transl Psychiatry. 8, 6 (2018).
Kushida, M. et al. Effects of the 1975 Japanese diet on the gut microbiota in younger adults. J Nutr Biochem. 64, 121–127 (2019).
Nagpal, R. et al. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine. 47, 529–542 (2019).
Saji, N. et al. Cilostazol May Decrease Plasma Inflammatory Biomarkers in Patients with Recent Small Subcortical Infarcts: A Pilot Study. J Stroke Cerebrovasc Dis. 6, 1639–1645 (2018).
Toba, K. The guideline for comprehensive geriatric assessment. Nihon Ronen Igakkai Zasshi. 42, 177–180 (2005).
Mahoney, F. I., Barthel, D. W. & Callahan, J. P. Rehabilitation of the hemiplegic patient: A clinical evaluation. South. Med. J. 48, 472–480 (1955).
Lawton, M. P. & Brody, E. M. Assessment of older people: Self-maintaining and instrumental activities of daily living. Gerontologist 9, 179–186 (1969).
Folstein, M. F., Folstein, S. E. & McHugh, P. R. Mini-mental state”; A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12, 189–198 (1975).
Morris, J. C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 43, 2412–2414 (1993).
Rosen, W. G., Mohs, R. C. & Davis, K. L. A new rating scale for Alzheimer’s disease. Am J Psychiatry. 141, 1356–1364 (1984).
Raven J. Guide to using the coloured progressive matrices. London. H.K. Lewis. (1965).
Dubois, B., Slachevsky, A., Litvan, I. & Pillon, B. The FAB: a Frontal Assessment Battery at bedside. Neurology. 55, 1621–1626 (2000).
Wechsler D. Wechsler memory scale-revised. San Antonio, TX. Psychological Corporation. (1981).
Saji, N. et al. Comparison of arteriosclerotic indicators in patients with ischemic stroke: Ankle-brachial index, brachial-ankle pulse wave velocity, and cardio-ankle vascular index. Hypertens Res. 38, 323–328 (2015).
Saji, N., Toba, K. & Sakurai, T. Cerebral small vessel disease and arterial stiffness: Tsunami effect in the brain? Pulse. 3, 182–189 (2016).
Fried, L. P. et al. Frailty in older adults: evidence for a phenotype. The J Gerontol A Bio Sci Med Sci. 56, M146–156 (2001).
Vellas, B. et al. Overview of the mna–its history and challenges. J Nutr Health Aging. 10, 456–463 (2006).
Perneczky, R. et al. Mapping scores onto stages: mini-mental state examination and clinical dementia rating. Am J Geriatr Psychiatry. 14, 139–144 (2006).
Wardlaw, J. M. et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol. 12, 822–838 (2013).
Matsuda, H. et al. Automatic voxel-based morphometry of structural MRI by SPM8 plus diffeomorphic anatomic registration through exponentiated lie algebra improves the diagnosis of probable Alzheimer disease. AJNR Am J Neuroradiol. 33, 1109–1114 (2012).
Ito, K. et al. Prediction of outcomes in MCI with (123)I-IMP-CBF SPECT: a multicenter prospective cohort study. Ann Nucl Med. 27, 898–906 (2013).
Osborn, A. M., Moore, E. R. & Timmis, K. N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2, 39–50 (2000).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature. 473, 174–180 (2011).
Emoto, T. et al. Analysis of Gut Microbiota in Coronary Artery Disease Patients: a Possible Link between Gut Microbiota and Coronary Artery Disease. J Atheroscler Thromb. 23, 908–921 (2016).
Spychala, M. S. et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann Neurol. 84, 23–36 (2018).
Acknowledgements
This study was supported by research grants from the Research Funding of Longevity Sciences (26–20, 27–21, 28–15, 30–1, and 19–24), the National Center for Geriatrics and Gerontology, and the National Agriculture and Food Research Organization (NARO) Bio-oriented Technology Research Advancement Institution project (Advanced Integration Research for Agriculture and Interdisciplinary Fields). We thank Maki Yamamoto, Yukie Ohsaki, Saori Yoshimura, Hana Saito, and Ayaka Suzuki (NCGG), and Yuya Shinkawa (Kurume University) for their technical and secretarial assistance, and the BioBank, NCGG, for quality control of the clinical samples and data. We thank Rachel James and Lisa Giles, PhD, from Edanz Group (www.edanzediting.com/ac) for editing drafts of this manuscript.
Author information
Authors and Affiliations
Contributions
N.S. is the principal investigator and contributed to the concept, drafting, and design of the protocol. K.M., T.H., T.T., T.S., A.K., S.N., K.T. and T.S. contributed to the design of the study, analysis of the data, and review of the manuscript’s intellectual content.
Corresponding author
Ethics declarations
Competing interests
N.S. has received grants from the NARO Bio-oriented Technology Research Advancement Institution project (Advanced Integration Research for Agriculture and Interdisciplinary Fields) and the BMS/Pfizer Japan Thrombosis Investigator Initiated Research Program. N.S., S.N. and T.S. have received research grants from the Research Funding of Longevity Sciences from the National Center for Geriatrics and Gerontology. N.S., K.T. and T.S. have received research funds for Comprehensive Research on Aging and Health from the Japan Agency for Medical Research and Development (AMED). T.T. has received grants from the NARO Bio-oriented Technology Research Advancement Institution project (Advanced Integration Research for Agriculture and Interdisciplinary Fields).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saji, N., Murotani, K., Hisada, T. et al. The relationship between the gut microbiome and mild cognitive impairment in patients without dementia: a cross-sectional study conducted in Japan. Sci Rep 9, 19227 (2019). https://doi.org/10.1038/s41598-019-55851-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-55851-y
This article is cited by
-
QNetDiff: a quantitative measurement of network rewiring
BMC Bioinformatics (2024)
-
Sodium oligomannate alters gut microbiota, reduces cerebral amyloidosis and reactive microglia in a sex-specific manner
Molecular Neurodegeneration (2024)
-
Association between erythrocyte membrane fatty acids and gut bacteria in obesity-related cognitive dysfunction
AMB Express (2023)
-
A peripheral signature of Alzheimer’s disease featuring microbiota-gut-brain axis markers
Alzheimer's Research & Therapy (2023)
-
Cerebral Small Vessel Disease: a Review of the Pathophysiological Mechanisms
Translational Stroke Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.