Abstract
The fruit fly species, Ceratitis rosa sensu stricto and Ceratitis quilicii, are sibling species restricted to the lowland and highland regions, respectively. Until recently, these sibling species were considered as allopatric populations of C. rosa with distinct bionomics. We used deep Next Generation Sequencing (NGS) technology on intact guts of individuals from the two sibling species to compare their transcriptional profiles and simultaneously understand gut microbiome and host molecular processes and identify distinguishing genetic differences between the two species. Since the genomes of both species had not been published previously, the transcriptomes were assembled de novo into transcripts. Microbe-specific transcript orthologs were separated from the assembly by filtering searches of the transcripts against microbe databases using OrthoMCL. We then used differential expression analysis of host-specific transcripts (i.e. those remaining after the microbe-specific transcripts had been removed) and microbe-specific transcripts from the two-sibling species to identify defining species-specific transcripts that were present in only one fruit fly species or the other, but not in both. In C. quilicii females, bacterial transcripts of Pectobacterium spp., Enterobacterium buttiauxella, Enterobacter cloacae and Klebsiella variicola were upregulated compared to the C. rosa s.s. females. Comparison of expression levels of the host transcripts revealed a heavier investment by C. quilicii (compared with C. rosa s.s.) in: immunity; energy production; cell proliferation; insecticide resistance; reproduction and proliferation; and redox reactions that are usually associated with responses to stress and degradation of fruit metabolites.
Similar content being viewed by others
Introduction
The Natal fruit fly, Ceratitis rosa Karsch, is a polyphagous Afro-tropical pest species with a host range of over 100 wild and cultivated plants1. In Africa, C. rosa is found in southern and eastern Africa2,3, including the Indian Ocean islands of Mauritius and La Réunion, where it is an invasive and devastating quarantine pest4,5,6. Ceratitis rosa occurs in R1 and R2 morphotypes7 which have recently been described as the sibling species C. rosa sensu stricto and Ceratitis quilicii8. In Kenya, C. rosa is confined to coastal areas2,3 while C. quilicii is confined to central highland regions (1,533–1,771 m above sea level)9. The spatial geographic segregation of these sibling species appears to be defined by differential thermal developmental requirements of the two species6,10,11.
Physiologically, the midgut of organisms, including C. rosa sensu lato represents the physiological contact between the organism and its environment; it is possible that microbial (gut microbiome) and host transcript expression profiles of the midgut would be informed by the environments in which these flies exist. Despite our limited knowledge of the gut microbiome of these insects12, available information suggests a wide range of insect-microbe interactions (from symbionts to facultative bacteria) in nature13. While symbionts and their hosts are interdependent, vertically or environmentally-acquired facultative bacteria14 are not essential for host survival, but can influence host fitness and ecological adaptation to the environment15. Amongst fruit flies, the gut microbiome of Ceratitis capitata is largely dominated by free-living Pectobacterium, Enterobacter and Klebsiella species from the Enterobacteriaceae family, that are stable throughout the life cycle of the fly and between geographical regions16,17,18,19,20. Unlike vertically-transmitted endosymbionts, these extracellular bacteria are transmitted horizontally21, and have only been shown recently to have clear impact on the germ line and reproduction via modulation of oogenesis, maternal-to-zygotic-transition in offspring and phenotypic variation in mutants mediated by fly molecular factors22. Many chronic infectious agents have subtle deleterious effects on hosts23. Also, there is potential for host transcriptional profiles to be influenced by the gut microbiome as well as other environmental factors, which can thus define species-specific adaptation to the local environment. The recent delineation of the C. rosa morphotypes into distinct sibling species necessitates further characterisation of their genotypes to identify inherent molecular differences in transcripts of both the gut microbiome and the host that will allow the two morphologically identical species to be distinguished from each other.
In this study, we conducted deep Next Generation Sequencing (NGS) of intact guts from the sibling species, C. rosa s.s and C. quilicii. We isolated host and microbial transcripts from the transcriptome and identified microbe-specific and host-specific transcripts that were differentially expressed in the two species.
Results
De novo assembly and mapping statistics of the transcriptome from C. rosa s.s and C. quilicii transcripts
High quality (83–103 million) reads were obtained with at least 93.46 and 41.49% Q30 and GC contents from C. rosa s.s. and C. quilicii libraries (male and female combined), respectively (Fig. 1). We successfully assembled the reads, de novo, into 36170 transcripts with an overall N100, median and average length of 738, 336 and 552 nucleotides, respectively. Our TransDecoder ORF analysis24 identified 15463 transcripts as the best candidates (with eclipsed ORFs removed) (Fig. 2). Most of the ORFs were internal Coding Domain Sequences (CDS) without isoforms. Complete CDs constituted about 18% of the transcripts (Fig. 2). Approximately 41.0–71.3% of the reads were successfully mapped to our transcripts and at least 43.6% of our read mappings were unique to specific transcripts (Fig. 3).
Nature of open reading frames (ORFs) isolated from the assembly using the TransDecoder program24.
OrthoMCL separation of host and microbiome transcripts
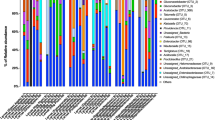
When results from the OrthoMCL analysis25 of the de novo assembled gut transcripts were mapped against the orthoMCL database26 var 5 it was revealed that most of our gut transcripts had fruit fly orthologs with fewer orthologs from bacteria; most orthologs were complete CDS (Fig. 4). On further analysis, 72.3% of the host transcripts were homologous to C. capitata fruit flies (Table 1) while the bacterial transcripts were homologous to 13 species of bacteria (Table 2). RNA-Seq differential analysis amongst the bacterial transcripts from C. rosa s.s and C. quilicii and from male vs. female flies revealed significant upregulation of transcripts associated with eight species of bacteria in female C. quilicii compared with corresponding C. rosa s.s females (Fig. 5). Induction of bacteria-associated transcripts was similar amongst the remaining comparison categories (P > 0.05).
Differential expression of host gut transcripts between male and female C. rosa s.s. and C. quilicii libraries
RNA-Seq differential analysis of host gut-specific transcripts revealed High proportion of significantly induced host transcripts in C. quilicii compared with C. rosa s.s for both genders (Fig. 6). In male C. quilicii (compared with male C. rosa s.s.), GSEA GO term enrichment analysis27,28,29 revealed significant induction of putative pathways associated with: innate immune responses against microbes; apoptosis-related cellular stress responses linked to endoplasmic reticulum (ER); negative regulation of immunity; catalytic transfer of hexose sugar across membranes; and intra–organismal carbohydrate catabolism (Table 3). Further to this, STRING database enriched pathway analysis30 revealed induction of putative pathways associated with: phagocytosis; ER protein processing; glycolysis; folate biosynthesis (crucial for DNA replication and cell division); plant terpenoids; and limonene and pinene degradation (Table 4). Volcano plot analysis of individual gene sequences associated with these observations identified further transcripts that were upregulated in male C. quilicii compared to male C. rosa s.s (Fig. 6) and that the majority of these were primarily associated with: energy production (mitochondrial Acyl-CoA synthetase, glycogen phosphorylase, L-lactate dehydrogenase, glucosidase, NADH dehydrogenase and nuclear valosin); immunity (CD109, protein TsetseEP, proclotting enzyme antigen, tectonin-2, toll, streptogramin A acetyltransferase); cell growth and proliferations (carboxypeptidase D, signal transducer and transcription activator, transposon, selenide, replication polyprotein, BAG domain-containing protein samui, myc, replicase polyprotein, and protein P1); synthesis and transport of protein and other macromolecules (solute carrier organic anion transporter, RNA1, polyprotein, 50 S ribosomal protein, protein translocase, odorant-binding protein, alkaline phosphatase, B-cell receptor-associated protein, nuclear pore complex protein, inorganic phosphate cotransporter, trehalose transporter and protein transport protein); and resistance to insecticides (cytochrome P450). Only transcripts of pathways associated with urine nucleotide biosynthesis (major energy carriers) were more highly expressed in male C. rosa s.s. compared with male C. quilicii (Tables 3 and 4). Volcano plot analysis also revealed that most of the transcripts that were more highly expressed in male C. rosa s.s were of viral (capsid protein alpha, RNA-directed RNA polymerase, threonine-tRNA ligase and microtubule-associated protein) or bacterial (cytidylate kinase) origin. The rest of the transcripts induced in male C. rosa s.s were largely associated with energy metabolism (NADPH, carbamoyl-phosphate synthase, protein melted, fructose-1,6-bisphosphatase).
Volcano plot showing host transcripts that were differentially expressed in the gut tissues of C. rosa s.s and C. quilicii. The transcripts expression level data were supported by at least 100 reads in each category (i.e. in C. rosa s.s or C. quilicii) or five CPM (Counts Per Million) by edge R analysis44,45. The red dots indicate points-of-interest, i.e. the best BLAST hits53 on the manually annotated and reviewed Swiss-Prot database54, that displayed both large magnitude fold-changes (x axis) and high statistical significance (−log10 of p value, y axis). Points with a fold-change less than 2 (log2 = 1) and/ or a False Detection Rate (FDR) corrected p value of less than 0.05 are shown in black, and indicate transcripts that did not change significantly in expression between the two species. Panel A, B, C = the transcripts that had the most, medium and least difference in their expression between the two species.
When female C. rosa s.s and C. quilicii were compared, the pathways induced in C. quilicii females compared with C. rosa s.s females were associated with: yolk formation; maintenance of the cellular redox environment; control of viability and duration of the adult phase of the life cycle; and regulation of notch signaling pathways that affect cell differentiation and responses to insufficient oxygen (hypoxia) (Fig. 6). In addition to the upregulation of the general metabolic pathways, and limonene and pinene degradation pathways that were observed in male C. quilicii, pyrimidine metabolism pathways were also upregulated in female C. quilicii (Table 4). Analysis of the volcano plot of individual gene expression profiles (Fig. 6) of female C. quilicii (compared with female C. rosa s.s.) revealed upregulation of pathways associated with: some common energy production regulators (glucosidase and NADH dehydrogenase); immunity (CD109, protein TsetseEP, proclotting enzyme antigen and streptogramin A acetyltransferase); cell growth and proliferations (replication polyprotein, myc, replicase polyprotein, and protein P1); synthesis and transport of protein and other macromolecules (solute carrier organic anion transporter, protein translocase and inorganic phosphate cotransporter and trehalose transporter); and resistance to insecticide (cytochrome P450). Transcripts that were more highly expressed in female C. rosa s.s (compared with female C. quilicii) included pathways associated with: energy metabolism (glyceraldehyde-3-phosphate and glucosylceramidase dehydrogenase); immunity (peptidoglycan-recognition protein); transcription (eukaryotic translation initiation factor, RNA polymerase and coatomer); proteolysis (trypsin theta, proteolysis); and egg formation/reproduction (chorion protein, outer membrane protein A, vitellogenin and vigilin). Only transcripts for pathways associated with urine nucleotides biosynthesis (major energy carriers) were more highly expressed in male C. rosa s.s compared with male C. quilicii (Table 4). Volcano plot analysis revealed that most of the transcripts that were more highly expressed in male C. rosa s.s. were of viral (capsid protein alpha, RNA-directed RNA polymerase, threonine-tRNA ligase and microtubule-associated protein) or bacterial (cytidylate kinase) origin. The rest were associated with energy metabolism (NADPH, carbamoyl-phosphate synthase, protein melted, fructose-1,6-bisphosphatase). Up-regulation of bacteria/virus-specific (cytidylate kinase and RNA-directed RNA polymerase) transcripts was also common in female C. rosa s.s. The comparison made in this study of expression profiles of 11 randomly selected DE genes from male and female datasets, revealed a Pearson correlation coefficient of R = 0.8146 and R = 0.9338 (Text S1) for the genes evaluated, which is indicative of a valid transcriptome.
Discussion
The goals of this study were (1) to establish the identity of naturally-occurring gut microbes (gut microbiome) associated with the sibling species, C. rosa s.s. and C. quilicii and (2) to use host transcriptional profiles to identify host species-specific transcripts that define the identity of C. rosa s.s. and C. quilicii sibling species. The C. rosa s.s. (ex Kibarani, Msambweni district) and C. quilicii (ex Kithoka, Imenti North district) colonies used in this study were reared on carrot-based artificial diets with no antibiotics. Oviposition was done on apple mango domes and the insects reared in rooms with similar conditions of their places of origin (28 ± 1 °C, 50 ± 8% RH for C. rosa s.s. and 23 ± 1 °C, 65 ± 5% RH for C. quilicii). This was essential so as to minimize the effects of environmental differences on the colonies. Ideally, sample collections were to be of similar generation age, but we realized that the two colonies had different reproductive rates. The fecundity rate of C. quilicii was way higher than that of C. rosa s.s. hence C. quilicii colony was at the 59th generation while the C. rosa s.s. colony was at the 36th generation.
Using OrthoMCL analyses we identified, transcripts of 13 distinct extracellular bacterial species, some of which (Pectobacterium, Enterobacter and Klebsiella species from the family Enterobacteriaceae) have previously been identified as stable free-living bacteria dominating the gut microbiome of C. capitata across its geographical distribution16,17,18,19,20. Others like Enterobacterium buttauxella are known to proliferate in a stable way throughout the life cycle of C. capitata31. Surprisingly, we also detected phytopathogenic Pectobacterium spp. in C. quilicii, which may have colonized and ultimately evolved alternative associations, perhaps involving insect mediation of plant pathogen dissemination32. Another plausible explanation would be it could have originated from the carrot-based diet since the diet was made devoid of antibiotics. Kahala et al.55 demonstrated that carrots infected with this bacterium appeared mainly in shop samples during autumn, winter, and spring, causing spoilage gradually. Others, such as Enterobacter cloacae and Klebsiella variicola have also been reported colonizing the gut of C. capitata and Zeugodacus cucurbitae in nature33,34. The reason for the presence of some bacterial species in C. quilicii, and not in C. rosa s.s, and the reverse scenario for the presence of virus transcripts in C. rosa s.s. and not C. quillicii, is not obvious but may reflect the prevalence of these bacteria/virus species in the original environments of the two species, respectively. It may also relate to the original host plant from which the flies were collected (although they had both been in laboratory culture for > 30 generations), the relative robustness of their immune responses to particular pathogens, or endemic/mutually beneficial host-parasite stability. These hypotheses are possible because they are based on documented evidence for the influence of environmental and host factors on extracellular gut bacteria and a broad range of somatic host functions in D. melanogaster35,36. Furthermore, common chronic, noninvasive associations with particular bacterial lineages are often beneficial, or even required, for the development and reproduction of hosts37. Beneficial effects include developmental interactions that prime the immune system and improve tolerance to thermal stress; these benefits might account for the differences we have observed in C. rosa s.s and C. quilicii which are geographically and thermally divergent.
Our transcriptomic analyses of transcripts revealed a greater investment in physiological processes associated with immunity, energy production, cell proliferation, insecticide resistance and degradation of the fruit metabolites in male C. quilicii compared with male C. rosa s.s. Given that our microbial analysis (above) revealed the presence of various bacteria within the guts of C. quilicii, it is possible that the immune responses observed in C. quilicii might be in response to these bacteria which could, in turn, reflect the relative abundance of these bacteria in the environmental biota, including within host plants. Higher demand for energy and proliferation in C. quilicii than in C. rosa s.s., suggests a higher intrinsic rate of growth and thus fitness. The presence of insecticide resistance genes shows that C. quilicii also has enhanced resistance to insecticides (and probably other xenobiotics) and/or it is under constant selection pressure for insecticide resistance. In contrast, C. rosa s.s harbours viral transcripts that may again reflect the abundance of these viruses in their environmental biota and/or a weaker immunity against viruses. In addition to the differences observed in male flies, comparisons of female C. quilicii and C. rosa s.s also revealed upregulation of pathways and genes essentially associated with reproduction and proliferation, but also with redox which is usually associated with responses to stress. Additionally, the presence of viral transcripts in female C. rosa s.s similar to those in male C. rosa s.s suggests either a potential environmental source for the viruses or their vertical transmission. Despite the discernible differences drawn from this study on the two sibling species in gene expression levels and microbiota of the mid-gut tissues and contents, the study is based on only laboratory colonies. Consequently, colony reared insects can only provide limited, but acceptable insight on phenotype of the flies in their natural habitats that should later be validated downstream. Therefore, the differentially expressed genes can be used as diagnostic markers for the two sibling species. Further studies with wild populations should be undertaken.
Conclusion
In conclusion, this study identified potential natural gut microbes and host transcriptional profiles that could be used to define and differentiate between the sibling species, C. rosa s.s. and C. quilicii. We identified stable, free-living bacterial species (and others) in C. quilicii with potential for stable proliferation throughout the life cycle of the fruit fly. In C. quilicii we also identified greater investment in several physiological processes that distinguished this species from C. rosa s.s. which, in contrast, harboured several viruses that were not present in C. quilicii. Whether the extracellular gut microbiome we identified in C. quilicii are responsible for a potential reduction in reproductive fitness remains to be determined. Recent findings suggest that horizontally transmitted extracellular gut Acetobacter species can influence the D. melanogaster germ line by enhancing oogenesis linked to aldehyde dehydrogenase in the host22.
Material and Methods
Test insects
The C. rosa s.s. colony used in this study was established from individuals collected from guava fruits from Kibarani, Msambweni district (S 04°19'62.8”; E 039°32'41.1”; 34 m a. s. l), a coastal region of Kenya. The flies were reared in Plexiglass cages (65 cm × 35 cm × 65 cm) and the colony maintained at 28 ± 1 °C, 50 ± 8% RH and photoperiod of L12: D12. The C. quilicii colony was established from individuals collected from mango fruits from Kithoka, a highland region in Imenti North district (N 00°05'58.9”; E 037°40'39.5”; 1,425 m a. s. l) of the central region in Kenya. The C. quilicii was reared in Plexiglass cages and the colony maintained at at 23 ± 1 °C, 65 ± 5% RH and photoperiod of L12: D12. Both species were reared in a sterile environment and on carrot-based artificial diets38 with no antibiotic, in the icipe animal rearing and quarantine unit, Nairobi, Kenya. At the time of the bioassay, the C. quilicii colony was at the 59th generation and the C. rosa s.s. colony was at the 36th generation. Noteworthy, the establishment of the two colonies was initiated concurrently, however, the C. rosa s. s. colony took way much longer to be established at the icipe laboratories because the species is adapted to lowland temperate conditions. Because of this, the reproduction rates of C. quilicii was way higher and faster facilitating more generations as compared to its counterpart C. rosa s. s. which was slow.
Extraction of C. rosa s.s. and C. quilicii RNA
Experimental insects were collected two days post emergence and maintained exclusively on water. The entire gut and its contents were dissected from individual male and female C. rosa s.s and C. quilicii in 1X phosphate buffered saline (PBS) (pH 8.0) under a Leica (EZ4HD) stereoscope (n = 100 for each sex of each species); the guts for each sex/ species combination were then pooled and stored in liquid nitrogen until required for downstream analyses. Total RNA was extracted using the Isolate II RNA Mini Kit (Bioline, London, UK), following the manufacturer’s instructions, and the resulting RNA immediately stored at −80 °C. RNA yield was determined using a Nanodrop 2000/2000c Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and the size and preliminary integrity of the RNA determined by gel electrophoresis on 1.2% non-denaturing agarose followed by ethidium bromide staining. Visualization and documentation of the gel image was done using a KETA GL imaging system (Wealtec Corp, Meadowvale Way Sparks, Nevada, USA). The RNA (50 uL) was subsequently lyophilized and preserved in RNAstable tubes (Biomatrica, Inc, USA) as per the manufacturer’s instructions until the RNA was required for NGS.
RNA sequencing of the C. rosa s.s. and C. quilicii transcriptomes
The integrity of RNA from each sample was assessed individually using a Bioanalyzer (Agilent Technologies, USA), and cDNA was generated using an Illumina TruSeq RNA Sample Preparation Kit (Illumina, Hayward, CA, USA). This technique is based on rRNA depletion and not polyA + selection which means that both prokaryotic and eukaryotic transcripts are retained in the libraries. Forward-Reverse (FR) strand-specific libraries were prepared from each sample and were sequenced by Macrogen Inc. (Seoul, South Korea) using an Illumina HiSeq. 2000 (Illumina, Hayward, CA, USA) platform with two 101 nt paired-end reads. Low quality reads, reads with less than 101 base pairs, and adapter sequences were removed using Illumina build software (Illumina, Hayward, CA, USA) as part of the sequence clean-up. This generated final fastq formatted raw data for each library. Overall, eight fastq files were generated, a pair each for each sibling species and sex (i.e. (C. rosa s.s and C. quilicii, male or female) × 2).
Identification and validation of transcripts that were differentially expressed (DE) in C. rosa s.s. and C. quilicii
The quality of the RNA sequence reads in each file was assessed using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and the high quality data used to clean reads using fastx quality trimmer (http://hannonlab.cshl.edu/fastx_toolkit/) software. Since both the C. rosa s.s. and C. quilicii reference genomes were not available (unpublished), we could not employ established genome-guided and gene-set based strategies to evaluate the expression profiles of the transcripts. We thus employed a de novo transcript assembly-based strategy to generate the transcripts from the transcriptomes. We then separated them into microbial and host transcripts and then identified which transcripts (microbial or host) were differentially expressed in the different libraries. Finally, we established which metabolic pathways were predominant and functionally associated with the differential transcript expression profiles using geneset enrichment analysis. Briefly, all the transcriptomes were combined, assembled de novo into transcripts and the quality of the assembled transcripts assessed using the short reads Trinity assembly program24,39. We isolated the longest transcripts (most representative of the respective genes) with open reading frames that could be translated into peptides that were at least 100 amino acids long using the TransDecoder program24. To separate the microbial transcripts from the host transcripts amongst these long transcript sets, we mapped the transcripts using the OrthoMCL25 ortholog against the orthologs (OrthoMCL) database var 526. All sources of known transcripts from available microbe databases were identified using OrthoMCL. This separated the transcripts into those that represented microbial species and those that were host transcripts based on differences in their codon usage bias. The database contained 810, 686 ortholog groups clustered from 1, 398, 546 proteins obtained from 150 genomes. The genomes consisted of four Amoebozoan, six Firmicutes, nine Euglenozoan, 11 Viridiplantae, 15 Alveolates, 16 Archaea, 19 Proteobacteria, 24 fungi and 29 Metazoan taxonomic groups, in addition to six and 11 other eukaryote and bacteria taxonomic groups respectively27. Furthermore, the identification of orthologous groups in prokaryotic genomes has permitted cross-referencing of genes from multiple species, facilitating genome annotation, protein family classification and studies on bacterial evolution40,41,42,43. The transcripts were then queried against the BLAST non-redundant database which served to; (i) confirm the microbe ID’s and (ii) identified putative ID’s of the novel transcripts that could not be identified in the microbial database. To identify transcriptional expression profiles for both the microbial transcripts and the host transcripts that were differentially expressed between the libraries (sibling species), the cleaned reads from individual transcriptomes in the microbial or host transcript sets were individually mapped using CLC genomic workbench version 9.5 (CLC Bio, Aarhus, Denmark). The resultant mapping read counts for each library on each transcript set were used to determine differential expression of the respective microbial or host (male or female) transcripts using edgeR analysis44,45 software. To minimize detection of false positives for differential expression, a conservative regime was adopted against both libraries. Within this regime, we considered transcripts to be differentially expressed (DE) between treatments if: 1) they had at least a two-fold difference in expression level; 2) the false detection rate (FDR) was corrected p < 0.05; 3) they were supported by at least 100 read mappings in either categories (i.e. in C. rosa s.s, C. quilicii or microbial transcripts); and 4) they had five CPM (Counts Per Million). Fold changes as a ratio of CPM values between microbial or host transcripts from the C. rosa s.s and C. quilicii libraries were defined. Moreover, the metabolic pathways and networks that underwent differential functional enrichment (expression) between the two hosts (C. rosa s.s vs. C. quilicii) or between genders were identified using secondary canonical gene set enrichment analysis (GSEA) approaches within the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt)46.
Validation of transcriptome expression profiles using qRT-PCR
Since our data were derived from a single transcriptome for each treatment (i.e. species and gender), we employed a quantitative reverse transcription Polymerase Chain Reaction (qRT-PCR) technique as an independent tool to determine whether our RNA-Seq analysis results could potentially be independently replicated; we compared fold changes in randomly selected DE host transcripts that were achieved using the two approaches. This strategy has been employed successfully for validation of RNA-seq expression levels from single transcriptomes in other studies47,48,49,50. We did the validation using eight independent biological replicates (n = 8 for each of the different species and gender combinations) obtained from dissected flies generated under similar experimental conditions to those described for the transcriptome samples. Briefly, we prepared cDNAs (from 1 µg Total RNA) from the midgut of each replicate using High Capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s protocol. Quantitative RT-PCR was done on each replicate and specifically on 11 randomly selected DE genes using gene-specific primers (as detailed in Supplementary Information S1). The reaction mix consisted of 3 µg cDNA template that was amplified from each biological replicate; there were three independent technical replicates each with 7.5 µL of Fast SYBR® Green master mix (Applied Biosystems, Carlsbad, CA) and 0.5 picomoles of each of the specific primers for the various genes of interest. Reactions were made in the Strategene MX3005P real time qPCR machine (Agilent Technologies, California, USA). All qRT-PCR results were normalized to two genes that were expressed by Bestkeeper51 in the two sibling species, which were also quantified from each biological replicate. REST software52 was used for pairwise gene expression analysis, including an internal multiple-tests correction. Fold changes in transcript expression levels were established by comparing levels of expression of the transcripts in the midguts of C. rosa s.s with those in the midguts of C. quilicii. We conducted Pearson correlation analysis of the fold changes that were obtained from qRT-PCR with those that had been obtained previously from the RNA-seq data to determine the validity of our transcriptomes. Separate validation of the microbial transcriptomes was not performed since they were inherently intertwined with the host transcriptomes and generally had lower expression levels which would be technically difficult to detect using qRT-PCR.
Ethics approval
All insect rearing, handling and experiments were performed using standard operating procedures at the ICIPE Animal Rearing and Quarantine Unit as approved by the National Commission of Science, Technology and Innovations, Kenya.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author upon request and will also be made available through open source platforms.
References
De Meyer, M. et al. Annotated check list of host plants for Afrotropical fruit flies (Diptera: Tephritidae) of the genus Ceratitis. Zoologische Documentatie Koninklijk Museum voor Midden Afrika 27, 1–92 (2002).
De Meyer, M. On the identity of the natal fruit fly Ceratitis rosa Karsch (Diptera, Tephritidae). Bull. Inst. R. Sci. Nat. Belg. Entomol 71, 55–62 (2001).
De Meyer, M., Copeland, R. S., Wharton, R. A. & McPheron, B. A. On the geographical origin of the medfly, Ceratitis capitata (Wiedemann) (Diptera: Tephritidae), In: Barnes B. (Ed.), Proc. 6th Int. Fruit Fly Symp. 45–53 (2002).
De Meyer, M. Distribution patterns and host-plant relationships within the genus Ceratitis MacLeay (Diptera: Tephritidae) in Africa. Cimbebasia 17, 219–228 (2001).
Orian, A. J. E. & Moutia, L. A. Fruit flies (Trypetidae) of economic importance in Mauritius. Rev. Agric. et Suc. de l’Ile Maurice 39, 142–150 (1960).
Duyck, P. F. & Quilici, S. Survival and development of different life stages of three Ceratitis spp. (Diptera: Tephritidae) reared at five constant temperatures. Bull. Entomol. Res. 92, 461–469 (2002).
Virgilio, M., Delatte, H., Quilici, S., Backeljau, T. & De Meyer, M. Cryptic diversity and gene flow among three African agricultural pests: Ceratitis rosa, Ceratitis fasciventris and Ceratitis anonae (Diptera, Tephritidae). Mol. Ecol. 22, 2526–2539, https://doi.org/10.1111/mec.12278 (2013).
De Meyer, M., Mwatawala, M., Copeland, R. S. & Virgilio, M. Description of new Ceratitis species (Diptera: Tephritidae) from Africa, or how morphological and DNA data are complementary in discovering unknown species and matching sexes. Eur. J. Taxon. 233, 1–23, https://doi.org/10.5852/ejt.2016.233 (2016).
Copeland, R. S. & Wharton, R. A. Year-round production of pest Ceratitis species (Diptera: Tephritidae) in fruit of the invasive species Solanum mauritianum in Kenya. Entomol. Soc. America 99, 530–535, 10.1603/0013-8746(2006)99[530:YPOPCS]2.0.CO;2 (2006).
Grout, T. G. & Stoltz, K. C. Developmental rates at constant temperatures of three economically important Ceratitis spp. (Diptera: Tephritidae) from southern Africa. Environ. Entomol. 36, 1310–1317 (2007).
Tanga, C. M. et al. Comparative analysis of development and survival of two Natal fruit fly Ceratitis rosa Karsch (Diptera, Tephritidae) populations from Kenya and South Africa. ZooKeys. 540, 467–487, https://doi.org/10.3897/zookeys.540.9906 (2015).
Weinert, L. A., Tinsley, M. C., Temperley, M. & Jiggins, F. M. Are we underestimating the diversity and incidence of insect bacterial symbionts? A case study in ladybird beetles. Biology Letters 3, 678–681, https://doi.org/10.1098/rsbl.2007.0373 (2007).
Baumann, P., Moran, N. A. & Baumann, L. In The Prokaryotes: Volume 1: Symbiotic Associations, Biotechnology, Applied Microbiology (eds Martin Dworkin et al.) 403-438 (Springer New York, 2006).
Kikuchi, Y. et al. Symbiont-mediated insecticide resistance. PNAS 109, 8618–8622, https://doi.org/10.1073/pnas.1200231109 (2012).
Ben-Yosef, M., Jurkevitch, E. & Yuval, B. Effect of bacteria on nutritional status and reproductive success of the Mediterranean fruit fly Ceratitis capitata. Physiol. Entomol. 33, 145–154, https://doi.org/10.1111/j.1365-3032.2008.00617.x (2008).
Behar, A., Yuval, B. & Jurkevitch, E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol. Ecol. 14, 2637–2643, https://doi.org/10.1111/j.1365-294X.2005.02615.x (2005).
Behar, A., Jurkevitch, E. & Yuval, B. Bringing back the fruit into fruit fly-bacteria interactions. Mol. Ecol. 17, 1375–1386, https://doi.org/10.1111/j.1365-294X.2008.03674.x (2008).
Marchini, D., Rosetto, M., Dallai, R. & Marri, L. Bacteria associated with the oesophageal bulb of the medfly Ceratitis capitata (Diptera:Tephritidae). Curr. Microbiol. 44, 120–124 (2002).
Lauzon, C. R., McCombs, S. D., Potter, S. E. & Peabody, N. C. Establishment and vertical passage of Enterobacter (Pantoea) agglomerans and Klebsiella pneumoniae through all life stages of the Mediterranean fruit fly (Diptera: Tephritidae). Anns. Entomol. Soc. Africa 102, 85–95, https://doi.org/10.1603/008.102.0109 (2009).
Behar, A., Yuval, B. & Jurkevitch, E. Community structure of the Mediterranean fruit fly microbiota: seasonal and spatial sources of variation. Israel J. Ecol. & Evol. 54, 181–191, https://doi.org/10.1080/15659801.2008.10639612 (2013).
Ben-Yosef, M., Aharon, Y., Jurkevitch, E. & Yuval, B. Give us the tools and we will do the job: symbiotic bacteria affect olive fly fitness in a diet-dependent fashion. Proc. Biol. Sci. 277, 1545–1552 (2010). 10.1098/rspb.2009.2102.
Elgart, M. et al. Impact of gut microbiota on the fly’s germ line. Nat. Commun. 7, 11280, https://doi.org/10.1038/ncomms11280 (2016).
Mateos, M. et al. Heritable endosymbionts of Drosophila. Genetics 174, 363–376, https://doi.org/10.1534/genetics.106.058818 (2006).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512, https://doi.org/10.1038/nprot.2013.084 (2013).
Li, L., Stoeckert, C. J. Jr. & Roos, D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189, https://doi.org/10.1101/gr.1224503 (2003).
Chen, F., Mackey, A. J., Stoeckert, C. J. Jr. & Roos, D. S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–368, https://doi.org/10.1093/nar/gkj123 (2006).
Conesa, A. & Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832, https://doi.org/10.1155/2008/619832 (2008).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676, https://doi.org/10.1093/bioinformatics/bti610 (2005).
Gotz, S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435, https://doi.org/10.1093/nar/gkn176 (2008).
Franceschini, A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–815, https://doi.org/10.1093/nar/gks1094 (2013).
Aharon, Y. et al. Phylogenetic, metabolic, and taxonomic diversities shape mediterranean fruit fly microbiotas during ontogeny. Appl. & Environ. Microbiol. 79, 303–313, https://doi.org/10.1128/aem.02761-12 (2013).
Nadarasah, G. & Stavrinides, J. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol. Rev. 35, 555–575, https://doi.org/10.1111/j.1574-6976.2011.00264.x (2011).
Cox, C. R. & Gilmore, M. S. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Inf. & Imm. 75, 1565–1576, https://doi.org/10.1128/iai.01496-06 (2007).
Hadapad, A. B., Prabhakar, C. S., Chandekar, S. C., Tripathi, J. & Hire, R. S. Diversity of bacterial communities in the midgut of Bactrocera cucurbitae (Diptera: Tephritidae) populations and their potential use as attractants. Pest Man. Sci. 72, 1222–1230, https://doi.org/10.1002/ps.4102 (2016).
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A. & Kopp, A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genetics 7, e1002272, https://doi.org/10.1371/journal.pgen.1002272 (2011).
Buchon, N., Broderick, N. A., Chakrabarti, S. & Lemaitre, B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & Develop. 23, 2333–2344, https://doi.org/10.1101/gad.1827009 (2009).
Brummel, T., Ching, A., Seroude, L., Simon, A. F. & Benzer, S. Drosophila lifespan enhancement by exogenous bacteria. PNAS 101, 12974–12979, https://doi.org/10.1073/pnas.0405207101 (2004).
Ekesi, S., Mohamed, S. A. & Chang, C. L. A liquid larval diet for rearing Bactrocera invadens and Ceratitis fasciventris (Diptera: Tephritidae). Int. J. Trop. Ins. Sci. 34, S90–S98, https://doi.org/10.1017/S1742758414000113 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnol. 29, 644–652, https://doi.org/10.1038/nbt.1883 (2011).
Galperin, M. Y. & Koonin, E. V. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 10, 571–578 (1999).
Forterre, P. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18, 236–237 (2002).
Natale, D. A., Galperin, M. Y., Tatusov, R. L. & Koonin, E. V. Using the COG database to improve gene recognition in complete genomes. Genetica 108, 9–17 (2000a).
Tatusov, R. L., Koonin, E. V. & Lipman, D. J. A genomic perspective on protein families. Science 278, 631–637 (1997).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, https://doi.org/10.1093/bioinformatics/btp616 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297, https://doi.org/10.1093/nar/gks042 (2012).
Wang, J., Duncan, D., Shi, Z. & Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–83, https://doi.org/10.1093/nar/gkt439 (2013).
Benoit, J. B. et al. A novel highly divergent protein family identified from a viviparous insect by RNA-seq analysis: a potential target for tsetse fly-specific abortifacients. PLoS Genetics 10, e1003874, https://doi.org/10.1371/journal.pgen.1003874 (2014).
Attardo, G. M. et al. The homeodomain protein ladybird late regulates synthesis of milk proteins during pregnancy in the tsetse fly (Glossina morsitans). PLoS neglected tropical diseases 8, e2645, https://doi.org/10.1371/journal.pntd.0002645 (2014).
Telleria, E. L. et al. Insights into the trypanosome-host interactions revealed through transcriptomic analysis of parasitized tsetse fly salivary glands. PLoS Neglected Trop. Dis. 8, e2649, https://doi.org/10.1371/journal.pntd.0002649 (2014).
Bateta, R. W. et al. Tsetse fly (Glossina pallidipes) midgut responses to Trypanosoma brucei challenge. Parasites & Vectors (2017 In Press).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Letters 26, 509–515 (2004).
Pfaffl, M. W., Horgan, G. W. & Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36 (2002).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410, https://doi.org/10.1016/s0022-2836(05)80360-2 (1990).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Kahala, M. et al. Molecular characterization of spoilage bacteria as a means to observe the microbiological quality of carrot. Journal of Food Protection 75(3), 523–532, https://doi.org/10.4315/0362-028X.JFP-11-185 (2012).
Acknowledgements
The authors are grateful to Mr John Kiilu for rearing the C. rosa s.s and C. quilicii colonies. We are also grateful to Ms Maureen Adhiambo for her technical assistance and to Mr Brian Mwashi for editing the images. This work was supported by grants from the Integrated Biological Control Applied Research Programme-Fruit Fly Component, DCI-FOOD/2014/346–739. Additional support included ICIPE core funding and funding from UK Aid (UK Government), the Swedish International Development Cooperation Agency, the Swiss Agency for Development and Cooperation, and the Kenyan Government.
Author information
Authors and Affiliations
Contributions
F.M.K., S.E., S.M. and A.R.M. conceived the study. F.M.K., S.M., C.M.T. and S.E. provided the resources necessary for the assays to be performed. F.M.K. and S.E. designed the experiments. F.L.O.O. optimized and performed all the assays. P.O.M. did the transcriptome assembly and bioinformatics analysis. E.A.O. did the qPCR data validation. F.M.K., P.O.M., F.L.O.O., E.A.O. and M.R. contributed to data analysis and interpretation. F.M.K. and C.M.T. supervised the experiments. S.M., A.R.M. and C.M.T. participated in discussions about the results. F.M.K., P.O.M. and F.L.O.O. drafted the manuscript. All the co-authors read and revised the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khamis, F.M., Mireji, P.O., Ombura, F.L.O. et al. Species-specific transcriptional profiles of the gut and gut microbiome of Ceratitis quilicii and Ceratitis rosa sensu stricto. Sci Rep 9, 18355 (2019). https://doi.org/10.1038/s41598-019-54989-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54989-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.