Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) are a major cause of healthcare and community- associated infections due to their ability to express a variety of virulence factors. We investigated 209 MRSA isolates obtained from 1 January to 31 December 2016 using a combination of phenotypic and genotypic methods to understand the genetic backgrounds of MRSA strains obtained in a General hospital in Kuwait. Antibiotics susceptibility was performed with disk diffusion, and MIC was measured with Etest strips. Molecular typing was performed using SCCmec typing, spa typing, and DNA microarray for antibiotic resistance and virulence genes. The isolates were susceptible to vancomycin, teicoplanin, rifampicin, ceftaroline, and linezolid but were resistant to gentamicin, tetracycline, erythromycin, fusidic acid, chloramphenicol and ciprofloxacin. Molecular typing revealed six SCCmec types, 56 spa types and 16 clonal complexes (CC). The common SCCmec types were type IV (39.5%), type III (34.4%), type V (25.8%) and type VI (3.8%). The dominant spa types were t860 (23.9%), t945 (8.6%), t127 (6.7%), t688 (6.7%), t304 (6.2) and t044 (5.7%). The other spa types occurred sporadically. Genes for PVL was detected in 59 (28.2%) of the isolates. CC8-ST239-MRSA-III + SCCmer (23.3%) was the most prevalent clone, followed by CC6-MRSA-IV (8.3%), CC80-MRSA-IV [PVL+] (5.8%), CC5-MRSA-VI + SCCfus (5.0%), CC30-MRSA-IV[PVL+] (4.1%), CC1-MRSA-V + SCCfus [PVL+] (4.1%), CC5-MRSA-V + SCCfus (4.1%) and CC22-MRSA-IV[PVL+] (4.1%). The study revealed that despite the emergence of MRSA with diverse genetic backgrounds over the years, ST239-MRSA-III remained the dominant clone in the hospital. This warrants reassessment of infection prevention and control procedures at this hospital.
Similar content being viewed by others
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an opportunistic pathogen that can cause mild to invasive, life-threatening infections1, MRSA was first reported in the United Kingdom, soon after the use of methicillin in the healthcare system in the 1960’s2, but have since been reported in many countries3,4,5. Methicillin-resistance is mediated by mecA gene which encodes penicillin-binding protein 2a (PBP 2a) that results in resistance to beta-lactam antibiotics such as methicillin, cloxacillin and oxacillin5,6. The mecA gene is located on a mobile genomic island known as staphylococcal cassette chromosome mec (SCCmec) and is inserted in the chromosome of methicillin-resistant staphylococci5. The SCCmec genetic element differ in size and structural organisation, and on the basis of their size and structural differences, 13 SCCmec types designated types I-XIII have been reported6,7.
MRSA isolates have been characterised as either hospital or healthcare- hospital-associated MRSA (HA-MRSA) or community-associated (CA-MRSA) on the basis of their SCCmec types. HA-MRSA harbour SCCmec types I, II and III while the CA-MRSA isolates carry SCCmec types IV, V and VI6,8. SCCmec typing9,10, and other molecular typing methods including pulsed-field gel electrophoresis, multilocus sequence typing, Staphylococcal protein A (Spa) typing, DNA microarray and whole genome sequencing5,6,11,12,13, have been employed to investigate the clonal distributions of MRSA strains in different countries, and have revealed a diversity in their genetic backgrounds in the different geographical locations.
The epidemiology of MRSA strains is constantly changing and their prevalence and characteristics are known to vary between hospitals in the same country or wards in the same hospital3,14,15. Consequently, it is important to study MRSA isolates from local healthcare facilities with unique patient populations to obtain data that can aid empirical treatment of infections and to compare them with those obtained in other healthcare facilities in the country. The Farwaniya Hospital, Kuwait is a university-affiliated hospital with 1,200 beds, which provide services that include medical, surgical, orthopaedic, obstetric and gynaecologic, paediatrics, and two intensive care units16,17. A previous study conducted on MRSA obtained at the Farwaniya hospital from 1996 to 2001 using pulsed-field gel electrophoresis, and coagulase gene typing revealed the presence of a dominant and persistent multiresistant MRSA clone16, that was later identified as ST239-MRSA-III clone18. In addition, the majority of MRSA isolates acquired by patients admitted to the Intensive Care unit of the hospital in 2005–2007 were ST239-MRSA-III19. confirming the persistence of ST239-MRSA-III in the hospital. Recent studies on MRSA obtained in other hospitals in Kuwait have revealed the presence of diverse CA-MRSA clones20,21. This study was conducted on MRSA isolates obtained from patients admitted to the Farwaniya hospital in 2016 to determine their antibiotic resistance patterns and clonal distribution and compare the results to those obtained in 1996–2001 for the purpose of assisting in patients’ management and control of infections, and to compare the MRSA clones to those in other Kuwait hospitals.
Results
The 209 isolates were obtained from 209 patients comprising 143 males (68.4%) and 66 females (31.6%) located at the male medical wards (N = 60), male surgical wards (N = 31), female medical wards (N = 4), female surgical wards (N = 4), Obstetrics & gynaecology ward (N = 24), Intensive care units (N = 21), CCU (N = 6), Paediatrics ward (N = 9), Paediatric intensive care unit (6), Casualty (N = 6), Orthopaedics (N = 5), Urology (N = 6), ENT (N = 6) and Out Patient department (N = 13). The location for three patients was not specified.
All of the 209 MRSA isolates were susceptible to rifampicin, linezolid and vancomycin (MIC ≤ 2 µg/ml). The distribution of MIC values for vancomycin and teicoplanin are summarized on Table 1 and the MIC values for each isolates is presented in a Supplementary Table 1. In total, 202 (96.8%) were sensitive to teicoplanin with MIC ≤ 2 µg/ml, six (3.4%) isolates had MIC: 3 µg/ml and one isolate had MIC:4. µg/ml. Most of the isolates were susceptible to vancomycin and teicoplanin with MIC of 1.5 µg/ml.
The isolates were resistant to fusidic acid (N = 133; 63.4%), gentamicin (N = 114; 54.5%), tetracycline (N = 114; 54.5%), erythromycin and clindamycin (N = 109; 52.2%), ciprofloxacin (N = 104; 49.8%) and trimethoprim (N = 45; 21.5%), Thirty (14.4%) and 79 MRSA isolates expressed inducible and constitutive resistance to clindamycin, respectively. Nineteen (9.0%) and four (1.9%) isolates were resistant to chloramphenicol and high-level mupirocin respectively. In total, 164 (78.5%) of the isolates expressed multiresistance (resistance to three or more classes of antibiotics) while 45 (21.5%) isolates were non-multiresistant (resistant to less than three classes of antibiotics (Table 2). Most of the isolates were resistant to six (28.7%), classes of antibiotics. This was followed by isolates that were resistant to five, four and three classes of antibiotics. Nineteen (9.0%) were resistant only to beta-lactam antibiotics (Table 2).
Molecular characterization of MRSA in isolates
SCCmec typing revealed the presence of six SCCmec types with the majority belonging to SCCmec type IV (N = 75; 39.5%) followed by SCCmec types III (N = 72; 34.4%), SCCmec type V (N = 52; 25.8%), SCCmec type V1 (N = 5; 2.4%), SCCmec type I (N = 2; 1%) and SCCmec type II (N = 1;0.5%).
Spa typing of the 209 isolates revealed 56 spa types. Spa type t860 was the dominant spa type and was detected in 50 (23.9%) of the isolates. This was followed by t945 (N = 18; 8.6%), t127 (N = 14; 6.7%), t688 (N = 14; 6.7%), t304 (N = 13; 6.2%) and t044 (12; 5.7%). The spa types that occurred sporadically were t003, t002, t005, t008, t018, t019, t021, t032, t037, t042, t045, t148, t105, t1120, t11836, t14700, t1247, t12398, t16185, t1839, t16202, t148, t306, t314, t355, t359, t362, t363, t376, t3841, t425, t535, t5414, t657, t6845, t690, t701, t713, t7583, t790, t8154, t8168, t852, and t084. The spa gene could not be amplified in three isolates.
DNA microarray analysis of MRSA isolates
A total of 120 MRSA isolates representing the 56 spa types were selected for DNA microarray analysis. The selection was based on their spa types, and clinical samples. Care was taken to include different spa types from all clinical samples. The results yielded 16 clonal complexes (CCs) and one singleton. The distribution of MRSA clonal complexes and their genotypes are shown in Supplementary Table 2). The most prevalent CC was CC8/ST239 (N = 32; 26.6%) followed by CC5 (21; 17.5%), CC1 (11; 9.1%), CC6 (11; 9.1%), CC22 (11; 9.1%), CC80 (8; 6.6%) and CC30 (7; 5.8%), CC97 (N = 4; 3.3%), CC15 (N = 3; 2.5%), CC152 (N = 2; 1.6%), CC88 (N = 2; 1.6%), CC361 (N = 2; 1.6%), CC2250/2277 (N = 1;0.8%), CC45 (N = 1; 0.8%), CC121 (N = 1; 0.8%), CC59 (N = 1;0.8%) and a singleton ST2867.
Thirty of the 32 CC8 isolates belonged to ST239-MRSA-III + SCCmer, (N = 28), ST239-MRSA-III + ccrC (N = 1) and ST239-MRSA-III + SCCmer + ccrC (N = 1) clone making ST239-MRSA-III + SCCmer the commonest genotype. The ST239-MRSA-III isolates belonged to six spa types consisting of t860 (N = 16), t945 N = 10,) and one each of t037, t713, t425 and t1247. The other common genotypes were CC6-MRSA-IV, WA MRSA-51 (N = 10), CC80-MRSA-IV[PVL+] European CA-MRSA clone (N = 7), CC5-MRSA-VI + SCCfus (N = 6), CC5-MRSA-V + SCCfus, WA MRSA-14/109 (N = 5), CC1-MRSAV + SCCfus [PVL+] (N = 5), CC22-MRSA-IV[PVL+] (N = 5) and CC30-MRSA-IV[PVL+] (N = 5). The remaining genotypes occurred in one to four isolates.
Distribution of MRSA isolates among clinical sources
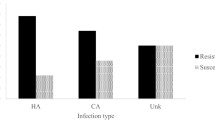
We examined MRSA isolates analysed by DNA microarray according to their clinical sources to establish any specific association between clones and clinical sources and infection types. The most common clinical samples that yielded MRSA isolates were wound (N = 52), blood (N = 26), pus (N = 22), and the respiratory samples, sputum (N = 5) and tracheal aspirates (N = 13). The other specimens yielded fewer MRSA isolates. The well-known HA-MRSA clone, ST239-MRSA-III, was the dominant isolate in axilla, groin, and was the major isolate in nasal and wound samples but was isolated sporadically from blood, skin and respiratory samples (sputum and tracheal aspirates). In contrast, blood, wound and respiratory samples yielded higher proportions of diverse clones carrying CA-MRSA genotypes (that is carrying SCCmec IV, V, VI) with none of the clones dominating in the clinical samples.
Antibiotic resistance genotypes
All isolates were resistant to benzyl penicillin and harboured blaZ and mecA. All of the gentamicin- resistant isolates were positive for aacA-aphD that encodes resistance to gentamicin, kanamycin, amikacin and tobramycin, and aphA3 which mediates resistance to kanamycin and neomycin. The aphA3 was also detected in two isolates of CC361 (n = 1) and CC59 (N = 1) that were resistant to kanamycin but susceptible to gentamicin. Fifty isolates, belonging to CC8-ST239 (n = 29), CC6 (N = 10), CC22 (7), CC15 (N = 3), and CC5 (N = 1), harboured aadD which encodes resistance to kanamycin, neomycin and tobramycin.
The fusidic acid-resistant isolates were positive for fusC (N = 26) or fusB (N = 10). While fusB was only detected in isolates of CC80 (N = 7) and CC5 (N = 3), fusC was detected in isolates with diverse genetic backgrounds.
Erythromycin resistance was associated with different resistance determinants including ermA (rRNA methyltransferase A), ermC (rRNA methyltransferase C), linA (lincosaminide nucleotidyltransferase), msr(A) (macrolide efflux pump) and mph(C) (macrolide phosphotransferase II). ermA was the most prevalent erythromycin resistance determinant. It was detected in 28 ST239 and one CC5 isolates. This was followed by ermC which was detected in 26 isolates of diverse genetic backgrounds. ermB was detected in a single CC59 isolate. Both mph(C) and msr(A) occurred together in eight isolates of CC1-ST772-MRSA- (N = 1), CC1-MRSA-V + SCCfus (N = 2), CC8-MRSA-IV (N = 1), CC5-MRSA-IV-Paediatric clone (N = 1), CC30-MRSA-IV[PVL+] (N = 2) and CC361-MRSA-IV (N = 1). linA was detected in three CC15 and one ST239 isolates. A single CC97 isolate was positive for vga(A), an ABC transporter that confers resistance to streptogramins.
Tetracycline resistance was detected in 55 of the 120 isolates analysed by microarray. The tetracycline resistance determinants, tetK and tetM, were detected in 50 of the 55 isolates respectively. Five CC1-MRSA-V+ SCCfus [PVL+] isolates were negative for both tetK and tetM although they were phenotypically tetracycline-resistant. tetM was detected in 37 isolates consisting of 30 ST239, six CC5-MRSA-VI + SCCfus and a CC5-MRSA-VI-paediatric clone. tetK was detected in 17 isolates with different genetic backgrounds. Both tetK and tetM occurred together in two ST239 isolates.
The 13 chloramphenicol-resistant isolates harboured cat (chloramphenicol acetyl transferase) (N = 7) or fexA (chloramphenicol/florfenicol exporter) which confers resistance to chloramphenicol and florfenicol (N = 6). The fexA was detected in t688 isolates belonging to CC5.
All trimethoprim – resistant isolates were positive for dfrS1 (dihydrofolate reductase gene). Similarly, all high-level mupirocin-resistant isolates were positive for mupA (isoleucyl-tRNA synthetase) that mediates high-level mupirocin resistance.
Microarray also detected resistance determinants to antibiotics that were not tested phenotypically. These included fosB that is associated with fosfomycin resistance, sat (streptothrin acetyltransferase) that is associated with strepthtothrin resistance, Sdrm, a putative transport protein whose function is unknown. The qacA and qacC (multidrug efflux proteins) that mediate resistance to quaternary ammonium compounds. fosB was detected in 73 of the 120 isolates making it one of the common resistance determinants in a variety of genetic backgrounds.
Detection of virulence determinants
Microarray analysis revealed the presence of different virulence determinants including regulatory genes (accessory gene regulators (agr), toxins including, Panton-Valentine leucocidin (PVL), haemolysins, leucocidins, Staphylococcal enterotoxins (SE), toxic shock syndrome toxins TSST-1), epidermal cell differentiation inhibitors, capsular polysaccharides (cap), biofilm associated factors (icaA/ C/D) and immune evasion cluster. All isolates were positive for haemolysins genes, hl, hla, hlb, hllll and hld, biofilm-encoding genes, icaA/C/D and clumping factor A (clfA) and clumping factor B (clfB). The isolates belonged to four accessory gene regulator (agr) types; agr I to agr IV. The leading agrI carrying isolates (N = 64) belonged to CC6, CC8, CC22, CC45, CC59, CC97, CC152 and CC361. The agr II isolates belonged to CC5, CC15, ST772 and ST2867, while agr III isolates were of CC1, CC30, CC80 and CC88. A single CC121 isolate carried agr IV. CC1 isolates, except ST772-MRSA-V, CC6, ST239, CC15, CC30, CC45, CC59, CC80, CC88, CC121 and CC361 were positive for capsular polysaccharide type 8 gene, cap8.The rest of the isolates carried cap5.
Genes for PVL was detected in 37 (30.8%) of the 120 isolates that belonged to CC1 (N = 9), CC5 (N = 4), CC22 (N = 5), CC30 (N = 7), CC59 (N = 1), CC80 (N = 7), CC88 (N = 1), and CC152 (N = 2).
One hundred and three (85.8%) of the 120 isolates harboured at least one Staphylococcal enterotoxins (SE) gene. The most prevalent SE genes were egc (N = 46), sea (N = 45), seg (N = 45), sei (N = 43), sek (N = 39), seq (N = 39), seb (N = 32), sej (N = 15), sed (N = 14) and seh (N = 8). Both sec and sel were detected in four isolates each. Most of the 103 isolates harboured two or more SE genes. Only five isolates carried single SE genes. Thirty-eight isolates carried two SE genes, 26 isolates carried three SE genes with the commonest combination being seg, sei and egc found in 18 of the isolates. Four SE genes were detected in 15 isolates and 19 isolates carried five or six SE genes. Four isolates consisting of three ST772-MRSA-V and one CC5-MRSA-V + PVL harboured six SE genes (sea, sed, seg, sei, sej and egc). Eighteen isolates belonging to CC15 (N = 3) and CC97 (N = 4), CC80 (N = 7), CC152 (N = 1), CC2250/2277 (N = 1) and ST2867 (N = 2) did not harbour SE genes. The genes for toxic shock syndrome toxin, tst, was detected in six isolates belonging to CC22-MRSA-IV[tst + ] (N = 3), CC30-MRSA-[VI + fus] +PVL (N = 2) and CC5-MRSA-IV[Paediatric clone] (N = 1). Eight isolates belonging to CC80 were positive for etD that codes for exfoliative toxin D.
Genes for epidermal cell differentiation inhibitors type A, edinA, was detected in two isolates belonging to CC5-IV-MRSA, and edinB, that codes for epidermal cell differentiation inhibitor type B, was detected in eight CC80-MRSA-IV, two CC152-MRSA-V + fus, and two ST2867-MRSA-V isolates.
The MRSA isolates varied in the carriage of genes for the immune evasion clusters (IEC), sak (staphylokinase), chp (chemotaxis inhibiting protein) and scn (staphylococcal complement inhibitor). Fifty-five (45.8%) of the 120 isolates carried all three IEC determinants, sak. chp/scn. These included 28 of the 30 ST239 – MRSA-III isolates, CC5-MRSA-IV, WA-MRSA-121, CC5-MRSA-IV (Paediatric clone), CC5-MRSA-V, WA MRSA-11/34/35/90/108, CC22-IV-MRSA, CC30-MRSA-IV, CC45-MRSA-[VI + fus] and CC88-MRSA-IV. Forty-six isolates carried sak and Scn while isolates of CC59 and CC5-MRSA-VI, new paediatric clone harboured chp and sak. The CC15-MRSA-V isolates harboured chp and scn. ST772-MRSA-V isolates harboured only scn while an ST239-MRSA-III + ccrC isolate lacked any of the IEC genes.
Discussion
Due to constant changes in the epidemiology of MRSA strains it is necessary to conduct regular surveillance to determine clonal MRSA populations circulating in healthcare facilities to understand the evolution and dissemination of specific MRSA clones causing infections for the application of suitable infection control and preventive measures. The results of the present study have provided updates on the distribution and types of MRSA clones that circulated in the hospital in 2016. The isolates belonged to diverse genetic backgrounds with 60.3% and 39.7% of the MRSA isolates carrying the CA-MRSA and HA-MRSA genotypes respectively which is consistent with global trends5,6, and with recent reports that CA-MRSA are increasing in Kuwait hospitals20,21. The MRSA isolates belonged to six SCCmec types, 56 spa types and 16 clonal complexes (CC), with CC8, CC5, CC1, CC22, CC6, CC80, CC30 and CC97 as the common CCs highlighting the significant changes in the clonal composition of MRSA in the hospital since the last study in 1996–200116. The reasons for the proliferation of the diverse MRSA clones observed in this study is not fully clear. However, the admission of an increasing number of patients already colonized with community genotypes of MRSA into the hospital may offer partial explanation. It is also worth noting that this observation is consistent with observations in different centres where CA-MRSA clones are overtaking the traditional HA-MRSA clones5,6,14.
The study showed that 30 (25%) of the 120 isolates analysed by DNA microarray were ST239-MRSA-III making ST239-MRSA-III the dominant MRSA clone in the hospital in 2016 as was the case in 1996–200116. However, whereas MRSA isolates obtained in 1996–2001 belonged to a single dominant clone, ST239-MRSA-III18, those obtained in 2016 belonged to diverse genetic backgrounds. Significantly, although ST239-MRSA-III was the dominant genotype in this study as was the case in 1996–200116, and 2005–200719, their spa types were different. Whereas in 1996–2001, the dominant spa types were t037 and t421, the spa types of the 2005–2007 isolates were t421 and t94519, and those isolated in 2016 were dominated by t860 and t945.The introduction of t860 and its replacement of t037 and t421 subtypes demonstrates the constant evolution of the ST239-MRSA–III clone which may explain its successful persistence in the hospital. Similarly, ST239-III-MRSA was the common HA-MRSA clone in Saudi Arabia22,23, Iran24, and China25. In contrast, CA-MRSA clones are the dominant MRSA clones among recent MRSA isolates in other hospitals in Kuwait18,20,21.
The PVL-positive CC22-IV-MRSA, comprising t852, t005, and t11836, was the most prevalent CC22-MRSA-IV variant followed by the Middle Eastern variant in this study. In contrast, the tst-positive CC22-MRSA-IV, also known as the UK-EMRSA-15/Middle Eastern variant, was the most common variant in the other hospitals in Kuwait21,26. It is uncertain whether this observation signals a shift in the distribution of CC22-MRSA-IV variants in Kuwait. However, CC22-MRSA-IV [PVL+]/t852 was also reported as the dominant CC22-MRSA-IV variant in Qatar27.
The CC97 (ST97-MRSA-V) isolates were first reported in this hospital in 2007 as a cause of an outbreak in a neonatal intensive care and special care units28. However, until now, no further CC97-MRSA-V isolates were detected in the hospital following the termination of the outbreak. Curiously, the current CC97-MRSA-V isolates, except one (t359), belonged to spa t267, and were resistant to gentamicin, kanamycin, and fusidic acid similar to the resistance profile of the outbreak isolates even though they were isolated many years apart. Unfortunately, the spa type for the outbreak strains were not determined which makes it difficult to establish whether the same clone reappeared in the hospital in 2016. However, the spa t267 of the current isolates is different from spa t2297 and t1234 that were reported previously among CC97-MRSA-V in other hospitals in Kuwait18,20.
We noticed an isolate belonging to CC59 for the first time in Kuwait in this study. CC59 is a predominant CA-MRSA clone in Taiwan29, but has also been isolated in Hong Kong30, Japan31, and Western Australia31. However, it is rare in the Arabian gulf region. Hence, this report of CC59 in Kuwait suggests its recent importation into the country. The isolate in this report shares characteristics with the Taiwan (ST59/952-VT /PVL) and Western Australian (ST59-IV) WA-MRSA-118 clones suggesting a common origin.
We observed that the axilla, groins and nose were colonised mostly by the HA-MRSA clone, ST239-MRSA-III, whereas the other sites were dominated by heterogeneous CA-MRSA clones. This is consistent with previous observations that the groin, axilla and nose are often colonised by ST239-MRSA-III strains which usually serve as sources of MRSA transmission1,5,14. Also, the dominance of CA-MRSA isolates in wound, pus and skin samples is consistent with earlier reports that CA-MRSA cause predominantly skin and soft tissue infections1,4,5,8,14. Furthermore, the isolation of CA-MRSA isolates from blood, and respiratory samples confirms their capacity to cause different types of infections other than skin and soft tissue infections1,6,14.
A high proportion of the isolates were resistant to multiple antibiotics including fusidic acid (63.4%), gentamicin (54.5%), tetracycline (54.5%), erythromycin (52.2%), ciprofloxacin (49.8%) and trimethoprim (21.5%). High prevalence of fusidic acid resistance is common among MRSA obtained in Kuwait hospitals. In recent studies, fusidic acid resistance was detected in 16.5% of MRSA obtained in the Maternity hospital21, and in 41.2% of MRSA obtained in Kuwait hospitals between 2011 and 201532. The higher prevalence reported in the present study highlights an increasing trend for fusidic acid resistance among MRSA isolated in Kuwait hospitals.The high prevalence of fusidic acid resistance among MRSA isolates was attributed in part to the emerging chimeric genetic elements comprising SCCmec elements and the fusidic acid resistance determinant, fusC (SCCfusC)20 as also seen in this study (Table 1). The chimeric genetic element possibly facilitates the transmission of fusidic acid resistance in isolates belonging to different genetic backgrounds. The increasing prevalence of resistance to gentamicin, tetracycline, erythromycin, ciprofloxacin seen in this study have also been previously reported in MRSA otained from hospitals in Kuwait16,18,19,20.
We detected genes for multiple virulence factors including adhesins, haemolysins, enterotoxins and immune evasion clusters. However, the isolates differed in the prevalence of genes for PVL, enerotoxins, agr and capsular polysacharide (cap).The isolates belonged to cap5 (38.3%) and cap8 (61.7%) in agreement with previous observations that most clinical S. aureus isolates harbour cap5 or cap833. The higher prevalence of cap8 in this study mirrors the results of a previous study that showed that 57% S. aureus obtained from food handlers in Kuwait restaurants carried cap834. Similarly, cap8 was prominent among S.aureus obtained from patients in Gabon35. This study also revealed that cap type was associated with defined genetic backgrounds.
The agrI was the leading agr type among the isolates in this study followed by agrII and agrIII. Similarly, most (50%) of S. aureus from food handlers in Kuwait restaurants belonged to agrI34. AgrI was also the dominant agr type among MRSA isolates obtained in Egypt36, India37, Iran38, Nigeria39, Turkey40, and China41. The study further revealed that the agr types were linked to specific clonal complexes as have been observed in S. aureus isolated from human and food sources in China41.
We identified genes coding for PVL in 30.8% (37/120) of the isolates belonging to different genetic backgrounds. This was higher than the 14.6% prevalence of genes for PVL in S.aureus in the same hospital in 2009–201017. The higher prevalence of PVL- gene- positive isolates in this study may reflect the diversity of current clones in the hospital.
We found genes for staphylococcal enterotoxins (SE) in 85.1% of the isolates with most of the isolates harbouring two to six SE genes.Whereas the prominent SE genes in our study were egc (38.3%), sea (37.5%), seg (37.5%), sei (35.8%), sek (32.5%) and seq (32.5%), sea was the most common SE gene detected in MRSA from patients in Turkey40, Iran38, and Nigeria39. Since the carriage of SE genes as well as genes for agr and cap are genotype specific, the dominance of these factors may reflect the dominance of specific genotypes in the different geographical regions.
In conclusion, the evidence gathered from typing our MRSA isolates have revealed the expansion of the population of MRSA clones over time with the healthcare-associated MRSA, ST239-MRSA-III, as the dominant clone despite the emergence of diverse clones with CA-MRSA genotypes. These findings warrant a review of infection control and prevention procedures for the application of effective control measures that will limit antibiotic usage leading to improved patients’ care.
Materials and Methods
Bacterial isolates
A total of 209 single patient MRSA isolates were obtained in the Microbiology laboratory of Farwaniya Hospital during a period of 12 months, from January 1 2016, to 31 December 2016. The isolates were collected from cultures of various clinical specimens as part of routine diagnostic care and submitted for molecular typing at the MRSA Reference Laboratory located at the department of Microbiology, Faculty of Medicine, Kuwait University. Only a single isolate from a patient is submitted for typing. Repeat isolates from the same patient are excluded. The isolates were obtained from wound (N = 52; 24.9%), blood (N = 25; 12%), pus (N = 22; 10.5%), Nasal (N = 6;2.8%), Groins (N = 7;3.3%), axilla (N = 5;2.4%), sputum (N = 5; 2.4%), eye (N = 4;1.9%), ear (N = 3;1.4%), fluids (N = 3;1.4%), high vaginal swabs (N = 2; 0.9%) and an unidentified swab (N = 1; 0.4%). All isolates were identified as S. aureus by their cultural characteristics including growth on blood agar (5% sheep blood), growth on mannitol salt agar, Gram stain, positive tube coagulase test and DNase tests. The isolates were stored at −80 °C in 40% glycerol broth (v/v in brain heart infusion broth) and sub cultured onto brain heart infusion agar for purity prior to investigations.
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was performed by the disk diffusion method according to the guidelines of the Clinical Laboratory Standards Institute (CLSI)42, with the following antimicrobial disks (Oxoid): benzyl penicillin (10U), cefoxitin (30 μg), amikacin (30 μg), kanamycin (30 μg), tobramycin (10 μg), mupirocin (200 μg and 5 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg), Spectinomycin (25 μg), chloramphenicol (30 μg), tetracycline (10 μg), trimethoprim (2.5 μg), fusidic acid (10 μg), rifampicin (5 μg), ciprofloxacin (5 μg), teicoplanin (30 μg), and linezolid (30 μg). Minimum inhibitory concentration (MIC) for cefoxitin, mupirocin, vancomycin and teicoplanin were determined with Etest strips (AB BioMerieux, Marcy l′Etoile, France) according to the manufacturer’s instructions. The interpretation of the MIC values was based on the antibiotic breakpoint concentration recommended by the CLSI42. Susceptibility to fusidic acid was interpreted according to the British Society to Antimicrobial Chemotherapy43. S. aureus strains ATCC25923 and ATCC29213 were used as a quality control strain for disk diffusion and MIC determination respectively.
SCCmec typing and spa typing
All MRSA isolates were characterized using SCCmec typing and spa typing. SCCmec typing was performed as described by Zhang et al.10, Spa typing was performed as described by Harmsen et al.13.
DNA microarray
The S. aureus genotyping Kit 2.0 (Alere Technology, Jena, Germany) was used following protocols provided by the manufacturer. The method detects 334 target sequences including genes encoding species markers, PVL, SCCmec, capsule (cap), and accessory gene regulator (agr) group, enterotoxins, adhesins, and antibiotic resistance4. DNA microarray was used to assign the MRSA isolates to clonal complexes (CC) as described previously11.
References
Tong, S. Y. C., Davis, J. S., Elchenberger, E., Holland, T. L. & Fowler, V. G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and managements. Clin Microbiol Rev 28, 603–650 (2015).
Jevons, M. P. “Celbenin”-resistant staphylococci. Br Med J 1, 124–125 (1961).
Chambers, H. F. & Deleo, F. R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7, 629–641 (2009).
Monecke, S. et al. A Field Guide to Pandemic, Epidemic and Sporadic Clones of Methicillin-Resistant Staphylococcus aureus. PLoS ONE 6(4), e17936, https://doi.org/10.1371/journal.pone.00117936 (2011).
Hiramatsu, K. et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother 45, 117–136 (2013).
Lakhundi, S. & Zhang, K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31, e00020–18, https://doi.org/10.1128/CMR.00020-18 (2018).
International Working Group on the Classification of Staphylococcus Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53, 4961–4967 (2009).
David, M. Z. & Daum, R. S. Community – associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23, 616–687 (2010).
Oliveira, D. C. & de Lencaster, H. Multiplex PCR strategy for rapid identification of structural types of and variants of the mec elements in methicillin – resistant Staphylococcus aureus. Antimicrob Agents Chemother 46, 2155–2161 (2002).
Zhang, K. et al. Novel multiplex PCR assay for characterization and concomitant subtyping of Staphylococcal cassette chromosome mec type I to V in Methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43, 5026–5033 (2005).
Monecke, S., Slickers, P. & Ehricht, R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol 53, 237–251 (2008).
Enright, M. C. et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38, 1008–1015 (2000).
Harmsen, D. et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41, 5442–5448 (2003).
Laupland, K. B. et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: a multinational population-based surveillance study. Clin Microbiol Infect 9, 465–471 (2013).
Raji, A. et al. High genetic diversity of Staphylococcus aureus in a tertiary care hospital in southwest Nigeria. Diagn Microbiol Infect Dis 77, 367–369 (2013).
Udo, E. E., Al-Sweih, N., Mohanakrishnan, S. & West, P. W. Antibacterial resistance and molecular typing of methicillin-resistant Staphylococcus aureus in a Kuwaiti General hospital. Med Princ Pract 15, 39–45 (2006).
Alfouzan, W., Al-Haddad, A., Udo, E., Mathew, B. & Dhar, R. Frequency and clinical association of Panton-Valentine leukocidin-positive Staphylococcus aureus isolates: A study from Kuwait. Med Princ Pract 22, 245–249 (2013).
Boswihi, S. S., Udo, E. E., & Al-Sweih, N. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992–2010. PLoS ONE, 11(9), https://doi.org/10.1371/journal.pone.0162744 (2016).
AlFouzan, W., Dhar, R. & Udo, E. E. Genetic lineages of methicillin-resistant Staphylococcus aureus acquired during admission to an Intensive Care Unit of a general hospital. Med Princ Pract 26, 113–117 (2017).
Boswihi, S. S. et al. Emerging variants of methicillin-resistant Staphylococcus aureus genotypes in Kuwait hospitals. PloS ONE 13(4), e0195933, https://doi.org/10.1371/journal.pone.0195933 (2018).
Udo, E. E. & Al-Sweih, N. Dominance of community-associated methicillin-resistant Staphylococcus aureus clones in a maternity hospital. PLoS One. 12(6), e0179563, https://doi.org/10.1371/journal.pone.0179563 (2017).
Senok, A., Ehricht, R., Monecke, S., Al-Saedan, R. & Somily, A. Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: emergence of new clonal complexes in Saudi Arabia. New Microbes New Infections 14, 13–18 (2016).
Monecke, S. et al. Characterization of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol 12, 1–9 (2012).
Goudarzi, M. et al. Genetic diversity of methicillin resistant Staphylococcus aureus strains isolated from burn patients in Iran: ST239-SCCmec III/t037 emerges as the major clone. Microb Pathog 105, 1–7 (2017).
Peng, H., Liu, D., Ma, Y. & Goa, W. Comparison of community-and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012-2017. Scientific Reports 8, 17919, https://doi.org/10.1038/s41598-018-36206-5 (2018).
Udo, E. E., Boswihi, S. S. & Al-Sweih, N. High prevalence of toxic shock syndrome toxin-producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. New Microbes New Infections 12, 24–30 (2016).
El-Mahdy, T. S., El-Ahmady, M. & Goering, R. V. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated over a 2-year period in a Qatari hospital from multinational patients. Clin Microbiol Infect 20, 169–173 (2014).
Udo, E. E., Aly, N. Y., Sarkhoo, E., Al-Sawan, R. & Al-Asar, A. S. Detection and characterisation of an ST97-SCCmec-V community-associated methicillin-resistant Staphylococcus aureus clone in a neonatal intensive care unit and special care baby unit. J Med Microbiol 60, 600–604 (2011).
Takano, T. et al. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence Type 59 in Taiwan. Antimicrob Agents Chemother 52, 837–845 (2008).
Ho, P. L. et al. Molecular epidemiology and household transmission of community-associated methicillin-resistant Staphylococcus aureus in Hong Kong. Diagn Microbiol Infect Dis 57, 145–151 (2007).
Coombs, G. W. et al. Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob Agents Chemother 54, 1914–1921 (2010).
Udo, E. E. & Boswihi, S. S. Antibiotic Resistance Trends in Methicillin-Resistant Staphylococcus aureus isolated in Kuwait Hospitals: 2011-2015. Med Princ Pract 26, 485–490 (2017).
Von Eiff, C., Friedrich, A. W., Peters, G. & Becker, K. Prevalence of genes encoding for members of the staphylococcal leukotoxins family among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis 49, 157–162 (2004).
Udo, E. E., Al-Mufti, S. & Albert, M. J. The prevalence of antimicrobial resistance and carriage of virulence genes in Staphylococcus aureus isolated from food handlers in Kuwait City restaurants. BMC Res Notes 2, 108 (2009).
Schaumburg, F. et al. Virulence factors and genotypes of Staphylococcus aureus from infection and carriage in Gabon. Clin Microbiol Infect 17, 1507–1513 (2011).
El-Baz, R., Rizk, D. E., Barwa, R. & Hassan, R. Virulence characteristics and molecular relatedness of methicillin-resistant Staphylococcus aureus harbouring different staphylococcal cassette chromosome. Microb Pathog 113, 385–395 (2017).
Rajan, V., Schoenfelder, S. M., Ziebuhr, W. & Gopal, S. Genotyping of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) in a tertiary care center in Mysore, South India: ST2371-SCCmec IV emerges as the major clone. Infect Genet Evol 34, 230–235 (2015).
Cheraghi, S. et al. Analysis of virulence genes and accessory gene regulator (agr) types among methicillin-resistant Staphylococcus aureus strains in Iran. J Glob Antimicrob Resist 10, 315–320, https://doi.org/10.1016/j.jgar.2017.06.009 (2017).
Kolawole, D. O. et al. Characterization of colonizing Staphylococcus aureus isolated from surgical wards’ patients in a Nigerian university hospital. PLoS One 8(7), e68721, https://doi.org/10.1371/journal.pone.0068721 (2013).
Tekeli, A., Ocal, D. N., Ozmen, B. B., Karahan, Z. C. & Dolapci, I. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus bloodstream Isolates in a Turkish University Hospital Between 2002 and 2012. Microb Drug Resist 22, 564–569 (2016).
Luo, K. et al. Molecular characterisation of antimicrobial resistance and virulence determinants of Staphylococcus aureus isolates derived from clinical infection and food. J Clin Lab Anal 32(7), e224456, https://doi.org/10.1002/jcla.22456 (2018).
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty-second Information supplement M100-S25.CLSI, Wayne, PA. USA. (2015).
British Society to Antimicrobial Chemotherapy (BSAC) http://bsac.org.uk/susceptibility.BSAC (2013).
Acknowledgements
We are grateful to Mrs Bindu Mathew and Bobby Noronha for technical assistance. There was no external funding for this study.
Author information
Authors and Affiliations
Contributions
Conceived and designed experiments: E.E.U., W.A. Analysed data: E.E.U., W.A., A.M., A.A. Wrote manuscript: W.A., E.E.U., A.M., A.A. Supervision: E.E.U.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alfouzan, W., Udo, E.E., Modhaffer, A. et al. Molecular Characterization of Methicillin- Resistant Staphylococcus aureus in a Tertiary Care hospital in Kuwait. Sci Rep 9, 18527 (2019). https://doi.org/10.1038/s41598-019-54794-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54794-8
This article is cited by
-
Recombination-mediated dissemination of Methicillin-resistant S. aureus clonal complex 1 in the Egyptian health care settings
Annals of Clinical Microbiology and Antimicrobials (2023)
-
The global prevalence of fusidic acid resistance in clinical isolates of Staphylococcus aureus: a systematic review and meta-analysis
Antimicrobial Resistance & Infection Control (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.