Abstract
In this study, to investigate the secondary function of Rpl10a in zebrafish development, morpholino antisense oligonucleotides (MOs) were used to knock down the zebrafish ribosomal protein L10a (rpl10a). At 25 hpf (hours post-fertilization), embryos injected with the rpl10a MO showed an abnormal morphology, including short bodies, curved tails, and small yolk sac extensions. We observed pigment reductions, edema, larger yolk sacs, smaller eyes and smaller yolk sac extensions at 50 hpf. In addition, reductions in the expression of primordial germ cell (PGC) marker genes (nanos1 and vasa) were observed in rpl10a knockdown embryos. A rescue experiment using a rpl10a mRNA co-injection showed the recovery of the morphology and red blood cell production similar to wild-type. Moreover, the CRISPR-Cas9 system was used to edit the sequence of rpl10a exon 5, resulting in a homozygous 5-bp deletion in the zebrafish genome. The mutant embryos displayed a morphology similar to that of the knockdown animals. Furthermore, the loss of rpl10a function led to reduced expression of gata1, hbae3, and hbbe1 (erythroid synthesis) and increased tp53 expression. Overall, the results suggested that Rpl10a deficiency caused delays in embryonic development, as well as apoptosis and anemia, in zebrafish.
Similar content being viewed by others
Introduction
Ribosomal proteins (RPs) function in protein synthesis. Many lines of evidence suggest other roles for RPs, including DNA repair, apoptosis, translational regulation, developmental control, and red blood cell development1. Several RP mutations are associated with Diamond-Blackfan anemia (DBA) patients and include RPS19, RPS7, RPS10, RPS17, RPS24, RPS26, RPL5, RPL11, RPL26, and RPL35A2,3,4,5,6,7,8,9. The mutation of some RPs (e.g., RPL5, RPL11, RPL23) may cause a failure in ribosome biogenesis and the accumulation of free RPs that can trigger Tp53-mediated apoptosis; defects in ribosome biogenesis cause aberrations in red blood cells and thereby activate tp53 in the erythroid lineage10. Thus, proper ribosome formation is a checkpoint for cell cycle progression11. Additionally, rpl11 mutation or loss of Rpl11 function activates a Tp53-dependent checkpoint response to prevent abnormal embryonic development12.
Ribosomal protein L10a (Rpl10a) is in the L1P family of ribosomal proteins and is encoded by the rpl10a gene. A previous study showed that the Rpl10a protein might play an important role during embryogenesis and organogenesis13. Recombinant Rpl10a protein was also shown to be involved in shrimp ovary development both in vitro and in vivo14,15. Although Rpl10a has secondary functions in embryogenesis and ovary development, the effects of Rpl10a on primordial germ cells, or PGCs, during early embryogenesis have not been reported. PGCs are the precursors to reproductive gametes (spermatozoa and oocytes). The vasa gene has been identified as a marker of PGCs in zebrafish16, and nanos1, a germplasm gene, is essential for PGC migration and viability17. In Drosophila, PGCs deficient in Nanos activity showed abnormal development, shifts to gonad failure, a reduction in egg number, and morphological abnormalities18. These results in fruit flies were similar to results from zebrafish17.
Therefore, we knocked down and knocked out the rpl10a gene using morpholinos and CRISPR-Cas9, respectively, to investigate the function of the rpl10a gene in embryogenesis, germ cell development, and erythropoiesis. Quantitative RT-PCR was performed to determine the fold changes in gata1 and tp53 gene expression in rpl10a-deficient embryos. The expression of the zebrafish embryonic globin genes, hemoglobin alpha embryonic-3 (hbae3) and hemoglobin beta embryonic-1 (hbbe1), which were highly expressed at 48 hpf19, were used as markers of erythroid cells. Moreover, histology was used to support the morphological defects and the effects of rpl10a knockdown on PGC marker genes, including nanos1 and vasa gene expression, which were investigated using qRT-PCR and whole-mount in situ hybridization.
Results
Zebrafish rpl10a knockdown and knockout construction
The zebrafish rpl10a gene is located on chromosome 8 and contains six exons, resulting in a 648 bp cDNA sequence. The rpl10a gene encodes a protein of up to 216 amino acids. To investigate the effect of Rpl10a deficiency, we knocked down the rpl10a gene using MO injection to inhibit protein translation. The MO target sites are shown in the diagram in Fig. 1a. The injection of MOsp, with specificity to exon 5, altered the splicing and resulted in the exclusion of some of exon 5 from the mature mRNA. Moreover, the expression of the rpl10a gene decreased, as detected by RT-PCR (Fig. 1b). DNA sequencing confirmed the deletion in the splice site of exon 5 after MOsp injection. We found that only 33 bps of exon 5 were deleted (Fig. 1c), and 11 amino acids were predicted to be deleted (Fig. 1d).
(a) Schematic of the zebrafish rpl10a gene structure. White boxes represent the untranslated regions, while the translated region is shown with black boxes. The start codon (gray arrowhead) and stop codon (asterisk) positions are presented. The white arrowheads are marked at the position of the MOsp detection primer, and the arrowheads indicate the direction of PCR polymerization. The positions of the designed MOaug and MOsp are indicated. (b) RT-PCR analysis of rpl10a in MO-injected (MOaug, MOsp), rescued (MOsp + mRNA) and wild-type embryos; rpl38 was amplified as a control. M was shown as a 100 bp ladder. A smaller PCR product (551 bp) was observed from MOsp-injected embryos because 33 bp of exon 5 was skipped; the wild-type product was 584 bp (primers were designed to obtain products smaller than the full-length gene). (c) The nucleotide sequences of rpl10a cDNA are presented. The hyphens displayed 33 bps of exon 5 that were deleted after MOsp injection. The underline shows the position of primer sequences. The start codon and stop codon appeared in double underline and bold text, respectively. (d) The amino acids were predicted using the translation tool from www. ExPASy.com. The 11 amino acids were predicted to be removed after deletion. Hyphens showed deleted amino acids, and the stop codon is shown in an asterisk.
We used the CRISPR-Cas9 genomic editing system using the crRNA-tracrRNA-Cas9 complex to edit exon 5 of the rpl10a gene in zebrafish. From the domain prediction using ScanProsite, we found that the Rpl10a protein contains 1 domain, namely, the ribosomal protein L1 signature. This domain displays RNA chaperone activity with the amino acid sequence IKQIPRILGPGLNKAGKFPS, and these sequences were encoded by exon 5.
Additionally, the heteroduplexes of F0 mutant fish were detected via heteroduplex mobility assay (HMA) after CRISPR-Cas9 injection (Fig. 2a). The 115 embryos slowly died off after injection and only 18 became adult. They were screened for mutations transmitted through the germline. We selected progeny with positive germline transmission based on their patterns. The HMA results of the 5-bp deletion mutants showed a different pattern of heteroduplexes and homoduplexes in the heterozygous mutant (Fig. 2b). The heteroduplexes migrated more slowly than the homoduplexes due to structural distortion. The mutant genotype was confirmed by sequencing (Fig. 2c). Among the 18, eight mutated F0 were found to transmit a lesion into their F1 progenies, 6 adults were wild-type and the other 4 could not give F1 progeny. The number of F1 founders obtained from the F0 generation was 40 fishes. The F1 progenies had 6 mutation patterns, including a 3-bp deletion, a 5-bp deletion, an 11-bp deletion, a 15-bp deletion, a 17-bp deletion, and a 7-bp insertion. Amino acid changes were predicted based on the gene mutations (Fig. 2d). These patterns corresponded to 215, 134, 136, 121, 132 and 141 amino acids in the order mentioned above in F1 generation fish. For further analysis, the 5-bp deletion was used as a model to study the loss of the function of rpl10a gene.
(a) Pattern of heteroduplexes and homoduplexes observed after crRNA-tracrRNA-Cas9 complex (CRISPR-Cas9) injection. (b) HMA showing different heteroduplex and homoduplex patterns in the 5-bp deletion mutant compared to the wild-type (WT). (c) Schematics of the mutations obtained from genome editing. The crRNA target site and PAM sequences are presented with gray highlighting and red letters, respectively. Deleted nucleotides are shown with hyphens, and inserted nucleotides are in blue letters. (d) Predicted amino acid sequences after rpl10a gene knockout by CRISPR-Cas9 starting from amino acid position 190. The deleted sequences and stop codons are marked with hyphens and stars, respectively.
Morphological defects in rpl10a dysfunction
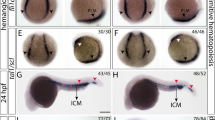
The phenotype of the knockdown embryos (morphants) was compared to that of wild-type embryos at approximately 25 hpf. The rpl10a MO-injected embryos showed morphological abnormalities, including shorter body lengths, bent tails, and smaller yolk sac extensions. We observed a reduction in pigmentation, edema, larger yolk sacs, smaller eyes, and smaller yolk sac extensions at 50 hpf (Fig. 3a). Most rpl10a morphants died by 3–7 days post-fertilization (dpf). The non-injected embryos had no phenotypic changes. Rescued embryos using rpl10a mRNA co-injection showed morphologies similar to that of the wild-type. The phenotype in each group was evaluated and calculated as percentages of normal and abnormal phenotypes (Fig. 3b). All MO-injected embryos had a similar phenotype. However, some of them showed a moderate phenotype in the MO-injected group. In the case of MOsp + mRNA, injected embryos showed a recovery phenotype of approximately 89%.
(a) Effect of Rpl10a deficiency on embryonic development. Lateral views of rpl10a MO-injected embryos and rescue phenotype observed at 25 and 50 hpf. At 25 and 50 hpf, smaller eyes, yolk sac extensions, bent tails, and developmental delays were observed in rpl10a knockdown. At 50 hpf, deficient embryos displayed smaller eyes with reduced pigmentation and edema (arrowheads). The MOsp + mRNA injected embryos displayed rescue phenotypes. (b) The phenotype of rpl10a MOaug (n = 90), MOsp (n = 105), and MOsp + mRNA (n = 54) injected embryos was evaluated compared to wild-type (n = 107) at 25 hpf. The morphologic abnormalities include a thinner yolk sac extension, a shorter body length, and a bent tail, whereas the normal wild-type embryos showed a normal yolk sac extension and a straight tail. The result was presented in the percentage of normal (black bar) and abnormal (white bar) embryos in each group.
The 5-bp deletion embryos were also used as a model to confirm the effect of Rpl10a deficiency on early embryogenesis. Embryos carrying a homozygous 5-bp rpl10a mutation showed a severe phenotype, and all mutant embryos died at 3–5 dpf. The phenotypes of homozygotes were more severe when compared with MO-injected embryos at 25 hpf. The heterozygous phenotype was difficult to distinguish; however, few embryos slightly differed from wild-type. Additionally, the homozygous 5 bp deletion at 48 hpf was more severe, including the curved tail, enlarged yolk sac, heart edema, depigmentation, and smaller eyes (Fig. 4a). The homozygous mutant fish at 3 dpf displayed smaller eyes, loss of pigmentation, bent tails, and edema. These morphologies were abnormal due to developmental interruption, leading to death. In addition, the rpl10a defect led to swelling of the yolk sac. This phenomenon indicated that nutrients in yolk sac accumulation and retaining. To confirm this without metabolic assessments of embryos, we performed H&E to stain the fluid in the yolk sac (Fig. 4b). Eosin stains the extracellular matrix and cytoplasmic parts. It is an acidic dye that binds to proteins and lipids in the cytoplasm. In this result, the pink spots in the yolk sac and yolk extension were proteins, lipids, and nutrients. The results indicated an accumulation of nutrients in the yolk sac due to low metabolism in the mutant fish. Moreover, the eosinophilic cytoplasmic filaments in muscle cells were decreased (Fig. 4b).
(a) Phenotypes of a 5-bp homozygous deletion (−/−) embryo, heterozygous deletion (+/−) and wild-type (+/+) embryos at 48 hpf are presented. Homozygous mutant embryos displayed severe phenotypes, including smaller eyes, heart and yolk sac edema, smaller yolk extension, curved tail and depigmentation. (b) At 3 dpf, the altered phenotypes and H&E staining were also performed.
Reduction in hemoglobin level with rpl10a defects
Typically, the loss of ribosomal function leads to anemia in patients. We investigated the effects of rpl10a defects on reducing hemoglobin level at 48 hpf. Low o-dianisidine staining signals were detected in the rpl10a knockdown. The embryos rescued by mRNA co-injection showed increased staining similar to that of the wild-type (Fig. 5a). Additionally, the 5-bp rpl10a deletion mutants showed results corresponding to those of the knockdown. The heterozygous mutants showed reduced hemoglobin staining. A reduction in hemoglobin content was observed in the homozygous mutants (Fig. 5b). As shown in Fig. 5c, the normal hemoglobin staining level was decreased to 22% in rpl10a knockdown. After rescue using rpl10a mRNA co-injection, the number of hemoglobin recovered and showed a normal level of 50%. In addition, the reduction of hemoglobin level in homozygous mutant embryos showed more severe than heterozygotes and knockdown embryos.
(a) Significant reductions in hemoglobin (Hb) staining in the embryos were observed after rpl10a gene knockdown (MOaug and MOsp), similar to rps19 knockdown, compared to the control; embryos co-injected with rpl10a transcript recovered at 48 hpf. (b) rpl10a embryos mutated using CRISPR-Cas9 knockout showed significant reductions in hemoglobin staining. (+/+: wild-type, +/−: heterozygous, −/−: homozygous mutant). The graph displayed the percentages of normal and abnormal levels of Hb staining in (c) knocked down rpl10a gene and mRNA rescue embryos; (d) 5 bp rpl10a mutant embryos. The dense Hb staining (orange dot) at the wild-type embryo yolk sac was considered normal while a significantly pale orange or slightly colored Hb staining in the yolk sac was evaluated as abnormal. The number of animals quantified in each group are shown on top of the bars.
Effect of rpl10a defects on expression of erythropoiesis-related genes
The fold change in the expression of the gata1 gene, which is involved in blood cell maturation, was reduced in MO-injected embryos at 24 hpf (Fig. 6a). Additionally, gata1 expression was downregulated in mutant embryos. However, MOsp-injected embryos and homozygous mutant fish showed no significant reduction. Higher expression of the tp53 gene was detected in all rpl10a-deficient embryos and homozygous mutants than in the wild-type embryos (Fig. 6b). The trend of these expression results was similar to that with rps19 MOaug injection. In addition, the fold change in tp53 expression levels declined after rescue with rpl10a mRNA. To check the expression of erythroid markers at the embryonic stage, hbae3 and hbbe1 were analyzed in the rpl10a knockdown and knockout embryos (Fig. 6c). The qPCR results showed that these genes, which are involved in erythroid synthesis, had low expression in Rpl10a-deficient embryos, suggesting that it might play a role in anemia.
Quantitative RT-PCR results showing the fold changes in the expression of the (a) gata1 and (b) tp53 genes in rpl10a MOaug, MOsp, homozygous mutant, and MOsp + mRNA injected embryos at 24 hpf compared to wild-type and rps19 knockdown embryos as controls. (c) Fold changes in the expression of hbae3 and hbbe1 mRNA in Rpl10a-deficient embryos and mutant samples at 48 hpf. Wild-type embryos were used as controls. Each group used 20 pooled embryos (replicates = 4). The data were analyzed for statistical significance by one-way ANOVA followed by Tukey’s multiple comparison test. (**p-value > 0.01).
Reduced expression of PGC marker genes in rpl10a-defective embryos
The fold changes in the expression of PGC marker genes after knocking down the rpl10a gene are presented compared to WT and rps19 knockdown at 25 hpf. The expression levels of nanos1 and vasa transcripts were reduced considerably in rpl10a-MOaug embryos by 0.42- and 0.37-fold from WT, respectively. The relative expression levels of nanos1 and vasa also significantly decreased compared to those associated with Rps19 deficiency. However, the expression of nanos1 in Rps19-deficient embryos differed slightly from that of the WT. The homozygous mutant embryos showed significant decreases in vasa and nanos1 mRNA expression levels. Furthermore, the vasa and nanos1 transcription levels were upregulated when rescued with rpl10a mRNA injection (Fig. 7). The qPCR results revealed that the PGC marker genes were reduced when the rpl10a gene was defective.
Fold change in the expression levels of nanos1 and vasa in rpl10a MOaug, MOsp, homozygous mutant and MOsp + mRNA injected embryos compared to WT and rps19-deficient embryos as controls. Each group used 20 pooled embryos (replicates = 4). The data were analyzed for statistical significance by one-way ANOVA followed by Tukey’s multiple comparison test. (*p-value > 0.05, **p-value > 0.01).
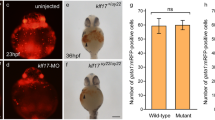
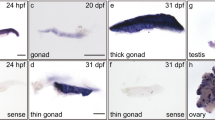
PGC marker genes in zebrafish embryos were also used to study the effect of rpl10a knockdown on the germline using whole-mount in situ hybridization. The vasa and nanos1 genes were selected as PGC markers after rpl10a gene knockdown at 25 hpf, and rps19 knockdown morphants were also analyzed as controls. The results showed that after rpl10a gene knockdown, the nanos1 and vasa transcripts localized in PGCs were reduced. Although most rpl10a morphants exhibited low PGC marker gene expression, some displayed no difference in expression from the wild-type. The rps19 morphants seemed similar to the wild-type. Although the decreased intensity referred to the reduction of PGC numbers, we did not count the number of PGCs but evaluated the level of intensity instead. The signals from nanos1 and vasa gene expression were divided into 2 groups to estimate the effect of the knockdown. Representative in situ hybridization samples are shown in Fig. 8a. The results showed that nanos1 had lower expression in rpl10a MOaug- and MOsp-injected embryos than it did in the other groups (Fig. 8b). The embryos rescued with rpl10a mRNA could recover nanos1 expression, similar to the wild-type group. There was little difference in vasa gene expression in rpl10a MOsp and the rescue experiment or control groups (Fig. 8c). These results indicated that rpl10a gene knockdown reduced PGC marker gene expression, especially for nanos1. Moreover, there was no decrease in PGC marker gene expression in rps19 MOaug morphants. The whole-mount in situ hybridization results also showed the same trends in expression as did the qPCR results.
(a) Whole-mount in situ hybridization showing the expression of PGC marker genes, including nanos1 and vasa, after morpholino injection at 25 hpf. The reduction of nanos1 and vasa expression intensity was obviously observed in MOaug-injected embryos. The nanos1 expression was also decreased in MOsp-injected embryos and it was recovered when mRNA was co-injected. The expression levels of nanos1 (b) and vasa (c) were graded as the normal and abnormal percentage. The mean pixel intensity of nanos1 and vasa gene expression of the normal embryos were 24.7 ± 8.7 and 22.6 ± 8.2, respectively. An abnormal level of the gene expression was taken as one with the value less than the mean minus one standard deviation of the gene expression of the wild-type, i.e., less than 16 for nanos1 and 14.4 for vasa gene. The number of animals quantified in each group are shown on top of the bars.
Discussion
Haploinsufficiency of RPs caused by loss-of-function mutations in diploid organisms results in typical phenotypic characteristics during development. The first identified RP was the Drosophila minute gene, which was reported by Kongsuwan et al.20 and described by Marygold et al.21. Minute mutants showed developmental retardation, short and thin bristles, irregular eyes, and reduced viability. An RPS6 mutation in Drosophila resulted in abnormal blood cell differentiation, slow development, and larval lethality due to the growth inhibition of larval organs22. Our results showed that Rpl10a insufficiency showed a severe phenotype and led to a delay in embryonic development in zebrafish. These altered phenotypes might occur because of changing translation or cell cycle arrest stimulation23. In addition, swelling of the yolk sac indicated nutrient accumulation and retention in the yolk sac. The nutrient accumulation might suggest that mutant fish have low metabolism. Moreover, the decrease in eosin staining in muscle cells indicated that the muscle has degenerated and resulted in the movement problem in mutant fish.
In addition, using morpholinos to knock down several RPs in zebrafish embryos seems to have more effects than haploinsufficiency does. For example, rps19 knockdown triggered anemia, which may be caused by apoptosis in the erythropoietic system24. Moreover, rpl11 knockdown directly resulted in head abnormalities and Tp53 induction12. The effects of RP knockdown lead to Tp53 buildup, cell death, and developmental retardation25. Many ribosomal protein deficiencies lead to inpatient treatment for anemia2,3,4,6,7,8. The rpl35, rpl35a, and rplp2 gene morphants showed severe hemoglobin reductions in blood cells. Similar to rps19, gene knockdown affected the erythropoietic system and DBA anemia24. Haploinsufficiency of RPs can cause abnormal phenotypes during development, alterations in cell development, lethality, growth inhibition, and anemia23. The Rpl10a deficiency might reduce the red blood cells caused by apoptosis of the erythropoietic system, similar to the result of Rps19 deficiency.
Usually, RP mutations in either the small or large ribosomal subunits caused failure in the binding of the 40 S and 60 S subunits. An insufficiency of RPs caused decreases in translation efficiency. The failure of the 40 S or 60 S subunit led to a reduction in protein translation; therefore, the position of erythroid progenitor cells downstream of the protein corresponded to erythroid defects6,26. Our findings indicated that Rpl10a-deficient zebrafish had reduced hemoglobin levels but increased the expression of tp53. While Torihara et al. (2011) indicated that in Rps19 deficiency, the Tp53-independent pathway is more important for the anemia phenotype, the morphological abnormalities were dependent on Tp53 activation27. However, the exact role of Tp53, leading to anemia in Rpl10a-deficient zebrafish, is still unknown. Additionally, ribosomal protein deficiency might be a proapoptotic defect due to defects in ribosome assembly. This outcome leads to increased Tp53 activity and the stimulation of a proapoptotic phenotype6. Ribosomal protein deficiency led to abnormal morphology and development in zebrafish23. Dysfunctions in rpl10a gene resulted in morphological abnormalities and growth retardation. Rpl10a-deficient zebrafish died between 3–7 dpf. Early embryonic lethality might be caused by Tp53 activation and apoptosis, although there is no actual evidence for cell death and rescue of cell death upon loss or gain of rpl10a function.
In addition, the knockdown of zebrafish nanos1 by morpholino injection led to fewer PGCs and defects in migration17. In our study, we observed that the expression levels of nanos1 and vasa mRNA were significantly reduced in rpl10a morpholino-injected embryos. This outcome suggests that PGC reductions and migration defects occurred. The loss of Rpl10a might cause PGC to die due to apoptosis. The expression of tp53 was upregulated in Rpl10a-deficient embryos. On the other hand, nanos1 and vasa gene expression were decreased. Defect of the rpl10a gene caused PGC marker genes to decrease, and these genes played an important role in the survival of PGCs. Therefore, the PGC numbers could be reduced. However, the function of rpl10a in reproductive organ development requires further study.
In summary, our findings show that rpl10a knockdown and knockout zebrafish will be useful models to investigate the additional functions of ribosomal proteins in several pathways and organs in the future.
Methods
Animals
Zebrafish (wild-type AB line) were raised and maintained according to standard laboratory conditions in the Bio-resource Division at the Frontier Science Research Center, University of Miyazaki, Japan. The embryos were raised in E3 embryo medium at 28.5 °C. All experimental procedures were performed in accordance with relevant guidelines and regulations and approved by the University of Miyazaki.
Morpholino injections
Two types of MOs were obtained from Gene Tools, LLC (OR, USA), to knock down the rpl10a gene. The AUG MO (MOaug: 5′-GACCTTGCTCATTTTGGCGTGATAT-3′) contained 13 bp of the 5′ UTR, the translation start site, 2 bp of exon 1 and 7 bp of exon 2 to block Rpl10a protein translation. Splice MOs were designed to target pre-mRNA splicing at exon 5/intron 5 (MOsp: 5′-ATCACAAATATAGACATACCTTCTT-3′). The rps19 MOaug (5′-CACTGTTACACCACCTGGCATCTTG-3′) was used to develop an anemia phenotype described previously24. MOs (MOaug at 0.5 µg/µl; MOsp at 5 µg/µl) were injected into one- or two-cell-stage embryos (wild-type AB line) using an IM-30 Electric Micro-injector (Narishige, Japan). The morphology was observed at 25 and 50 hpf.
In vitro mRNA synthesis for rescue experiments
The full-length coding sequence of rpl10a (GenBank accession number NM_199636.1) was amplified using the specific primers shown in Table 1. The purified PCR product and the pCS2+ vector (provided by Dr. Kunio Inoue, Kobe University, Japan) were digested with the BamHI and EcoRI restriction enzymes (NEB, US), ligated and transformed into DH5α competent E. coli cells (Takara, Japan). The recombinant rpl10a-pCS2+ plasmid was linearized using the NotI restriction enzyme. Then, mRNA was transcribed using an mMESSAGE mMACHINE SP6 Transcription kit (Ambion, CA, USA). A mixture of MOsp (5 µg/µl) and synthesized rpl10a mRNA (400 ng/µl) was used for rescue experiments.
rpl10a CRISPR injection
The crRNA was designed to target exon 5 of rpl10a gene (GenBank accession number NW_001879347) using the CRISPRdirect website (http://crispr.dbcls.jp/) and synthesized by the Integrated DNA Technology (IDT) company. A mixture of crRNA (25 pg), tracrRNA (100 pg), and Cas9 protein (400 pg) was injected into 115 zebrafish embryos at the 1-cell stage. Five embryos at 24 hpf were taken for the heteroduplex mobility assay (HMA), according to Kotani et al.28. The primer sequences for the HMA assay are shown in Table 1.
Semi-quantitative RT-PCR and qRT-PCR
Total RNA was extracted from the wild-type, rpl10a MO-injected and homozygous mutants using TRIzol reagent (Invitrogen, CA, USA). The samples were randomly collected at 24 and 48 hpf. cDNA synthesis was performed using a High Capacity cDNA Transcriptase kit (ABI, CA, USA). Semi-quantitative analysis of rpl10a and rpl38 was carried out using an Expand™ High Fidelity PCR System kit (Roche, Mannheim, Germany). The quantitative real-time PCR analysis was carried out to assess apoptosis-, erythropoiesis-, and germ cell-related mRNAs using 5 ng of cDNA as a template, specific primers at a concentration of 300 nM, and 1x FastStart SYBR Green Master Mix (ABI, CA, USA). The fold changes in tp53, gata1, hbae3, hbbe1, nanos1 and vasa gene expression were determined using the 2−ΔΔCT method. Gene expression was normalized to that of the rpl38 gene. The primer sequences for PCR are shown in Table 1.
Histology
The embryos at 3 dpf were selected for histological analysis. The tails of the embryos were cut from individuals in the fish water with tricaine to confirm their genotypes. Then, the embryos were fixed in 4% paraformaldehyde in PBS for 24 h and then washed with PBS. After rinsing, the embryos were placed in a tissue processor and embedded in paraffin blocks. The paraffin blocks were sectioned using a rotary microtome and placed on a slide. The sections were deparaffinized using toluene, rehydrated with an ethanol gradient and stained with hematoxylin and eosin. The morphology was observed using an Olympus BX53 microscope (Olympus, Tokyo, Japan).
Hemoglobin staining
Embryos at 48 hpf were stained using o-dianisidine to detect active hemoglobin29. Dechorionated embryos were incubated with staining buffer containing 0.6 mg/mL o-dianisidine, 0.01 M sodium acetate pH 4.5, 0.65% H2O2 and 40% ethanol for 7 min in the dark.
Whole mount in situ hybridization
Digoxigenin-labeled RNA probes were synthesized against vasa and nanos1 mRNA. Each PCR product was amplified from wild-type zebrafish cDNA pool using specific primers shown in Table 1. The PCR fragments were purified using a MagExtractor PCR & Gel Clean-up kit (Toyobo, Japan) and ligated into the pGEM®-T Easy vector (Promega, WI, USA). The ligation mixture plasmid was transformed into DH5α competent E. coli cells. In vitro transcription was performed using a T7 or SP6 RNA polymerase kit (Roche, Mannheim, Germany) with linearized plasmid as a template. The zebrafish embryos at 24 hpf were dechorionated and fixed in 4% (w/v) paraformaldehyde (PFA) in 1x PBS overnight at 4 °C. The embryos were dehydrated with 50% methanol in PBS and in 100% methanol. Subsequently, the embryos were rehydrated in 75, 50, and 25% (v/v) methanol in PBS and washed with PBST (1x PBS pH 7.4 + 0.1% Tween 20) for 5 min each. Then, the samples were permeabilized by incubating with 10 µg/mL proteinase K for 10 min. The embryos were soaked in the hybridization buffer for 2 h at 56 °C for prehybridization, and incubated with each RNA probe (30–50 ng) for 18 h. The embryos were rinsed with 2x SSCT in 50% formaldehyde for 1 h at 56 °C. The RNase treatment (RNase A and RNase T1) step was performed over 10 min to remove endogenous RNA. The solution was removed and incubated with 2x SSCT in 50% formaldehyde for 1 h at 56 °C, followed by incubation with 2x SSCT, 0.2x SSCT, and PBS. The nonspecific binding sites were saturated by soaking in blocking buffer at 4 °C for 2 h. Afterward, the embryos were incubated with anti-DIG antibody (1:3,000) in blocking buffer overnight at 4 °C with gentle shaking. On the last day, the samples were washed with PBST and alkaline reaction buffer. An NBT/BCIP substrate was added to detect alkaline phosphatase until the color developed, and the reaction was stopped by rinsing with PBST.
The expression levels of the PGC marker genes were roughly graded into two groups, including abnormal and normal expression. The expression level of the nanos1 and vasa was measured using Fiji analysis software (https://fiji.sc). The image of individual embryos was captured under microscopy. Then, the images were inverted and converted to 8-bit grayscale. A region of interest (ROI) was manually outlined and measured around expression signal at yolk sac extension area of each embryo. Then the ROI with the same shape and the same area was moved to the region that contained no signal. This intensity was determined as a background. The intensity value of the background region was subtracted from the stained region. The intensity of ISH signal was calculated as pixel intensity individually. Due to the variability of the expression signal, the level of intensity was scored as normal and abnormal. An Abnormal level of the gene expression was taken as one with the value less than the mean minus one standard deviation of the gene expression of the wild-type. While the normal level was the mean pixel intensity of the gene expression of the wild-type ± SD. The percentage of normal and abnormal expression of the nanos1 and vasa gene of each group were then determined.
References
Wool, I. G. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21, 164–165 (1996).
Draptchinskaia, N. et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21, 169 (1999).
Gazda, H. T. et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 79, 1110–1118 (2006).
Cmejla, R., Cmejlova, J., Handrkova, H., Petrak, J. & Pospisilova, D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 28, 1178–1182 (2007).
Choesmel, V. et al. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum. Mol. Genet. 17, 1253–1263 (2008).
Farrar, J. E. et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 112, 1582–1592 (2008).
Gazda, H. T. et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 83, 769–780 (2008).
Doherty, L. et al. Ribosomal Protein Genes RPS10 and RPS26 Are Commonly Mutated in Diamond-Blackfan Anemia. Am. J. Hum. Genet. 86, 222–228 (2010).
Gazda, H. T. et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum. Mutat. 33, 1037–1044 (2012).
Dutt, S. et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117, 2567–2576 (2011).
Pfister, A. S. & Kühl, M. Of Wnts and Ribosomes. Prog. Mol. Biol. Transl. Sci. 153, 131–155 (2018).
Chakraborty, A., Uechi, T., Higa, S., Torihara, H. & Kenmochi, N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One 4, e4152 (2009).
Fisicaro, N. et al. Identification of genes downregulated in the thymus by cyclosporin-A: Preliminary characterization of clone CSA-19. Mol. Immunol. 32, 565–572 (1995).
Wonglapsuwan, M., Miyazaki, T., Loongyai, W. & Chotigeat, W. Characterization and biological activity of the ribosomal protein L10a of the white shrimp: Fenneropenaeus merguiensis De Man during vitellogenesis. Mar. Biotechnol. 12, 230–240 (2010).
Palasin, K., Makkapan, W., Thongnoi, T. & Chotigeat, W. Stimulation of ovarian development in white shrimp, Fenneropenaeus merguiensis De Man, with a recombinant ribosomal protein L10a. Aquaculture 432, 38–45 (2014).
Yoon, C., Kawakami, K. & Hopkins, N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124, 3157–3165 (1997).
Köprunner, M., Thisse, C., Thisse, B. & Raz, E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 15, 2877–2885 (2001).
Deshpande, G., Calhoun, G., Yanowitz, J. L. & Schedl, P. D. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell 99, 271–281 (1999).
Brownlie, A. et al. Characterization of embryonic globin genes of the zebrafish. Dev. Biol. 255, 48–61 (2003).
Kongsuwan, K. et al. A Drosophila Minute gene encodes a ribosomal protein. Nature 317, 555 (1985).
Marygold, S. J. et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8, R216 (2007).
Watson, K. L., Konrad, K. D., Woods, D. F. & Bryant, P. J. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA 89, 11302–11306 (1992).
Warner, J. R. & McIntosh, K. B. How common are extra-ribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 (2009).
Uechi, T. et al. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum. Mol. Genet. 17, 3204–3211 (2008).
Costa, L. D. et al. Diamond-Blackfan anemia, ribosome and erythropoiesis. Transfus. Clin. Biol. 17, 112–119 (2010).
Yadav, G. V., Chakraborty, A., Uechi, T. & Kenmochi, N. Ribosomal protein deficiency causes Tp53-independent erythropoiesis failure in zebrafish. Int. J. Biochem. Cell Biol. 49, 1–7 (2014).
Torihara, H. et al. Erythropoiesis failure due to RPS19 deficiency is independent of an activated Tp53 response in a zebrafish model of Diamond-Blackfan anaemia. Br. J. Haematol. 152, 648–654 (2011).
Kotani, H., Taimatsu, K., Ohga, R., Ota, S. & Kawahara, A. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in zebrafish. PLoS One 10, e0128319 (2015).
Detrich, H. W. et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 92, 10713–10717 (1995).
Acknowledgements
We thank Dr. Saya Okui and Mrs. Yukari Nakajima for providing technical assistance. We are sincerely grateful to the Royal Golden Jubilee Graduate Program of the Thailand Research Fund (TRF) (4.J.PS/52/B.1). This work was supported by JSPS KAKENHI Grants 2591003 (N.K.) and JP15K09656 (T.U.).
Author information
Authors and Affiliations
Contributions
N.K., T.U., K.P., M.Y. and W.C. participated in the conception, experiment designing and T.U., K.P. and M.Y. carried out the experiments, statistical analysis. W.C., N.K., T.U., M.Y. and K.P. participated in manuscript preparation. Y.H., N.C. and N.S. provided advice about technical issues and carried out the histology experiment. W.C., N.K., K.P. and T.U. participated in writing the final versions of the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Palasin, K., Uechi, T., Yoshihama, M. et al. Abnormal development of zebrafish after knockout and knockdown of ribosomal protein L10a. Sci Rep 9, 18130 (2019). https://doi.org/10.1038/s41598-019-54544-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54544-w
This article is cited by
-
CRISPR/Cas9-Mediated Transgenesis of the Masu Salmon (Oncorhynchus masou) elovl2 Gene Improves n-3 Fatty Acid Content in Channel Catfish (Ictalurus punctatus)
Marine Biotechnology (2022)
-
Teratogenic and developmental toxic effects of etridiazole on zebrafish (Danio rerio) embryos
Applied Biological Chemistry (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.