Abstract
Infants undergo extensive developments during their first year of life. Although the biological mechanisms involved are not yet fully understood, changes in the DNA methylation in mammals are believed to play a key role. This study was designed to investigate changes in infant DNA methylation that occurs between 6 and 52 weeks. A total of 214 infant saliva samples from 6 or 52 weeks were assessed using principal component analyses and t-distributed stochastic neighbor-embedding algorithms. Between the two time points, there were clear differences in DNA methylation. To further investigate these findings, paired two-sided student’s t-tests were performed. Differently methylated regions were defined as at least two consecutive probes that showed significant differences, with a q-value < 0.01 and a mean difference > 0.2. After correcting for false discovery rates, changes in the DNA methylation levels were found in 42 genes. Of these, 36 genes showed increased and six decreased DNA methylation. The overall DNA methylation changes indicated decreased gene expression. This was surprising because infants undergo such profound developments during their first year of life. The results were evaluated by taking into consideration the extensive development that occurs during pregnancy. During the first year of life, infants have an overall three-fold increase in weight, while the fetus develops from a single cell into a viable infant in 9 months, with an 875-million-fold increase in weight. It is possible that the findings represent a biological slowing mechanism in response to extensive fetal development. In conclusion, our study provides evidence of DNA methylation changes during the first year of life, representing a possible biological slowing mechanism. We encourage future studies of DNA methylation changes in infants to replicate the findings by using a repeated measures model and less stringent criteria to see if the same genes can be found, as well as investigating whether other genes are involved in development during this period.
Similar content being viewed by others
Introduction
In the first year of life, profound changes take place in human physical and neurodevelopmental functioning1. Infants learn to sit, stand up, walk, and say their first words2. This rapid growth occurs in an orderly and regulated sequence, laying the building blocks for future growth1. Although infant developmental milestones are well-known2, the underlying biological mechanisms driving this development in the first year of life are not yet fully understood. Epigenetics is believed to play an important role in mediating the developmental changes in mammalian development3, but to the best of our knowledge, few studies have been published specifically on the epigenetic role in infant development.
Here, DNA methylation is one type of epigenetic mechanism that regulates gene expression without altering the DNA sequence4; it is involved in many cellular processes and is known to be relatively stable. However, DNA methylation might change within an individual over time5. Embryonic and pluripotent stem cell DNA methylation is close to zero6 but changes extensively from fertilization to implantation7. Children from 2 to 16 years of age have been found to have increased levels of the age-related gene DNA methylation, with the greatest changes being found early, from 2 to 10 years of age8.
Two age-specific epigenetic occurrences have been studied: epigenetic drift9 and the epigenetic clock6,10. Epigenetic drift relates to the changes in DNA methylation over time, which differs among individuals11. Examples can be found in twin DNA methylation studies that find that DNA methylation differences increase over time in relation to age and different lifestyles9,12. The epigenetic clock, which was conceived by Horvath6 and Hannum13, is a means to calculate a person’s epigenetic age based on DNA methylation calculated from age-related CpG sites. Studies of age-related DNA methylation changes have mainly been of older populations, which are confounded by decade-long processes of environmental age-effecting exposures and aging itself; studies of younger populations have therefore been encouraged14.
Gene function and its epigenetic regulation are far from completely understood15, but DNA methylation has been associated with the regulation of gene expression16, and increases in DNA methylation have been associated with decreased gene expression17 although it depends on where the DNA methylation occurs in the gene. Gene DNA methylation can be loosely divided into a few different regions: the promoter regions (TSS1500 and TSS200), 5′ untranslated region (UTR), the 1st exon, the gene body, and 3′UTR. It is thought that the promoter regions initiate the transcription of a particular gene16, and it is widely recognized that the DNA methylation of this area is associated with decreased gene expression17.
To the best of our knowledge, there have been no epigenome-wide association studies (EWAS) of DNA methylation changes in infants during the first year of life. Hence, the objectives of the current study were three-fold: (1) to assess whether DNA methylation changes between 6 and 52 weeks (2), and if so, to describe the genes associated with these DNA methylation changes, and (3) to discuss the findings in association with infant development.
Materials and Methods
Ethical approval
This study was approved by the Regional Committees for Medical and Health Research Ethics (REK) in Norway (REK reference number: 2011/560/REK). Informed consent was obtained for infant participation from all mothers. The experiments were performed in accordance with relevant guidelines and regulations.
Participants
A subsample of 172 infants from the “Little in Norway” study18 was used for the current study; for the experiment, 274 saliva samples from infants 6 weeks old (n = 62), 52 weeks old (n = 30), or both (n = 61) were selected. The sociodemographic variables are shown in Table 1. The saliva samples were collected using the Oragene DNA assisted collection kit (OG-575) (DNA Genotek, 2018). Quality control (QC) and cell composition analyses removed 60 saliva samples, leaving 214 saliva samples from 153 infants for epigenetic analyses. The infants’ mothers volunteered information about their age, education level, and marital status. Data on fetal gender were collected from birth records.
Saliva samples and DNA methylation profiling
The 274 infant saliva samples were collected at 6 and 52 weeks using the OG-575 assisted collection kit. DNA methylation profiling was conducted at the Norwegian Sequencing Centre using the Infinium Human Methylation 450 K BeadChip array (Illumina, San Diego, CA, USA). DNA extraction was conducted using the Oragene prep-IT.L2P kit (DNA Genotek, Ottawa, ON, Canada), and the quantity was assessed using PicoGreen (Thermo Fisher, Waltham, MA, USA). The EZ-96 DNA Methylation-Gold Kit (Zymo Research) was used for the bisulfite conversion of 320–500 ng of the saliva DNA samples.
The DNA samples were randomly located on 96-well plates to minimize potential batch effects, and beta-mixture quantile normalization (BMIQ) was used to normalize the β-values19. During QC, 29,233 cross-reactive probes20, 4,232 probes with single nucleotide polymorphisms (SNPs) at the CpG site, 16,819 probes and 13 samples with unreliable measurements (detection p-values > 0.01), 9,675 probes located on the sex chromosomes, and 2,303 non-CpG probes were removed. In total, 18 samples were removed, leaving 256 for cell composition assessment.
Cell composition
Because the human body consists of over 250 different cell types and the epigenome is highly variable between these cell types21, various analyses of infant saliva cell composition were conducted. Previous research has shown that leukocytes and epithelial cells are both found in saliva samples that come from the oral cavities of children (mean age = 6.7 years)22, but to the best of our knowledge, no research has been done on infant saliva cellularity. To assess cellular composition, a small subsample of saliva samples (n = 8) from 6-week-old infants were examined under a microscope. The results showed platelet epithelial cells and bacteria, no immune cells, but as this was only a performed at 6 weeks on a small sample, leucocytes in the samples could not be excluded, the amount of leukocytes in all saliva samples was calculated using the leukocytes methylation for purity (LUMP) analysis23; the results showed that 42 of the 256 samples contained > 10% leukocytes. These DNA samples were excluded from the analyses. This choice of cutoff was made to exclude outliers, while keeping as may samples as possible. In total, 42 samples were removed, leaving 214 samples for the analyses.
Computational analyses
Sociodemographic data analyses were performed using SPSS version 25 (IBM, SPSS Statistics, New York, NY, USA). The raw methylation data preprocessing was done with RnBeads v.1.2.124 using the R programming language (http://www.r-project.org/). The methylation analysis was performed using MATLAB R2017B (The MathWorks Inc., Natick, MA, USA), and principal component analyses (PCA) were performed using Evince (Prediktera AB, Umeå, Sweden).
All statistical tests were done using two-sided student’s t-tests, assuming unequal variance, and any false discovery was corrected (q-value)25. Changes in differentially methylated regions (DMR) between 6 and 52 weeks were defined using the following criteria: q-value < 0.01, mean difference between groups > 0.2, and a minimum of two consecutive significant probes within a gene. All statistical analysis, t-distributed stochastic neighbor embedding (t-SNE), beta-histograms, sample scatter plots, and gene plots were generated using MATLAB version R2017b (The MathWorks, Inc., Natick, MA, USA). The PCA model was generated using Evince (Prediktera AB, Umeå, Sweden).
Results
Study population
The current study was the first to examine changes in DNA methylation between 6 and 52 weeks in infant saliva samples. The study population consisted of 153 Norwegian infants born mainly to mothers with a higher educational status and with stable living conditions26. The characteristic features of these infants at birth were as expected for infants born in Norway, except for the high maternal education status, which was overrepresented in this population27.
Infant saliva samples show homogenous DNA methylation
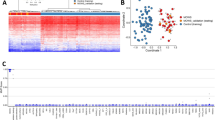
The DNA methylation QC analyses showed that there was minimal heterogeneity in methylation among the cell types assayed and that the methylation levels were more similar for the two time points for a single infant than between infants, as shown in Fig. 1, indicating that the samples were well suited for DNA methylation analyses. The distribution of the beta values across all the samples shows a clear bi-modal distribution, as shown in Fig. 2, which indicates minimal heterogeneity in methylation among the cell types assayed.
Sample scatter plots The sample-to-sample scatter plots display an anticipated cigar-like shape, indicating that the methylation levels for most probes are similar. The left two panels were generated using the same subject, with 6 weeks being displayed on the X-axis and 52 weeks on the Y-axis. The color indicates the probe density. These density plots show no large-scale methylation differences based on the time points, here with 6 weeks shown in blue and 52 weeks shown in red, but there is linear behavior, with most of the probes being located close to the diagonal. There is an increased number of probes that are differently methylated, that is, located far from the diagonal, indicating individual-specific methylation patterns that vary between the infants.
Unsupervised dimensional reduction analyses show separation between time points
The DNA methylation changes in infants between 6 and 52 weeks were analyzed using two separate algorithms (PCA) and t-distributed stochastic neighbor embedding (t-SNE), which are both agnostic to grouping issues. PCA is well-known algorithm used to investigate differences in data, while t-SNE is a newer, nonlinear dimensionality reduction algorithm that investigates the similarities in data28. The two different unsupervised dimensional reduction methods were applied to the samples using all 423,315 probes. Both algorithmic plots showed clear separations between the two time points, as shown in Fig. 3A,B. For PCA, the first three components showed clear separations between the two time points, as shown in Fig. 3A, and the algorithm gave similar results for t-SNE, as shown in Fig. 3B. These results indicate that there is a consistent time point difference and that this difference is greater than the individual differences.
Statistically significant DNA methylation changes found in 42 genes
Stringent analytical criteria were selected to avoid discovering false DNA methylation sites because this was the first time that analyses have been performed to investigate DNA methylation changes for this population. Statistically significant DMRs were found in 42 genes in a total of 101 (out of 423,315) probes. The distribution of the probes within each gene showed consistent increases or decreases in methylation. A total of 36 genes showed increased methylation, and six showed decreased methylation. See Supplementary Table S1 for a detailed description of the results.
Decreased DNA methylation was associated with only 6 genes
In total, six genes had decreased DNA methylation. CLU29 and XAF130 are apoptosis inhibitors, ORAOV1 regulates the cell cycle and apoptosis31, PAQR7 regulates progesterone receptors32, and EIF4E3 promotes messenger RNA transport and proliferation33, while RTP4 has largely unknown functions.
Increased DNA methylation associated with systemic biological processes
Change to ‘Of the 42 genes found to have statistically significant changes in DNA methylation, 13 genes that are associated with intracellular processes. The detailed results for these genes and their associated intracellular functions are presented in Table 2. Of the remaining genes, 24 were previously described as being associated with systemic biological functions 34 and all were found to have increased methylation. The genes and associated systemic functions are presented in Table 3’.
MCHR1
The melanin-concentrating hormone receptor 1 (MCHR1) gene is an example of one of the genes with increased DNA methylation from 6 to 52 weeks. MCHR1 has nine probes on the Illumina Infinium HumanMethylation450 BeadChip array. In these analyses, the results showed increased DNA methylation at two CpG sites: the 1st exon and the body. The mean DNA methylation increase for the two sites had an Δβ value of 0.21. DNA methylation changes in this gene have been associated with schizophrenia34 and bipolar disorder35. The changes in DNA methylation in the CpG site for the 1st exon, cg21342728, found in the current study, were described in the study of schizophrenia34. The DNA methylation changes for MCHR1 are shown in Fig. 4.
Methylation pattern by time point across multiple probes for MCHR1 The graph shows the β-value on the X-axis across multiple probes for each gene. The left Y-axis shows the genomic coordinates for each probe, while the right Y-axis displays the probe Id. The CpG column to the right indicates the CpG island and the gene column in the gene body where the probe is from. For each probe, the methylation level is illustrated by a box plot, where the box is the interquartile range, and the median is the dot for the 6-week group (blue) and the 52-week group (red). Significant probes are shown with * for q < 0.01 and a difference > 0.2 and ** for q < 0.001 and difference > 0.3. The significant methylation probes for MCHR1 are located in the 1st exon and in the body and are not found in a known CpG island.
SLC1A2
The solute carrier family 1 member 2 (SLC1A2) gene is another example of a gene with increased DNA methylation from 6 to 52 weeks. This gene has been found to have DNA methylation differences associated with schizophrenia and bipolar disorder36 and prematurity in infants 37. It has 32 probes on the Illumina Infinium HumanMethylation450 BeadChip array. The analyses showed a significant increase in methylation in the first three adjacent CpG sites in the promoter region (TSS1500) from 6 to 52 weeks. The mean DNA methylation increase of the three sites had an Δβ value of 0.29, and the largest difference was found at cg10159951, with a mean DNA methylation at 6 weeks of 0.12 and at 52 weeks of 0.45. The DNA methylation difference in the CpG site of the promoter island, cg25963980, has previously been associated with DNA methylation changes associated with infant prematurity 37. The DNA methylation changes for SLC1A2 are shown in Fig. 5.
Methylation pattern by time point across multiple probes for SLC1A2 The graph shows the β-value on the X-axis across multiple probes for each gene. The left Y-axis shows the genomic coordinates for each probe, while the right Y-axis displays the probe Id. The CpG column to the right indicates the CpG island and the gene column in the gene body where the probe is from. For each probe, the methylation level is illustrated by a box plot, where the box is the interquartile range, and the median is the dot for the 6-week group (blue) and the 52-week group (red). Significant probes are shown with * for q < 0.01 and difference > 0.2 and ** for q < 0.001 and difference > 0.3. The significant methylation probes for SLC1A2 are located in the promoter area (TSS1500).
Discussion
To the best of our knowledge, we conducted the first EWAS using Illumina450K analyses of infant saliva samples to study DNA methylation changes between 6 and 52 weeks of age. Two very different and unsupervised dimensionality algorithms—PCA and t-SNE—were used in the current study. The results showed clear separations between infant DNA methylation at the two studied time points. The greatest consistent difference is the time point and nonindividual differences. Because both the PCA and t-SNE showed the same separation in infant DNA methylation between the two time points, the findings indicate that the biological mechanisms associated with normal infant development in the first year of life are associated with DNA methylation changes.
To better understand which genes were associated with these changes, we analyzed the DNA methylation further and found that there were 42 genes across 101 probes with statistically significant DNA methylation changes. Of the 42 genes, 36 had increased DNA methylation, and six had decreased methylation levels. The effect of DNA methylation depends on where it occurs in the gene, and the DNA methylation changes in the current study occurred in different parts of the genes but most commonly were associated with decreased gene expression17.
The suggested decreased gene expression was unexpected as infants experience rapid developmental growth during the first year of life. However, infant development in the first year of life is relatively limited compared with the changes that occur during pregnancy38. The fetus develops from a single cell to a viable infant in 9 months, with an 875-million-fold increase in weight39,40; meanwhile, during the first year of life, the infant only experiences a three-fold increase in weight2. Therefore, although our findings are limited to buccal epithelial cells, they suggest a biological growth-slowing mechanism post-birth after the rapid fetal growth during pregnancy.
Our study has several strengths and limitations that need to be considered when interpreting the results. One important limitation for DNA methylation analyses is cell composition. It would have been better if the saliva sample at both 6 weeks and 52 weeks had been assessed. To compensate for this, LUMP scores were calculated for all saliva samples, finding leucocytes in the samples and using a cutoff of 10% for the analyses. However, if possible, further studies should consider assessing sample cell composition from all time points included in the analyses. Other limitations of the current study were that the population was made up of an overrepresentation of mothers with a high level of education compared with the general Norwegian population, and the study did not compare the characteristics of the whole “Little in Norway” cohort with the sample analyzed in the current study to assess selection bias and understand whether the findings are generalizable to the whole population.
One of the major strengths of the current study was that the saliva samples came from two time points, but still, these results cannot answer the question of whether DNA methylation fluctuates over the first year or if there is only an overall increase in DNA methylation. Therefore, to assess this, future studies should consider collecting more than two saliva samples over the first year of life. Another major strength was that the current study was based on the methodology used in cancer research because this field is at the forefront of epigenetic research, and human biology is the same for both cancer and biological development. Applying cutting-edge bioinformatical methods28 used in cancer research to examine biological mechanisms, 42 genes were found to have DNA methylation changes associated with early-life biological development. Of these, 14 had previously been found in different forms of cancers associated with epigenetic changes41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58, yet here, only two studies found epigenetic associations with development37,59. This might be because these genes are all associated with cancer, but it is just as likely that more cancer research is being conducted because of public and political efforts that increase cancer research funding.
The bioinformatic analysis of the epigenetic data commonly includes a determination of the significant differences at a single CpG site, considering them independently of each other and adjusting for false discovery rates60. Because this was the first study to investigate DNA methylation changes between 6 and 52 weeks, we wanted to avoid false positive results; therefore, we set stricter criteria for the significance of DMRs than what has been commonly used. If we had used other criteria, other significant DNA methylation changes would have been identified, but we wanted all of the discovered CpG sites to be correct and the probability of false positive findings to be small. Using this analytical model, we found 42 CpG sites with infant DNA methylation changes, but different analytical models for the analyses could reveal different results. Our choice of analytical method limited the possibility of correcting for confounders, such as gender. Future analyses should consider choosing a repeated measures model, so confounding variables can be adjusted for, this way confounding variables such as gender should be addressed. Hence, we encourage future studies of DNA methylation changes in infants to replicate the findings by using a repeated measures model and less stringent criteria to see if the same genes can be found, as well as investigating whether other genes are involved in development during this period.
Conclusion
In conclusion, the algorithmic analyses showed that infant DNA methylation displays clear differences between 6 and 52 weeks. To investigate these differences further, two-sided student’s t-tests were performed. These analyses found 42 genes associated with DNA methylation changes. Of these, 36 genes showed increased and six decreased DNA methylation. The methylation changes indicated an overall decrease in gene expression, which, in turn, might represent a slowing mechanism to reduce the extensive growth development that occurs during pregnancy. Future studies of DNA methylation changes in infants could use repeated measures models and less stringent criteria to see if the same genes can be replicated, and whether other genes are involved in development during this period.
Data availability
Data available upon request from vibeke.moe@psykologi.uio.no
References
Bornstein, M. H. et al. Child development in developing countries: introduction and methods. Child Developemnt 83, 16–31, https://doi.org/10.1111/j.1467-8624.2011.01671.x (2012).
Johnson, C. P. & Blasco, P. A. Infant growth and development. Pediatrics in review 18, 224–242 (1997).
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nature Reviews Genetics 14, 204–220, https://doi.org/10.1038/nrg3354 (2013).
Razin, A. CpG methylation, chromatin structure and gene silencing-a three-way connection. The EMBO journal 17, 4905–4908, https://doi.org/10.1093/emboj/17.17.4905 (1998).
Bjornsson, H. T. et al. Intra-individual change over time in DNA methylation with familial clustering. Jama 299, 2877–2883, https://doi.org/10.1001/jama.299.24.2877 (2008).
Horvath, S. DNA methylation age of human tissues and cell types. Genome biology 14, R115, https://doi.org/10.1186/gb-2013-14-10-r115 (2013).
Kawai, K. et al. Parental age and gene expression profiles in individual human blastocysts. Scientific reports 8, 2380, https://doi.org/10.1038/s41598-018-20614-8 (2018).
Gervin, K. et al. Intra-individual changes in DNA methylation not mediated by cell-type composition are correlated with aging during childhood. Clinical epigenetics 8, 110, https://doi.org/10.1186/s13148-016-0277-3 (2016).
Fraga, M. F. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America (PNAS) 102, 10604–10609, https://doi.org/10.1073/pnas.0500398102 (2005).
Zhang, Q. et al. Improved prediction of chronological age from DNA methylation limits it as a biomarker of ageing. bioRxiv https://doi.org/10.1101/327890 (2018).
Teschendorff, A. E., West, J. & Beck, S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Human molecular genetics 22, R7–r15, https://doi.org/10.1093/hmg/ddt375 (2013).
Segal, N. L., Montoya, Y. S., Loke, Y. J. & Craig, J. M. Identical twins doubly exchanged at birth: a case report of genetic and environmental influences on the adult epigenome. Epigenomics 9, 5–12, https://doi.org/10.2217/epi-2016-0104 (2017).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular cell 49, 359–367, https://doi.org/10.1016/j.molcel.2012.10.016 (2013).
Suarez, A. et al. The epigenetic clock and pubertal, neuroendocrine, psychiatric, and cognitive outcomes in adolescents. Clinical epigenetics 10, 96, https://doi.org/10.1186/s13148-018-0528-6 (2018).
Torres, S. M. et al. DNA repair variants, indoor tanning, and risk of melanoma. Pigment Cell Melanoma Res 26, 677–684, https://doi.org/10.1111/pcmr.12117 (2013).
Bibikova, M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295, https://doi.org/10.1016/j.ygeno.2011.07.007 (2011).
Yaniv, M. Chromatin remodeling: from transcription to cancer. Cancer genetics 207, 352–357, https://doi.org/10.1016/j.cancergen.2014.03.006 (2014).
Sanner, N., Smith, L., Wentzel-Larsen, T. & Moe, V. Early identification of social-emotional problems: Applicability of the Infant-Toddler Social Emotional Assessment (ITSEA) at its lower age limit. Infant Behavior and Development 42, 69–85, https://doi.org/10.1016/j.infbeh.2015.11.001 (2016).
Teschendorff, A. E. et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics (Oxford, England) 29, 189–196, https://doi.org/10.1093/bioinformatics/bts680 (2013).
Chen, Y. A. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209, https://doi.org/10.4161/epi.23470 (2013).
Tammen, S. A., Friso, S. & Choi, S. W. Epigenetics: the link between nature and nurture. Molecular aspects of medicine 34, 753–764, https://doi.org/10.1016/j.mam.2012.07.018 (2013).
Theda, C. et al. Quantitation of the cellular content of saliva and buccal swab samples. Scientific reports 8, 6944, https://doi.org/10.1038/s41598-018-25311-0 (2018).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nature communications 6, 8971, https://doi.org/10.1038/ncomms9971 (2015).
Assenov, Y. et al. Comprehensive analysis of DNA methylation data with RnBeads. Nature methods 11, 1138–1140, https://doi.org/10.1038/nmeth.3115 (2014).
Storey, J. D. A direct approach to false discovery rates. Journal of the Royal Statistical Society 64, 479–498 (2002).
Moe, V., von Soest, T., Fredriksen, E., Olafsen, K. S. & Smith, L. The Multiple Determinants of Maternal Parenting Stress 12 Months After Birth: The Contribution of Antenatal Attachment Style, Adverse Childhood Experiences, and Infant Temperament. Frontiers in psychology 9, 1987, https://doi.org/10.3389/fpsyg.2018.01987 (2018).
Statistics Norway. Befolkningens utdanningsnivå., <https://www.ssb.no/utniv/> (2017).
van der Maaten, L. J. P. & H., G. E. Visualizing High-Dimensional Data Using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008).
Rohne, P., Prochnow, H. & Koch-Brandt, C. The CLU-files: disentanglement of a mystery. Biomol Concepts 7, 1–15, https://doi.org/10.1515/bmc-2015-0026 (2016).
Wang, Y., Liu, P., Wang, X. & Mao, H. Role of Xlinked inhibitor of apoptosisassociated factor1 in vasculogenic mimicry in ovarian cancer. Mol Med Rep 16, 325–330, https://doi.org/10.3892/mmr.2017.6597 (2017).
Jiang, L. et al. Oral cancer overexpressed 1 (ORAOV1) regulates cell cycle and apoptosis in cervical cancer HeLa cells. Mol Cancer 9, 20, https://doi.org/10.1186/1476-4598-9-20 (2010).
Pang, Y., Dong, J. & Thomas, P. Characterization, neurosteroid binding and brain distribution of human membrane progesterone receptors delta and {epsilon} (mPRdelta and mPR{epsilon}) and mPRdelta involvement in neurosteroid inhibition of apoptosis. Endocrinology 154, 283–295, https://doi.org/10.1210/en.2012-1772 (2013).
Osborne, M. J. et al. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proceedings of the National Academy of Sciences of the United States of America 110, 3877–3882, https://doi.org/10.1073/pnas.1216862110 (2013).
Liu, J. et al. The association of DNA methylation and brain volume in healthy individuals and schizophrenia patients. Schizophr Res 169, 447–452, https://doi.org/10.1016/j.schres.2015.08.035 (2015).
Dempster, E. L. et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Human molecular genetics 20, 4786–4796, https://doi.org/10.1093/hmg/ddr416 (2011).
Shao, L. & Vawter, M. P. Shared gene expression alterations in schizophrenia and bipolar disorder. Biological psychiatry 64, 89–97, https://doi.org/10.1016/j.biopsych.2007.11.010 (2008).
Sparrow, S. et al. Epigenomic profiling of preterm infants reveals DNA methylation differences at sites associated with neural function. Translational psychiatry 6, e716, https://doi.org/10.1038/tp.2015.210 (2016).
Moore, K., Persaud, T. V. N. & Torchia, M. The Developing Human: Clinically Oriented Embryology 10th Edition edn, (Elsevier, 2015).
P B. Júlíusson, et al. Growth charts for Norwegian children. Tiddskriftet for Den Norske Legeforening, 281-286, https://doi.org/10.4045/tidsskr.09.32473 (2009).
Khadilkar, V. & Khadilkar, A. Growth charts: A diagnostic tool. Indian journal of endocrinology and metabolism 15(Suppl 3), S166–171, https://doi.org/10.4103/2230-8210.84854 (2011).
Kiehl, S. et al. Epigenetic silencing of downstream genes mediated by tandem orientation in lung cancer. Sci Rep 7, 3896, https://doi.org/10.1038/s41598-017-04248-w (2017).
Bhanvadia, R. R. et al. MEIS1 and MEIS2 Expression and Prostate Cancer Progression: A Role For HOXB13 Binding Partners in Metastatic Disease. Clinical cancer research: an official journal of the American Association for Cancer Research, https://doi.org/10.1158/1078-0432.ccr-17-3673 (2018).
Yu, M. H. & Zhang, W. TEAD1 enhances proliferation via activating SP1 in colorectal cancer. Biomed Pharmacother 83, 496–501, https://doi.org/10.1016/j.biopha.2016.06.058 (2016).
Alholle, A. et al. Genome-wide DNA methylation profiling of recurrent and non-recurrent chordomas. Epigenetics 10, 213–220, https://doi.org/10.1080/15592294.2015.1006497 (2015).
Zacharopoulou, N. et al. The Epigenetic Factor KDM2B Regulates EMT and Small GTPases in Colon Tumor Cells. Cell Physiol Biochem 47, 368–377, https://doi.org/10.1159/000489917 (2018).
Cimmino, L. & Aifantis, I. Fingerprinting acute leukemia: DNA methylation profiling of B-acute lymphoblastic leukemia. Cancer Discov 2, 976–978, https://doi.org/10.1158/2159-8290.cd-12-0435 (2012).
Tritz, R. et al. FAPP2 gene downregulation increases tumor cell sensitivity to Fas-induced apoptosis. Biochem Biophys Res Commun 383, 167–171, https://doi.org/10.1016/j.bbrc.2009.03.126 (2009).
Wu, W. et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology 144, 956–966.e954, https://doi.org/10.1053/j.gastro.2013.01.019 (2013).
Kober, P., Bujko, M., Oledzki, J., Tysarowski, A. & Siedlecki, J. A. Methyl-CpG binding column-based identification of nine genes hypermethylated in colorectal cancer. Mol Carcinog 50, 846–856, https://doi.org/10.1002/mc.20763 (2011).
Xu, Y., Zhao, S., Cui, M. & Wang, Q. Down-regulation of microRNA-135b inhibited growth of cervical cancer cells by targeting FOXO1. Int J Clin Exp Pathol 8, 10294–10304 (2015).
Martínez-Cardús, A., et al. Epigenetic Homogeneity Within Colorectal Tumors Predicts Shorter Relapse-free and Overall Survival Times for Patients With Loco-regional Cancer. Gasteroenterology (2016).
Lang, Z. Q., Wu, Y. Q., Pan, X. B., Qu, G. M. & Zhang, T. G. The identification of multifocal breast cancer-associated long non-coding RNAs. Eur Rev Med Pharmacol Sci 21, 5648–5654, https://doi.org/10.26355/eurrev_201712_14008 (2017).
Schluckebier, L. et al. XAF1 expression levels in a non-small cell lung cancer cohort and its potential association with carcinogenesis. Oncol Rep 38, 402–410, https://doi.org/10.3892/or.2017.5680 (2017).
Yu, J. et al. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene 36, 182–193, https://doi.org/10.1038/onc.2016.187 (2017).
Liu, D., Shen, X., Zhu, G. & Xing, M. REC8 is a novel tumor suppressor gene epigenetically robustly targeted by the PI3K pathway in thyroid cancer. Oncotarget 6, 39211–39224, https://doi.org/10.18632/oncotarget.5391 (2015).
Kettunen, E. et al. Asbestos-associated genome-wide DNA methylation changes in lung cancer. Int J Cancer 141, 2014–2029, https://doi.org/10.1002/ijc.30897 (2017).
Deb, M. et al. Clusterin gene is predominantly regulated by histone modifications in human colon cancer and ectopic expression of the nuclear isoform induces cell death. Biochim Biophys Acta 1852, 1630–1645, https://doi.org/10.1016/j.bbadis.2015.04.021 (2015).
Rizzi, F. & Bettuzzi, S. Clusterin (CLU) and prostate cancer. Adv Cancer Res 105, 1–19, https://doi.org/10.1016/s0065-230x(09)05001-5 (2009).
Agha, G. et al. Birth weight-for-gestational age is associated with DNA methylation at birth and in childhood. Clinical epigenetics 8, 118, https://doi.org/10.1186/s13148-016-0285-3 (2016).
Bock, C. Analysing and interpreting DNA methylation data. Nature reviews. Genetics 13, 705–719, https://doi.org/10.1038/nrg3273 (2012).
Chahdi, A. & Sorokin, A. Protein kinase A-dependent phosphorylation modulates beta1Pix guanine nucleotide exchange factor activity through 14-3-3beta binding. Mol Cell Biol 28, 1679–1687, https://doi.org/10.1128/mcb.00898-07 (2008).
Yu, H. W. et al. beta-PIX controls intracellular viscoelasticity to regulate lung cancer cell migration. J Cell Mol Med 19, 934–947, https://doi.org/10.1111/jcmm.12441 (2015).
Chibnik, L. B. et al. Alzheimer’s loci: epigenetic associations and interaction with genetic factors. Ann Clin Transl Neurol 2, 636–647, https://doi.org/10.1002/acn3.201 (2015).
Kajiyama, Y., Tian, J. & Locker, J. Regulation of alpha-fetoprotein expression by Nkx2.8. Mol Cell Biol 22, 6122–6130 (2002).
Ring, H. Z. et al. The human neuregulin-2 (NRG2) gene: cloning, mapping and evaluation as a candidate for the autosomal recessive form of Charcot-Marie-Tooth disease linked to 5q. Hum Genet 104, 326–332 (1999).
Funato, Y. & Miki, H. Nucleoredoxin, a novel thioredoxin family member involved in cell growth and differentiation. Antioxid Redox Signal 9, 1035–1057, https://doi.org/10.1089/ars.2007.1550 (2007).
Pomeroy, E. J. et al. Ras oncogene-independent activation of RALB signaling is a targetable mechanism of escape from NRAS(V12) oncogene addiction in acute myeloid leukemia. Oncogene 36, 3263–3273, https://doi.org/10.1038/onc.2016.471 (2017).
Ward, A., Hopkins, J., McKay, M., Murray, S. & Jordan, P. W. Genetic Interactions Between the Meiosis-Specific Cohesin Components, STAG3, REC8, and RAD21L. G3 (Bethesda) 6, 1713–1724, https://doi.org/10.1534/g3.116.029462 (2016).
Levine, D. M. et al. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett’s esophagus. Nat Genet 45, 1487–1493, https://doi.org/10.1038/ng.2796 (2013).
Koshino, I., Mohandas, N. & Takakuwa, Y. Identification of a novel role for dematin in regulating red cell membrane function by modulating spectrin-actin interaction. J Biol Chem 287, 35244–35250, https://doi.org/10.1074/jbc.M111.305441 (2012).
Hoshi, Y., Uchida, Y., Tachikawa, M., Ohtsuki, S. & Terasaki, T. Actin filament-associated protein 1 (AFAP-1) is a key mediator in inflammatory signaling-induced rapid attenuation of intrinsic P-gp function in human brain capillary endothelial cells. J Neurochem 141, 247–262, https://doi.org/10.1111/jnc.13960 (2017).
Rak, J. et al. Cytohesin 1 regulates homing and engraftment of human hematopoietic stem and progenitor cells. Blood 129, 950–958, https://doi.org/10.1182/blood-2016-06-720649 (2017).
Sharma, M. D. et al. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity 38, 998–1012, https://doi.org/10.1016/j.immuni.2013.01.013 (2013).
Finta, C. & Zaphiropoulos, P. G. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem 277, 5882–5890, https://doi.org/10.1074/jbc.M109175200 (2002).
Habano, W. et al. Analysis of DNA methylation landscape reveals the roles of DNA methylation in the regulation of drug metabolizing enzymes. Clinical epigenetics 7, 105, https://doi.org/10.1186/s13148-015-0136-7 (2015).
Vieira, O. V. et al. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin-Darby canine kidney (MDCK) cells. Proceedings of the National Academy of Sciences of the United States of America 103, 18556–18561, https://doi.org/10.1073/pnas.0608291103 (2006).
Villaescusa, J. C. et al. A PBX1 transcriptional network controls dopaminergic neuron development and is impaired in Parkinson’s disease. Embo j 35, 1963–1978, https://doi.org/10.15252/embj.201593725 (2016).
Gordon, J. A. et al. Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs 194, 146–150, https://doi.org/10.1159/000324790 (2011).
Demerath, E. W. et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Human molecular genetics 24, 4464–4479, https://doi.org/10.1093/hmg/ddv161 (2015).
Al Muftah, W. A. et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clinical epigenetics 8, 13, https://doi.org/10.1186/s13148-016-0177-6 (2016).
Karaca, E. et al. Rare variants in the notch signaling pathway describe a novel type of autosomal recessive Klippel-Feil syndrome. Am J Med Genet A 167a, 2795–2799, https://doi.org/10.1002/ajmg.a.37263 (2015).
Sutton, A. L., Zhang, X., Ellison, T. I. & Macdonald, P. N. The 1,25(OH)2D3-regulated transcription factor MN1 stimulates vitamin D receptor-mediated transcription and inhibits osteoblastic cell proliferation. Mol Endocrinol 19, 2234–2244, https://doi.org/10.1210/me.2005-0081 (2005).
Panni, T. et al. Genome-Wide Analysis of DNA Methylation and Fine Particulate Matter Air Pollution in Three Study Populations: KORA F3, KORA F4, and the Normative Aging Study. Environmental health perspectives 124, 983–990, https://doi.org/10.1289/ehp.1509966 (2016).
Wei, B. & Jin, J. P. TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships. Gene 582, 1–13, https://doi.org/10.1016/j.gene.2016.01.006 (2016).
Agrawal, P. B. et al. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am J Hum Genet 95, 218–226, https://doi.org/10.1016/j.ajhg.2014.07.004 (2014).
Shah, S. et al. Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome research 24, 1725–1733, https://doi.org/10.1101/gr.176933.114 (2014).
Shandilya, J., Medler, K. F. & Roberts, S. G. Regulation of AURORA B function by mitotic checkpoint protein MAD2. Cell Cycle 15, 2196–2201, https://doi.org/10.1080/15384101.2016.1200773 (2016).
Marsh, D. J. et al. Melanin-concentrating hormone 1 receptor-deficient mice are lean, hyperactive, and hyperphagic and have altered metabolism. Proceedings of the National Academy of Sciences of the United States of America 99, 3240–3245, https://doi.org/10.1073/pnas.052706899 (2002).
Shimomura, Y. et al. Isolation and identification of melanin-concentrating hormone as the endogenous ligand of the SLC-1 receptor. Biochem Biophys Res Commun 261, 622–626, https://doi.org/10.1006/bbrc.1999.1104 (1999).
Humbert, N. et al. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. Embo j 29, 376–386, https://doi.org/10.1038/emboj.2009.342 (2010).
Schneider, E., El Hajj, N., Muller, F., Navarro, B. & Haaf, T. Epigenetic Dysregulation in the Prefrontal Cortex of Suicide Completers. Cytogenet Genome Res 146, 19–27, https://doi.org/10.1159/000435778 (2015).
Fragoso, Y. D. et al. Expression in the human brain of retinoic acid induced 1, a protein associated with neurobehavioural disorders. Brain Struct Funct 220, 1195–1203, https://doi.org/10.1007/s00429-014-0712-1 (2015).
Elsea, S. H. & Girirajan, S. Smith-Magenis syndrome. Eur J Hum Genet 16, 412–421, https://doi.org/10.1038/sj.ejhg.5202009 (2008).
Aleksovska, K. et al. Systematic review and meta-analysis of circulating S100B blood levels in schizophrenia. PloS one 9, e106342, https://doi.org/10.1371/journal.pone.0106342 (2014).
He, Z., Wang, K. C., Koprivica, V., Ming, G. & Song, H. J. Knowing how to navigate: mechanisms of semaphorin signaling in the nervous system. Sci STKE 2002, re1, https://doi.org/10.1126/stke.2002.119.re1 (2002).
Sheldon, A. L. & Robinson, M. B. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem Int 51, 333–355, https://doi.org/10.1016/j.neuint.2007.03.012 (2007).
Edvardson, S. et al. Infantile neurodegenerative disorder associated with mutations in TBCD, an essential gene in the tubulin heterodimer assembly pathway. Human molecular genetics 25, 4635–4648, https://doi.org/10.1093/hmg/ddw292 (2016).
Geraghty, A. A. et al. A Low Glycaemic Index Diet in Pregnancy Induces DNA Methylation Variation in Blood of Newborns: Results from the ROLO Randomised Controlled Trial. Nutrients 10, https://doi.org/10.3390/nu10040455 (2018).
Guan, W. J. et al. Spinocerebellar ataxia type 35 (SCA35)-associated transglutaminase 6 mutants sensitize cells to apoptosis. Biochem Biophys Res Commun 430, 780–786, https://doi.org/10.1016/j.bbrc.2012.11.069 (2013).
Zilhao, N. R. et al. Epigenome-Wide Association Study of Tic Disorders. Twin research and human genetics: the official journal of the International Society for Twin Studies 18, 699–709, https://doi.org/10.1017/thg.2015.72 (2015).
Agoston, Z. et al. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development 141, 28–38, https://doi.org/10.1242/dev.097295 (2014).
Berry, D. C., Croniger, C. M., Ghyselinck, N. B. & Noy, N. Transthyretin blocks retinol uptake and cell signaling by the holo-retinol-binding protein receptor STRA6. Molecular and cellular biology 32, 3851–3859, https://doi.org/10.1128/mcb.00775-12 (2012).
Yamagata, Y. et al. Genome-wide DNA methylation profiling in cultured eutopic and ectopic endometrial stromal cells. PloS one 9, e83612, https://doi.org/10.1371/journal.pone.0083612 (2014).
Acknowledgements
The Fulbright PhD Scholarship Program, Eckbos Legat, The Research Council of Norway (Grant # 196156), RBUP East and South, Oslo, Norway, and biostatistician Christian Page for his advice on our work.
Author information
Authors and Affiliations
Contributions
E.W. drafted the manuscript. A.B. undertook the bioinformatics analyses. E.W. and A.B. interpreted the results. V.M. and L.S. designed the Little in Norway study and collected the data. E.W., A.B., L.S., V.M., and E.R.H. reviewed and revised the manuscript. E.W., A.B., L.S., V.M., and E.R.H. gave final approval for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wikenius, E., Moe, V., Smith, L. et al. DNA methylation changes in infants between 6 and 52 weeks. Sci Rep 9, 17587 (2019). https://doi.org/10.1038/s41598-019-54355-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-54355-z
This article is cited by
-
Mild internet use is associated with epigenetic alterations of key neurotransmission genes in salivary DNA of young university students
Scientific Reports (2023)
-
Disrupted methylation patterns at birth persist in early childhood: a prospective cohort analysis
Clinical Epigenetics (2022)
-
Regulation of oxytocin receptor gene expression in obsessive–compulsive disorder: a possible role for the microbiota-host epigenetic axis
Clinical Epigenetics (2022)
-
Birthweight DNA methylation signatures in infant saliva
Clinical Epigenetics (2021)
-
The regulation mechanisms and the Lamarckian inheritance property of DNA methylation in animals
Mammalian Genome (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.