Abstract
As we approach the end of the antibiotic era, newer therapeutic options, such as antimicrobial peptides (AMPs), are in urgent demand. AMP surface grafting onto biomaterials has been described as a good strategy to overcome problems associated with their in vivo stability. Helicobacter pylori is among the bacteria that pose greatest threat to human health, being MSI-78A one of the few bactericidal AMPs against this bacterium. Here, we report that MSI-78A grafted onto model surfaces (Self-Assembled Monolayers –SAMs), in a concentration of 30.3 ± 1.2 ng/cm2 determined by quartz crystal microbalance with dissipation (QCM-D), was able to kill, by contact, 98% of planktonic H. pylori in only 2 h. This fact was not verified against the control bacteria (Staphylococcus epidermidis), although the minimal inhibitory concentration (MIC) of MSI-78A in solution is much lower for S. epidermidis (2 μg/mL) than for H. pylori (64 μg/mL). Our results also demonstrated that, in opposite to other bacteria, H. pylori cells were attracted to ethylene glycol terminated (antiadhesive) surfaces, which can explain the high bactericidal potential of grafted MSI-78A. This proof of concept study establishes the foundations for development of MSI-78A grafted nanoparticles for gastric infection management within a targeted nanomedicine concept.
Similar content being viewed by others
Introduction
Helicobacter pylori (H. pylori) is a Gram negative bacterium that infects more than 50% of the worldwide population1. This bacterium is the etiological agent of several gastro-duodenal diseases, such as chronic gastritis and peptic ulcer disease, but it is also responsible for 75% of the global gastric cancer burden1,2. To date, gastric cancer is the 5th most common and the 3rd deadliest cancer worldwide3,4. Consequently, H. pylori eradication from infected people is the best option to circumvent the infection outcomes. The available therapeutic scheme is based on the combination of at least two antibiotics plus a proton-pump inhibitor but it fails in 10–40% of the cases5,6. The failure of the therapeutic regimen is mostly due to the high rates of bacterial resistance to antibiotics1,7,8. In fact, the World Health Organization has placed H. pylori among the 16 antibiotic-resistant bacteria that pose the greatest threat to human health4. Therefore, innovative strategies based on non-antibiotic drugs are urgently required for gastric infection management.
Antimicrobial peptides (AMPs) are low molecular weight peptides that are widely distributed in living organisms as part of their immune system9,10,11,12. AMPs are an appealing alternative to the conventional antibiotic therapies, presenting low tendency to induce bacterial resistance, once they induce selective damage to bacterial membranes through mechanisms that bacteria find difficult to evade11,13,14,15,16,17. Up until now, more than 5,000 AMPs have been discovered or synthesized17,18,19 but only a few have been described to have anti-H. pylori activity, namely: Odorranain-HP20 and Magainin-221,22. MSI-78, commercially known as Pexiganan, is a 22-amino acid peptide Magainin-2 analogue, constructed through a series of amino acid substitutions and deletions in order to make the naturally occurring Magainin-2 more active21,22, namely against H. pylori23. In in vivo settings, “unbound AMPs” can undergo proteolysis and peptide aggregation, leading to a decrease in activity11. An advocated strategy to bypass these drawbacks is to immobilize AMPs with anti-H. pylori activity, as commonly performed with other antimicrobial peptides11. It is thought that immobilization would confer protection against enzymatic degradation in vivo and prevent aggregation, therefore increasing the AMPs long-term stability, which would then enhance activity and avoid the toxicity-issues associated with the use of high AMP concentrations to achieve biological effect11,24. So far, MSI-78 encapsulation in nanoparticles for H. pylori infection treatment has been studied25 but no strategy concerning the AMP surface grafting onto nanoparticles for gastric infection management has been reported.
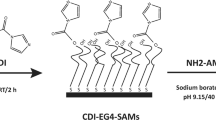
Planning the future development of a bioengineered non-antibiotic therapy against H. pylori based on surface-immobilized AMPs, this work aimed to access if, after surface-grafted, AMPs were able to retain its bioactivity against H. pylori. For that, model surfaces (Self-assembled monolayers - SAMs) were used in this proof of concept study. SAMs are easy to prepare, functionalize and control at molecular scale, being compatible with many surface characterization techniques used for AMP grafting detection26,27. Furthermore, SAMs have been previously used by us to demonstrate the specific recognition between H. pylori adhesins (BabA and SabA) and surface grafted glycans (Lewis b and sialyl-Lewis x)28, being afterwards this knowledge translated onto biocompatible polymers (chitosan microspheres)29. AMPs described in the literature as active against H. pylori were selected, namely: Odonorrain-HP20, MSI-78 (pexiganan)25 and MSI-78A21,22,23, which is derived from the MSI-78 by replacement of one amino acid (G13A). These AMPs were firstly screened in solution. Then, the most active AMP was synthesized with an extra cysteine (-SH) and with an aminohexanoic acid (ahx) spacer at either the N- or C-terminus. This modification allowed controlling both AMP orientation and exposure once grafted onto a surface. Moreover, the use of a maleimide-terminated polyethylene glycol (EG11) spacer also enhanced AMP exposure and permitted its covalent binding. The AMP immobilization strategy onto model surfaces was based on the thiol-maleimide chemistry and is described in Fig. 1.
AMP immobilization onto model surfaces (biotin-SAMs). Schematic representation, not to scale. (a) Mixed SAMs of biotin and tetraethylene glycol (EG4)-terminated thiols (biotin-SAMs) were previously prepared and thoroughly characterized by us28; (b) Neutravidin, which strongly binds to biotin moieties protruding from the SAMs surface, was used as a protein-bridge for binding the heterobifunctional biotin-polyethyleneglycol-11-maleimide spacer (EG11-MAL); (c) Biotin in one end of the spacer binds to the previously immobilized neutravidin on the biotin-SAMs surface; (d) The maleimide group (MAL) on the opposite terminal of the spacer allows the binding of the AMP (–SH) groups (thiol-maleimide chemistry).
Quartz Crystal Microbalance with Dissipation (QCM-D) was used to quantify the immobilized AMP mass on the engineered model surfaces. The in vitro activity of AMP-SAMs was tested using the highly pathogenic H. pylori J99 strain. Staphylococcus epidermidis ATCC 35984 strain was selected as control for bacterial adhesion onto SAMs, since its surface adhesion behaviour is well-known, namely its ability to adhere to most surfaces with exception of the non-fouling ones30,31.
Results
AMPs activity in solution
In Table 1, MIC and MBC values for the tested AMPs against the tested H. pylori strains are presented.
In our experimental settings, Odorranain-HP didn’t have antibacterial activity. MSI-78A had better antibacterial performance than MSI-78 and therefore, it underwent further modifications with an additional cysteine residue at the N- or C-terminus for later surface grafting. Of those, HS-MSI-78A had bacteriostatic (MIC) and bactericidal (MBC) effects in lower concentrations than MSI-78A-SH, being thus HS-MSI-78A the one selected for immobilization onto model surfaces (SAMs). For S. epidermidis, the adhesion control strain, the MIC for MSI-78A was set at 2 μg/mL.
AMP immobilization onto model surfaces (biotin-SAMs)
2.5% biotin-SAMs were selected for HS-MSI-78A immobilization, since model surfaces with this biotin/EG4 ratio were previously characterized and described as the ideal one for surface functionalization with ligands (more surface immobilized mass is achieved)28. The QCM-D technique allowed following in real time the AMP immobilization (Fig. 2) as well as to estimate the mass of surface immobilized peptide. The frequency shifts, Δf, are related to mass changes on the crystal surface, whereas dissipation shifts, ΔD, are related to the viscoelastic properties of the adsorbed layer32.
AMP immobilization onto 2.5% biotin-SAMs followed by QCM-D. ΔF- Frequency shifts; ΔD- Dissipation shifts. (1) neutravidin injection; high frequency shift correlates to large neutravidin size and high concentration. Dissipation variations also indicate protein adsorption onto 2.5% biotin-SAMs; (2) PBS rinsing; (3) EG11-MAL spacer injection; small shift in frequency is related to the PEG molecular weight and its low concentration. However, high dissipation shift indicates EG11-MAL linker incorporation onto model surface; 4) HS-MSI-78A; small shift in frequency correlates to the small size of the AMP. The decrease in the dissipation is related to AMP binding to the EG11-MAL linker, leading to a more compact (less fluid) surface.
After three independent experiments with duplicates, the peptide mass on the surface was calculated by applying the Sauerbrey equation to the 5th overtone, which was chosen as the most stable and reproducible overtone. This equation can be applied for systems where ΔD/Δf is lower than 4 × 10–6 Hz−1 and assumes that the adsorbed/bound film is rigid with no internal loss of energy, which is translated in low values of dissipation33. The immobilized mass of HS-MSI-78A was estimated to be 30.3 ± 1.2 ng/cm2, but this may be an overestimation, since this value may be taking into account water and other buffer constituents trapped between the adsorbed molecules34.
Antibacterial activity of the AMP-functionalized model surfaces (AMP-SAMs)
The antibacterial performance of AMP-SAMs (SAMs with the antimicrobial peptide HS-MSI-78A immobilized on its surface) was tested against the H. pylori J99 strain, a highly pathogenic human strain that is associated with poor patient prognosis35. AMP-SAMs activity was also evaluated against S. epidermidis ATCC 35984 strain, the control for surface adhesion behaviour. The viability of planktonic cells after exposure to the bioengineered model surfaces was evaluated by performing CFU counting. On the other hand, for surface adherent cells, a Live/Dead staining was used, which allowed distinguishing viable and non-viable bacterial cells (Fig. 3).
Figure 4 highlights the antibacterial performance of AMP-SAMs and control surfaces (Au, EG4, Biotin, EG11-MAL) against H. pylori J99.
Antimicrobial activity of AMP-SAMs against H. pylori J99 after: 2 h incubation in PBS (a) surface adherent cells (b) planktonic cells; 6 h incubation in recovery medium (c) surface adherent cells (d) planktonic cells. Results are expressed as average ± SD. *- Total adhesion (number of live + dead cells) in EG4-SAMs significantly different from all the other tested surfaces (p < 0.05); #- number of dead bacteria significantly different from live bacteria (p < 0.05); **/***-significantly different from Au (**p < 0.001; ***p < 0.001).
The 2 h adhesion was chosen based on previous assays, where it was determined that bacterial adhesion onto these model surfaces plateaus after 2 h27,36. No significant differences (p < 0.05) were observed for global adhesion (live and dead bacteria) among the tested surfaces, except for the EG4-SAMs, where adhesion was much lower in comparison to the other surfaces and also in accordance with prior findings (Fig. 4a)27. After 2 h, the majority of H. pylori adhered to control surfaces (Au, EG4, biotin and EG11-MAL) remained viable, in contrast to the 75% of non-viable H. pylori adhered to the AMP-SAMs (Fig. 4a). Regarding planktonic cells, no live bacteria were seen in the supernatants after 2 h of incubation with AMP-SAMs, but H. pylori viability was kept in control surfaces (Au, EG4, Biotin, EG11-MAL) (Fig. 4b). To further assess the antibacterial effect, namely to understand if surface-adherent bacteria were able to recover from exposure to AMP-SAMs, a set of surfaces was subsequently incubated in recovery media (MHB + 10%FBS) for 6 h (Fig. 4c). The incubation period for this recovery assay was based on the estimated duplication time for H. pylori (6 h)37. After 6 h in culture medium (Fig. 4c) and compared with 2 h (Fig. 4a), no significant increase was observed for overall adhesion (total number of live and dead bacteria). However, the number of viable and cultivable bacteria in the supernatants increased for all control surfaces (Fig. 4d). This increase may be translated as adherent live bacteria that were able to detach from the surfaces and proliferate in solution. More importantly, only a very small number of live bacteria that might be adhered to AMP-SAMs were released and able to thrive (Fig. 4d). Although MHB is the standard culture media for MICs determination in accordance with the CLSI guidelines, it is not the optimal recovery media for H. pylori, since this is a fastidous microrganism that requires nutrient enriched culture media38. To clarify if this could be interfering with the bacterial recovery rate, identical assays were performed using Brucella Broth culture medium. Similar results were obtained using both culture media, demonstrating that the use of MHB was not affecting bacterial recovery (data not shown).
It is noteworthy that, although in lower number, H. pylori cells still adhered to EG4-SAMs, which are described as “anti-adhesive” surfaces (Fig. 4a,c). To verify the non-fouling properties of the EG4-SAMs, and that there is no AMP release, further studies were performed using the S. epidermidis ATCC 35984 strain and results are represented in Fig. 5.
Antimicrobial activity of AMP-SAMs against S. epidermidis cells after: 2 h incubation in PBS (a) surface adherent cells (b) planktonic cells; after 4 h incubation in recovery medium (c) surface adherent cells (d) planktonic cells. Results are expressed as average ± SD. ** significantly different from Au (p < 0.0001); **** number of dead bacteria significantly different from live bacteria (p = 0.0013).
After 2 h of incubation, S. epidermidis did not adhere to EG4-SAMs, while AMP-SAMs promoted adhesion and subsequent killing of adhered bacteria (Fig. 5a). Yet, the number of total surface-adherent bacteria was remarkably lower than those observed for H. pylori J99 (Fig. 4a). After 2 h, the number of planktonic S. epidermidis cells was not affected by exposure to AMP-SAMs, with no significant differences being observed between AMP-SAMs and control surfaces (Fig. 5b). After 4 h in fresh culture media, there were no significant differences among the tested model surfaces, which is in agreement with the number of live bacteria after 2 h of adhesion, also similar between samples (Fig. 5c). After 4 h in fresh culture media (TSB), more S. epidermidis planktonic cells were observed in all samples, including AMP-SAMs (Fig. 5d). For both incubation times, high S. epidermidis adhesion was observed on gold, which made the quantification of adherent bacteria unfeasible.
Discussion
This work reports the proof of concept studies for the future development of bioengineered nanoparticles with surface grafted MSI-78A peptide as an antibiotic-free therapy against H. pylori.
First, different antimicrobial peptides previously reported as having anti-H. pylori effect were screened in solution. However, Odorranain-HP didn’t present antibacterial activity against the tested H. pylori strains panel (Table 1). This diverges from the previous work of Chen et al., where Odorranain-HP presented a 20 μg/mL MIC against H. pylori NTCC11637 strain20. A possible explanation might reside in the different experimental settings, namely the use of a different culture media. The remaining tested AMPs were active against H. pylori (Table 1). Of all, MSI-78A was the most effective. Therefore, the antimicrobial peptide MSI-78A was synthesized with an aminohexanoic acid (ahx) spacer and an extra cysteine (-SH) at the N- or C-terminus, envisioning its later surface immobilization in a controlled manner. Once the HS-MSI-78A demonstrated improved activity in solution (Table 1), it was selected for surface immobilization.
A key feature concerning the surface immobilization of the selected AMP is to retain its bioactivity, i.e., keeping its ability to bind to the desired target and maintaining minimum interactions with non-relevant matrix or assay components. To accomplish this, it is common to use highly hydrophilic PEG linkers, hence creating a microenvironment that decreases non-specific binding and provides additional mobility to the tethered AMPs39. Although the selected peptide was synthesized with a small ahx spacer, it was also used an EG11-unit spacer (EG11-MAL) for HS-MSI-78A immobilization. The use of this longer PEG allows to decrease peptide aggregation and subsequently boost the AMP binding, since it imparts water solubility that is transferred to the biotinylated molecule, ultimately improving AMP exposure from the surface39. A smaller EG-spacer (EG2-unit) was also tested but according to the performed QCM-D assays it demonstrated to be less efficient than the EG11-unit spacer, with lower mass of AMP immobilized on the surface (data not shown). In addition, to further justify the EG11-unit choice, it has been reported that antimicrobial peptides immobilized via EG spacers with longer arms (EGn; n > 8) yield higher antimicrobial activity than those tethered via shorter EG linkers40. The successful AMP immobilization onto nanostructured model surfaces (SAMs) was confirmed by the QCM-D assays (Fig. 2).
Then, the antibacterial activity of the engineered model surfaces functionalized with the selected AMP (AMP-SAMs) was evaluated. CFU counting assessed the viability of planktonic cells, while viability of surface adherent cells was evaluated using the Live/Dead Baclight KitTM. This dual staining kit allows to quantify and distinguish live from dead cells. It is composed of two fluorophores, SYTO9 and propidium iodide (PI), and is based on detection of membrane integrity: cells with a compromised membrane are considered dead or dying and will stain red (PI); whereas cells with an intact membrane will stain green (SYTO9). It is important to highlight that some bacterial cells incubated with SAMs stained yellow (Fig. 3). This may occur when SYTO9, which can be actively exported from the cytoplasm, is not completely replaced by PI, generating the yellow staining41. As SYTO9 and PI binding and releasing to/from nucleic acids are dynamic processes, it is possible that both green and red dyes were simultaneously retained within cells, originating the yellow staining. Since this indicates cells with damaged membrane, the yellow-stained bacteria were considered as dead cells41.
Regarding the antibacterial activity, after 2 h most of the H. pylori cells adhered to control surfaces (Au, EG4, biotin and EG11-MAL) were viable, in contrast to the 75% of non-viable H. pylori on AMP-SAMs surface (Fig. 4a). The excellent bactericidal effect of the AMP-SAMs was further demonstrated since after 2 h of incubation there were no live planktonic bacteria detected, while H. pylori viability was kept in the control surfaces (Au, EG4, Biotin, EG11-MAL) (Fig. 4b). After 6 h in culture medium (Fig. 4c) and compared with 2 h (Fig. 4a), there was no significant increase observed for the overall adhesion (total number of live and dead bacteria). This was not expected because 6 h is the estimated duplication time for H. pylori27. But since bacteria are coming from a potential stressful situation, namely incubation in PBS and exposure to abiotic model surfaces (SAMs), this might result in a slower growth/duplication rate, which could also explain some of the dead bacteria observed on the gold surface after 6 h. The AMP is covalently immobilized onto SAMs surface via the cysteine-maleimide reaction that ocurrs between the AMP and the EG11-spacer and thus, it is not likely that leaching from the surface occurs. Therefore, we hypothesize that AMP-SAMs have a contact-killing effect: first, H. pylori cells are attracted to the AMP-SAMs, interact with the exposed AMP and are killed; then, the non-viable bacteria are released from the surface, leaving the surface immobilized AMP available to interact with/kill other H. pylori cells. Dead bacteria must be released from the surface in order to obtain the herein reported high death rates. The assumption that the AMP is probably not being actively leached from the surface is further encouraged by the results obtatined with S. epidermidis: after 2 h of incubation, S. epidermidis planktonic cells were not affected by exposure to AMP-SAMs with no significant differences reported between AMP-SAMs and the control surfaces (Fig. 5b). Also, the number of planktonic S. epidermidis cells kept increasing even after 4 h incubation with AMP-SAMs (Fig. 5d).
Furthermore, H. pylori cells were not able to recover from exposure to AMP-SAMs, since after 6 h in recovery culture medium only a small number of the live bacteria that adhered to AMP-SAMs (Fig. 4b) were able to detach from the surface and thrive in solution (Fig. 4d). On the other hand, a higher number of bacteria in supernatants were observed for control surfaces (Fig. 4d), indicating that in more favourable conditions bacteria can easily detach from the surface and grow. This might also explain why EG4-SAMs, which had fewer H. pylori cells adhered at 2 h (Fig. 4a), generated a similar number of planktonic bacteria compared to the other surfaces (Fig. 4d) when transferred to culture medium.
EG4-terminated surfaces are described as non-fouling surfaces, i.e., resistant to cell and protein adhesion25,40,42. Although H. pylori adhesion to EG4-SAMs was much lower when compared with the other model surfaces (Fig. 4a,c), values were higher than expected. Therefore, S. epidermidis, whose low adhesive behavior to hydrophylic surfaces is well documented, was selected as adhesion control, while also enabled to establish the non-fouling nature of EG4-SAMs42,43. As anticipated, very few S. epidermidis cells adhered to EG4-SAMs (Fig. 5a). Moreover, the low number of S. epidermidis adhered to the control surfaces, with gold being the exception, may be related to the common underlying EG-nature of those SAMs (Fig. 5a,c). Overall, this highlights a distinct H. pylori adhesion behaviour towards EG-based SAMs, which might be related with H. pylori proficiency in binding to the highly hydrophilic gastric mucus layer44.
It was also observed a considerable difference between H. pylori adhesion to 2.5% biotin-SAMs (composed of 97.5% EG4-thiol) and “pure” EG4-SAMs (100% EG4-thiol). Biotin has been proposed to be a critical and limited nutrient during infections by human pathogens such as Mycobacterium tuberculosis45. Nonetheless, H. pylori is able to produce biotin, as biotin supplementation has no effect on its growth in a chemically defined media45. Also, analyses of the available H. pylori genomes showed that it encodes all the proteins required for assembly of the fused heterocyclic rings of biotin46,47. However, as biotin is a key protein-bound enzyme cofactor that plays essential roles in the transfer of CO2 in key central metabolic processes46, we hypothesize that the biotin moieties readily available on the surface of the bioengineered models may act as chemo attractant. This would justify the marked difference between H. pylori adhesion onto EG4-SAMs and 2.5% biotin-SAMs.
Theoretically, both encapsulation and immobilization processes are able to circumvent AMPs potential limitations, such as short half-life (proteolytic degradation). In fact, nanoparticles loaded with Pexiganan (MSI-78) were recently evaluated against H. pylori with promising in vitro and in vivo results25. Nonetheless, and although surface immobilization of AMPs has been widely studied for other purposes, such as hindering biofilm formation, very few work has been devoted to the immobilization of AMPs with anti-H. pylori activity within gastric infection scenario. Differently from encapsulation strategies, the surface immobilization of AMPs allows to control the peptide orientation, concentration and exposure, which may play a crucial role in boosting the performance of AMP-based strategies. Also, immobilization processes enable the use of lower AMP concentrations to achieve biological effect, since it is not dependent upon encapsulation processes that may result in low encapsulation yield and may diminish the possibility of toxicity-related issues. Moreover, functionalized surfaces with covalently bound AMP are not required to undergo any other event, such as material/composite degradation for peptide release and the chance of peptide aggregation is much lower during immobilization than when performing encapsulation. Another important aspect is how immobilization may tailor and improve the AMP specificity towards the gastric pathogen. The unbound AMP was 32 times more active in solution against S. epidermidis than against H. pylori, but once surface immobilized, higher selectivity towards H. pylori was achieved, being the effect of the AMP-functionalized surfaces against S. epidermidis residual.
The study herein reported demonstrates that the bioactivity of the peptide is retained after surface immobilization. In addition, the high bacterial eradication rates obtained after 2 h (>90%) using the surface grafted MSI-78A is a very promising result, once framed with the time of stomach emptying and the effective retention time of a possible oral strategy against H. pylori. This proof of concept study establishes the promising potential of immobilized AMPs as non-antibiotic alternatives to counteract H. pylori infection, encouraging translation of the obtained knowledge onto “real-world” bioengineered approaches.
Conclusion
This study demonstrates that not only MSI-78A can be surface-grafted without compromising its activity but also its immobilization onto non-adhesive surfaces boosts the antibacterial performance against H. pylori. Overall, the MSI-78A-functionalized model surfaces were highly effective against H. pylori. The gastric pathogen was killed in a short time span, since after 2 h only 2% of H. pylori cells remained viable in suspension. Furthermore, we also established that AMP-SAMs have higher activity towards H. pylori than to S. epidermidis. In summary, the herein reported proof-of-concept study validates the development of a bioengineered approach based on surface-grafted antimicrobial peptides aiming H. pylori eradication via non-antibiotic options.
Methods
Antimicrobial peptides (AMPs)
Tested AMPs and their respective sequences are listed in Table 2.
AMPs synthesis was performed as previously described13. The last two rows on Table 2 refer to MSI-78A derivatives on which an additional cysteine residue (represented by -SH instead of its standard single-letter code C, in order to emphasize its role as a thiol donor) was attached at either the N- or C-terminus of the bioactive sequence, through a 6-amino-hexanoic acid (ahx) spacer. Afterwards, AMPs were freeze dried and stored at −20 °C. Prior to use, AMPs were suspended in phosphate-buffered saline (PBS; pH~ 7.5) to a final 1 mg/mL concentration.
Bacteria
Helicobacter pylori
H. pylori human strains J99 and 26695 (provided by Department of Medical Biochemistry and Biophysics, Umeå University, Sweden), NCTC 11637 (ATCC 43504) and mouse-adapted SS1 strain (provided by Unité de Pathogenèse de Helicobacter, Institut Pasteur, France) were grown following the standard procedure. Briefly, H. pylori was cultured in Blood Agar (BA; Oxoid) supplemented with 10% of defibrinated horse blood (Probiologica) and with an antibiotic cocktail (polymyxin B, vancomycin, amphotericin B, trimethoprim; all from Sigma-Aldrich) under microaerophilic conditions (<5% O2; GenBox System, BioMérieux) at 37 °C for 48 h. Afterwards, some colonies were streaked onto fresh BA medium and kept in culture in the same conditions for another 48 h. Subsequently, bacterial inoculum was adjusted to 0.03 optical density at 600 nm (OD600), which corresponds to approximately 1 × 107 colony forming units (CFUs)/mL48. Initial inoculum was confirmed by inoculation of 10 μL drops of the bacterial suspension onto BA and CFUs were counted after 5 days of incubation in the same above-mentioned settings.
Staphylococcus epidermidis
S. epidermidis ATCC 35984 strain was selected as the control for bacterial adhesion onto SAMs. This choice was based on the fact that, although being remarkbly good at adhere, proliferate and establish biofilms, this strain is not capable of adherence to non-fouling surfaces30,31. Bacteria were grown on Trypticase Soy Agar (TSA; Merck Millipore) plates overnight at 37 °C (spreading) and then transferred to Trypticase Soy Broth (TSB; Merck Millipore). Incubation proceeded overnight at 37 °C, 150 rpm. Bacterial inoculum was then adjusted at OD600 to 0.33, which corresponds to approximately 1 × 108 CFUs/mL in TSB. This was optimized in order to have adherent cells onto model surfaces. The initial inoculum was confirmed by growing 10 μL drops of the suspension in TSA plates during 24 h at 37 °C.
AMPs activity in solution
Minimal Inhibitory concentration (MIC)
The MICs of the AMPs (Table 1) against H. pylori strains were determined using the microbroth dilution method in Mueller Hinton Broth (MHB) supplemented with 10% of inactivated Fetal Bovine Serum (FBS; Gibco) and in accordance with the Clinical Laboratory Standards Institute (CLSI) guidelines49. Briefly, AMPs were diluted in 0.01% of acetic acid with 0.2% Bovine Serum Albumin (BSA; Sigma) to allow AMP protonation and avoid its aggregation, respectively, following an already established protocol13. The selected AMPs concentrations ranged from 2 µg/mL up to 512 µg/mL. Incubation was performed at 37 °C in microaerophilic conditions for 72 h. The MIC was defined as the first AMP concentration that prevented visible (naked eye) bacterial growth. Two replicates of each AMP concentration were performed in three independent experiments.
Minimal bactericidal concentration (MBC)
The MBC was defined as the lowest AMP concentration that was able to diminish the viability of the initial bacterial inoculum by ≥99.9%. MBC determination followed the CLSI guidelines49 but briefly, a 50 µL sample was taken from the first three lowest concentrations where no bacterial growth was observed (concentration corresponding to the MIC), serially diluted in PBS until 10−6 and then spotted on BA. After 5 days of incubation at 37 °C in microaerophilic environment, the CFUs/mL were calculated. Two replicates of each AMP concentration were performed in three independent experiments.
AMP-functionalized model surfaces
AMP immobilization strategy is described in Fig. 1.
Gold substrates
Gold substrates (1 × 1 cm2) used in bacterial assays were obtained from Instituto de Engenharia de Sistemas e Computadores – Microsistemas e Nanotecnologias, Portugal (INESC-MN)50. For Quartz Crystal Microbalance with Dissipation (QCM-D) assays, gold-coated quartz crystal sensors (QSX301-Standard Gold, 4.95 Hz, 78 mm2 active sensor area) were obtained from Biolin Scientific.
Model surfaces: Biotin Self-Assembled Monolayers (biotin-SAMs)
1-Mercapto-11-undecyl tetra (ethylene glycol) (SH-(CH2)11-O-(CH2-CH2-O)4-H; EG4-thiol; 99%, SensoPath Technologies) and biotin-terminated tri(ethylene glycol) undecanethiol (SH-(CH2)10-CO-NH-(CH2)3-O-(CH2CH2O)2-(CH2)3-NH-Biotin; biotin-EG3-thiol, 99%, SensoPath Technologies) were prepared as pure solutions at 2 mM in absolute ethanol (Merck). Biotin-SAMs were prepared by immersing the gold coated surfaces (gold substrates or QCM-D crystals) in solutions containing 2.5% biotin-thiol (97.5% EG4-thiol) with 0.1 mM final concentration, as previously described by us (Fig. 1a)28. The whole process was performed under a dry nitrogen atmosphere inside a glove box. Incubation was performed at RT (~25 °C) during 20 h. After incubation, biotin-SAMs were rinsed with absolute ethanol, dried with a gentle nitrogen stream and used immediately.
AMP immobilization onto model surfaces (SAMs)
AMP immobilization onto model surfaces (biotin-SAMs) was followed in real time using a QCM-D device (Q-Sense E4 system, Biolin Scientific) at RT (~25 °C). Biotin-SAMs assembled onto QCM-D gold-coated crystals (section 2.4.1 and 2.4.2) were placed in the system. Afterwards, filtered PBS (0.22 μm pore size) was injected until a stable signal was obtained (baseline). Then, neutravidin (Invitrogen), 1 mg/mL in PBS, was incubated in static for 1 h (Fig. 1b). This neutravidin concentration allowed blocking all biotin moieties freely available on the biotin-SAMs surface, as previously determined by us. Next, rinsing was done with PBS followed by a 30 min static incubation of EZ-linkTM maleimide (MAL)-polyethylene glycol (EG11)-Biotin spacer (EG11-MAL; Thermo Fisher Scientific) (Fig. 1c) in a concentration of 1 mg/mL in PBS. The terminal MAL group of the heterobifunctional spacer reacts specifically and efficiently with sulfhydryl groups (-SH) of the added cysteine to the AMP establishing stable covalent bonds. After, rinsing was performed with PBS and the selected AMP (HS-MSI-78A) at 0.5 mg/mL in PBS was incubated for 45 min. Then, a final rinsing with PBS was done to remove unbound peptide (Fig. 1d). All solutions were injected at flow rate of 0.1 mL/min.
Once the changes in the system dissipation were considered low (<4), the mass of the surface immobilized AMP was estimated using the Sauerbrey Eq. (1) after 4 independent experiments and expressed in ng/cm2.
(1) Δm is the adsorbed mass, Δf is the frequency shift due to the adsorption, n is the overtone number and C is a constant characteristic of the sensor crystal (C = 17.7 ng·Hz−1·cm−2 for the 5 MHz crystals used).
In vitro antibacterial activity assays were performed with model surfaces (biotin-SAMs) functionalized with the HS-MSI-78A peptide (AMP-SAMs), following the incubation settings defined in the QCM-D experiments (based on 2.3 section results).
Antibacterial activity of the AMP-functionalized model surfaces (AMP-SAMs)
The antibacterial activity of AMP-functionalized surfaces (AMP-SAMs) was tested against H. pylori J99 strain (highly pathogenic and human strain) and S. epidermidis ATCC 35984 strain (control for bacterial adhesion onto surfaces). Bare gold surfaces (Au), EG4-SAMs (EG4), biotin-SAMs (Biotin) and biotin-SAMs + neutravidin + biotin-EG11-MAL linker (EG11-MAL) were used as surface controls.
H. pylori J99 and S. epidermidis ATCC 35984 were simultaneously incubated with surfaces (AMP-SAMs and controls) at 37 °C and 150 rpm under microaerophilic conditions. After 2 h of incubation, surfaces were divided into two groups. From the first group of surfaces, supernatants (planktonic cells) were removed, serially diluted and platted either in BA (H. pylori) or TSA (S. epidermidis) for CFU determination. H. pylori CFUs were determined after 5 days of incubation at 37 °C in microaerophilic conditions, while S. epidermidis CFUs counting were performed after 24 h of incubation at 37 °C. After supernatant removal, surfaces were rinsed with PBS and stained with Live/Dead Baclight KitTM (Invitrogen) according to the manufacturer’s protocol. Bacteria were visualized with an Inverted Fluorescence Microscope (Zeiss Axiovert 200 MOT) at 400x magnification. Bacterial quantification was manually determined from 15 photographs (5 random fields per sample) and expressed as bacteria/mm2. Aiming to determine if bacteria were able to recover from exposure to AMP-SAMs, the second group of surfaces was rinsed with PBS and transferred to MHB + 10%FBS (H. pylori) for more 6 h (8 h incubation total) or to TSB (S. epidermidis) for additional 4 h (6 h incubation total) (the selected incubation times were based on the growth kinetics of each bacteria). After the mentioned incubation periods, supernatants (planktonic cells) and surfaces were processed as mentioned for the 1st group. Three independent experiments were performed using samples in triplicate.
Statistical analysis
Statistical analysis was done with GraphPad Prism software (GraphPad Software Inc., version 6.0). Data was analysed by D’Agostino & Pearson omnibus normality test. Two-way ANOVA was used to determine statistical significance in the assays concerning surface adherent cells, followed by Dunnett’s multiple comparison tests. One-way ANOVA was used for statistical analysis of the assays regarding planktonic cells, followed by a Sidak’s multiple comparison test. A value of p < 0.05 was considered statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Malfertheiner, P. et al. Management of Helicobacter pylori infection-The Maastricht V/Florence Consensus Report. Gut. 66, 6–30 (2017).
Amieva, M. & Peek, R. M. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 150, 64–78 (2016).
Ferlay, J. et al. GLOBOCAN 2012, Cancer Incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 136, E359–386 (2015).
Dang, B. N. & Graham, D. Y. Helicobacter pylori infection and antibiotic resistance: a WHO high priority? Nat Rev Gastroenterol Hepatol. 14, 383–384 (2017).
Vakil, N. & Vaira, D. Treatment for H. pylori infection: New challenges with antimicrobial resistance. J. Clin Gastroenterol. 47, 383–388 (2013).
Ermis, F. & Senocak Tasci, E. Current Helicobacter pylori treatment in 2014. World J. Methodol. 5, 101–107 (2015).
Vakil, N. Helicobacter pylori treatment: a practical approach. Am. J. Gastroenterol. 101, 497–499 (2006).
Malfertheiner, P. et al. Management of Helicobacter pylori infection–the Maastricht IV/ Florence Consensus Report. Gut. 61, 646–64 (2012).
Oppenheim, J. J., Biragyn, A., Kwak, L. W. & Yang, D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 62, ii17–i21 (2003).
Gottler, L. M. & Ramamoorthy, A. Structure, Membrane Orientation, Mechanism, and Function of Pexiganan – A Highly Potent Antimicrobial Peptide Designed From Magainin. Biochim. Biophys. Acta. 1788, 1680–1686 (2009).
Costa, F., Carvalho, I. F., Montelaro, R. C., Gomes, P. & Martins, M. C. L. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomater. 7, 1431–1440 (2011).
Parreira, P., Duarte, M. F., Reis, C. A. & Martins, M. C. L. Helicobacter pylori infection: A brief overview on alternative natural treatments to conventional therapy. Crit. Rev. Microbiol. 42, 94–105 (2016).
Monteiro, C. et al. Antimicrobial properties of membrane-active dodecapeptides derived from MSI-78. Biochim. Biophys. Acta - Biomembr. 1848, 1139–1146 (2015).
Seo, M.-D., Won, H.-S., Kim, J.-H., Mishig-Ochir, T. & Lee, B.-J. Antimicrobial Peptides for Therapeutic Applications: A Review. Molecules. 17, 12276–12286 (2012).
Van Der Weerden, N. L., Bleackley, M. R. & Anderson, M. A. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell Mol Life Sci. 70, 3545–3570 (2013).
Barbosa, M., Vale, N., Costa, F. M. T. A., Martins, M. C. L. & Gomes, P. Tethering antimicrobial peptides onto chitosan: Optimization of azide-alkyne “click” reaction conditions. Carbohydr. Polym. 165, 384–393 (2017).
Zhao, X., Wu, H., Lu, H., Li, G. & Huang, Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS One. 8, e66657 (2013).
Bahar, A. A. & Ren, D. Antimicrobial peptides. Pharmaceuticals (Basel). 6, (1543–1575 (2013).
Ma, Y. et al. Peptidomics and genomics analysis of novel antimicrobial peptides from the frog, Rana nigrovittata. Genomics. 95, 66–71 (2010).
Chen, L. et al. An antimicrobial peptide with antimicrobial activity against Helicobacter pylori. Peptides. 28, 1527–1531 (2007).
Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 84, 5449–5453 (1987).
Jacob, L. & Zasloff, M. Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. Ciba Found. Symp. 186, 197–223 (1994).
Ge, Y. et al. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob. Agents Chemother. 43, 782–788 (1999).
Carmona-Ribeiro, A. M., Carrasco, L. D. & de, M. Novel formulations for antimicrobial peptides. Int J Mol Sci. 15, 18040–18083 (2014).
Zhang, X. L. et al. The synthetic antimicrobial peptide pexiganan and its nanoparticles (PNPs) exhibit the anti- Helicobacter pylori activity in vitro and in vivo. Molecules. 20, 3972–3985 (2015).
Love, J. C., Estroff, L. A., Kriebel, J. K., Nuzzo, R. G. & Whitesides, G. M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev. 105, 1103–1169 (2005).
Parreira, P. et al. Effect of surface chemistry on bacterial adhesion, viability, and morphology. J. Biomed. Mater. Res. A 99, 344–353 (2011).
Parreira, P. et al. Bioengineered surfaces promote specific protein-glycan mediated binding of the gastric pathogen Helicobacter pylori. Acta Biomater. 9, 8885–93 (2013).
Gonçalves, I. C. et al. Bacteria-targeted biomaterials: Glycan-coated microspheres to bind Helicobacter pylori. Acta Biomater. 33, 40–50 (2016).
Gelosia, A., Baldassarri, L., Deighton, M. & Van Nguyen, T. Phenotypic and genotypic markers of Staphylococcus epidermidis virulence. Clin. Microbiol. Infect. 7, 193–199 (2001).
Otto, M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 322, 207–228 (2008).
Briand, E., Humblot, V., Pradier, C.-M. M., Kasemo, B. & Svedhem, S. An OEGylated thiol monolayer for the tethering of liposomes and the study of liposome interactions. Talanta. 81, 1153–1161 (2010).
Azzaroni, O., Mir, M. M. & Knoll, W. Supramolecular architectures of streptavidin on biotinylated self-assembled monolayers. Tracking biomolecular reorganization after bioconjugation. J Phys Chem B. 111, 13499–13503 (2007).
Nielsen, S. B. & Otzen, D. E. Quartz crystal microbalances as tools for probing protein-membrane interactions. Methods Mol Biol. 974, 1–21 (2013).
Floch, P., Mégraud, F. & Lehours, P. Helicobacter pylori strains and gastric MALT lymphoma. Toxins (Basel). 9, (E132 (2017).
An, Y. H. & Friedman, R. J. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J Biomed Mater Res. 43, 338–348 (1998).
Chu, Y.-T., Wang, Y.-H., Wu, J.-J. & Lei, H.-Y. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells: implications for antibiotic resistance. Infect. Immun. 78, 4157–4165 (2010).
Walsh, E. J. & Moran, A. P. Influence of medium composition on the growth and antigen expression of Helicobacter pylori. J Appl Microbiol. 83, 67–75 (1997).
Shriver-Lake, L. C., Anderson, G. P. & Taitt, C. R. Effect of Linker Length on Cell Capture by Poly(ethylene glycol)-Immobilized Antimicrobial Peptides. Langmuir 33, 2878–2884 (2017).
Bagheri, M., Beyermann, M. & Dathe, M. Immobilization reduces the activity of surface-bound cationic antimicrobial peptides with no influence upon the activity spectrum. Antimicrob. Agents Chemother. 53, 1132–1141 (2009).
Stiefel, P., Schmidt-Emrich, S., Maniura-Weber, K. & Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 15, 36 (2015).
Cheng, G., Zhang, Z., Chen, S., Bryers, J. D. & Jiang, S. Inhibition of bacterial adhesion and biofilm formation on zwitterionic surfaces. Biomaterials. 28, 4192–4199 (2007).
Ista, L. K., Fan, H., Baca, O. & López, G. P. Attachment of bacteria to model solid surfaces’ oligo(ethylene glycol) surfaces inhibit bacterial attachment. FEMS Microbiol Lett. 142, 59–63 (1996).
Rhee, K. H., Park, J. S. & Cho, M. J. Helicobacter pylori: Bacterial strategy for incipient stage and persistent colonization in human gastric niches. Yonsei Medical Journal. 55, 1453–1466 (2014).
Reynolds, D. J. & Penn, C. W. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology. 140, 2649–2656 (1994).
Cronan, J. E. Biotin and Lipoic Acid: Synthesis, Attachment, and Regulation. EcoSal Plus. 3 (2013).
Bi, H., Zhu, L., Jia, J. & Cronan, J. E. A Biotin Biosynthesis Gene Restricted to Helicobacter. Sci. Rep. 6, 21162 (2016).
Gonçalves, I. C. et al. Bacterial-binding chitosan microspheres for gastric infection treatment and prevention. Acta Biomater. 9, 9370–9378 (2013).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Clinical and Laboratory Standards Institute (2015).
Martins, M. C. L., Ratner, B. D. & Barbosa, M. A. Protein adsorption on mixtures of hydroxyl- and methyl-terminated alkanethiols self-assembled monolayers. J. Biomed Mater Res A. 67, 158–171 (2003).
Acknowledgements
This article is a result of the project NORTE-01-0145-FEDER-00012, (NORTE 2020) and FCT/MCTES-through the UID/BIM/04293/2019, PTDC/CTM-BIO/4043/2014, UID/QUI/50006/2013 (LAQV-REQUIMTE) projects. Claudia Monteiro acknowledges FCT for the SFRH/BPD/79439/2011 grant.
Author information
Authors and Affiliations
Contributions
P. Parreira carried out AMP (MSI-78A) surface immobilization, QCM-D experiments, H. pylori antimicrobial assays, designed the study, data analysis and drafted the manuscript. C. Monteiro performed S. epidermidis antimicrobial assays and correspondent data analysis and also participated in the design of the study. C. Monteiro, V. Graça and J. Gomes determined the AMP’s MIC and MBC. S. Maia and P. Gomes performed the AMP synthesis and its characterization. I. C. Gonçalves and M.C.L. Martins conceived, designed and coordinated the study and helped draft the manuscript. The manuscript was written through contributions of all authors. All authors gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Parreira, P., Monteiro, C., Graça, V. et al. Surface Grafted MSI-78A Antimicrobial Peptide has High Potential for Gastric Infection Management. Sci Rep 9, 18212 (2019). https://doi.org/10.1038/s41598-019-53918-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-53918-4
This article is cited by
-
Peptide Flexibility and the Hydrophobic Moment are Determinants to Evaluate the Clinical Potential of Magainins
The Journal of Membrane Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.