Abstract
The composition of plant cell walls is important in determining cereal end uses. Unlike other widely consumed cereal grains barley is comparatively rich in (1,3;1,4)-β-glucan, a source of dietary fibre. Previous work showed Cellulose synthase-like genes synthesise (1,3;1,4)-β-glucan in several tissues. HvCslF6 encodes a grain (1,3;1,4)-β-glucan synthase, whereas the function of HvCslF9 is unknown. Here, the relationship between mRNA levels of HvCslF6, HvCslF9, HvGlbI (1,3;1,4)-β-glucan endohydrolase, and (1,3;1,4)-β-glucan content was studied in developing grains of four barley cultivars. HvCslF6 was differentially expressed during mid (8–15 DPA) and late (38 DPA) grain development stages while HvCslF9 transcript was only clearly detected at 8–10 DPA. A peak of HvGlbI expression was detected at 15 DPA. Differences in transcript abundance across the three genes could partially explain variation in grain (1,3;1,4)-β-glucan content in these genotypes. Remarkably narrow sequence variation was found within the HvCslF6 promoter and coding sequence and does not explain variation in (1,3;1,4)-β-glucan content. Our data emphasise the genotype-dependent accumulation of (1,3;1,4)-β-glucan during barley grain development and a role for the balance between hydrolysis and synthesis in determining (1,3;1,4)-β-glucan content, and suggests that other regulatory sequences or proteins are likely to be involved in this trait in developing grain.
Similar content being viewed by others
Introduction
(1,3;1,4)-β-Glucan is a plant cell wall polysaccharide that is found in a range of tissues in barley including the grain, where it constitutes approximately 70% dry weight (w/w)1 of endosperm primary cell walls. This polysaccharide accumulates throughout barley endosperm development. The four main phases that define endosperm development in barley and other cereals include the syncytial, cellularization, differentiation and maturation (also referred to as desiccation) phases, each of which are characterized by multiple morphological changes2,3. In barley, at 4–5 days after pollination (DAP), (1,3;1,4)-β-glucan is deposited in endosperm cell walls and is uniformly distributed by 10 DAP. (1,3;1,4)-β-Glucan increases in abundance between 16–36 DAP, coinciding with the grain filling and maturation stages4. A wide range of natural variation for this trait has been described in contemporary barley cultivars [~2.5–8% w/w;5,6] and wild barleys, Hordeum vulgare ssp. spontaneum [4.5–13.2% w/w7]. Barley has considerably higher grain (1,3;1,4)-β-glucan concentration (4–10% w/w) compared to other small grain cereals such as wheat (1% w/w) or rice (<0.06% w/w)8. In wheat, the sequence of deposition of cell wall polysaccharides differs from barley, mainly due to the early accumulation of arabinoxylan during endosperm cellularization9. While (1,3;1,4)-β-glucan is mainly detected in the central cell walls of wheat endosperm and decreases considerably in late endosperm development stages9, an even distribution of (1,3;1,4)-β-glucan abundance has been described in developing barley and Brachypodium grain10. In Brachypodium, the highest levels of (1,3;1,4)-β-glucan are detected during grain filling (17 DAP), showing only a small decrease upon the end of grain desiccation, when the polysaccharide represents up to 45% of the grain10,11. Apart from phenotypic differences in grain (1,3;1,4)-β-glucan concentration amongst members of the Poaceae family, it has been shown that the differential expression of the CslF6 gene, which encodes a (1,3;1,4)-β-glucan synthase12, can only partially explain variation in (1,3;1,4)-β-glucan content during grain development13,14,15. Additionally, recent studies suggest that HvCSLF6 has the capability to introduce both (1,3)- and (1,4)-β-linkages within this polysaccharide which generate cellotriosyl (DP3) and cellotetraosyl (DP4) units, affecting (1,3;1,4)-β-glucan solubility and downstream applications16,17. Differences in DP3/DP4 ratio are influenced by various environmental and genetic factors and also described across barley cultivars and grain compartments18.
During barley endosperm development, HvCslF6 and HvCslF9 are the most abundant transcripts of the HvCslF gene family. Expression of HvCslF9 at 3–8 DAP coincides with the initial appearance of (1,3;1,4)-β-glucan in endosperm cell walls4,13. However, the function of HvCslF9 is not entirely clear and it cannot be assumed that all members of the CslF gene family encode (1,3;1,4)-β-glucan synthases19. By contrast, HvCslF6 is well characterized as a (1,3;1,4)-β-glucan synthase that is expressed throughout grain development12. Variation in HvCslF6 transcript levels during late grain development coincide with differential accumulation of grain (1,3;1,4)-β-glucan in different barley cultivars15. Although it is unclear whether this variation is causative, it provides prima facie evidence that variation in HvCslF6 expression could lead to differences in (1,3;1,4)-β-glucan concentration in mature grain.
In addition to polysaccharide biosynthesis, multiple studies suggest that variation in grain (1,3;1,4)-β-glucan content is also influenced by polysaccharide remodelling and degradation pathways13,20,21,22. Two barley genes, HvGlbI and HvGlbII, which encode (1,3;1,4)-β-glucan endohydrolases, have been suggested to contribute to this trait, having an impact on malt (1,3;1,4)-β-glucan modification23,24,25. HvGlbI is the most abundant (1,3;1,4)-β-glucan endohydrolase expressed during endosperm differentiation, in embryonic tissue (scutellum), roots and during germination26, whereas HvGlbII is highly expressed in the aleurone of germinated grain, under gibberellic acid regulation13,26,27 and simulated malting conditions28.
While several naturally occurring polymorphisms have been identified in non-coding regions of HvCslF629,30,31, only three are located within exons and only one affects the protein sequence (A590T). This non-synonymous mutation was identified in the betaglucanless (bgl) mutant29, where it is associated with reduced (1,3;1,4)-β-glucan content and poor agronomic characteristics. However, as reported in a wide range of barley accessions with phenotypic extremes for this trait5,30, the A590T SNP is not diagnostic for grain (1,3;1,4)-β-glucan content. Additionally, Taketa et al.29 described three Sodium azide-induced alleles of bgl which are the result of non-synonymous mutations in highly conserved nucleotides and lead to a lack of (1,3;1,4)-β-glucan.
Two of the end user markets of barley have distinct preferences regarding grain (1,3;1,4)-β-glucan content. Barley cultivars with high (1,3;1,4)-β-glucan content are preferred by the food sector as this polysaccharide is not digested by humans and therefore contributes to total dietary fibre intake, acting against several human health conditions32,33,34. Conversely, the brewing and distilling sector require barley cultivars with low (1,3;1,4)-β-glucan content for an efficient malting process35,36. To date HvCslF6 is the main gene known to functionally contribute to the synthesis of grain (1,3;1,4)-β-glucan content. However, several studies5,29,30,31, have revealed a lack of variation in the HvCslF6 coding sequence (CDS), suggesting that another aspect of HvCslF6, such as the differential regulation of transcript abundance might contribute to variation in the observed phenotype. Previous quantitative trait loci (QTL) studies have suggested that in addition to HvCslF6, both HvCslF9 and HvGlbI could contribute to variation in the grain (1,3;1,4)-β-glucan5,37. To investigate this further we quantified transcript abundance of all three genes in a grain development series across several barley accessions with divergent grain (1,3;1,4)-β-glucan levels. In the same experiment we quantified (1,3;1,4)-β-glucan levels throughout grain development to assess the temporal deposition pattern of this polysaccharide. In parallel, to investigate sequence diversity of HvCsl6 in a wider collection of germplasm than previously examined, we used a combination of existing exome capture data and de novo sequencing of 3,000 bp of the HvCslF6 promoter.
Results
Variation in grain morphology does not explain (1,3;1,4)-β-glucan content
To identify whether any phenotypic measurements associated with grain morphology could indicate the amount of (1,3;1,4)-β-glucan in mature seeds, seven traits [seed weight, roundness, length, width, thickness, overall area and (1,3;1,4)-β-glucan content] were measured using 153 barley 2-row European elite barley cultivars (Table S1). Correlation coefficients between mature grain (1,3;1,4)-β-glucan content and grain phenotypic traits ranged from −0.006 to 0.108 (Table 1) indicating a lack of correlation between these traits.
The assessment of grain characteristics across 2-row cultivars (cvs): Dew, Imidis, Egmont and Gull used in the grain development series showed significant differences in average TGW, grain surface area and its components of width and length (p-value < 0.01) as shown in Fig. S1. The cvs Imidis and Egmont showed significantly higher average TGW (68.77 ± 0.20 and 59.95 ± 0.58, respectively) and larger grain area (26.57 ± 0.03 and 25.80 ± 0.10, respectively) compared to cvs Dew and Gull (p-value < 0.01, both). Significant differences in grain width (ranged 3.1–3.9 mm) and length (ranged 8.1–9.1 mm) were found across the four cvs (p-value < 0.01, both).
(1,3;1,4)-β-glucan accumulation is genotype dependent in barley, especially in later stages of grain development
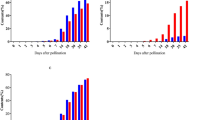
To assess changes in grain (1,3;1,4)-β-glucan content over time, the amount of this polysaccharide present in the grain was quantified at two mid (15 and 24–26 DPA) and two late (32 and 38 DPA) grain development stages in cvs Dew, Imidis, Egmont and Gull (Fig. 1). At 15 DPA, (1,3;1,4)-β-glucan was detected in all cvs and ranged from 1.58% (w/w) ± 0.02 to 3.46% (w/w) ± 0.69. A consistent increase in grain (1,3;1,4)-β-glucan levels was observed across all genotypes from 15 until 24–26 DPA, regardless of final (1,3;1,4)-β-glucan concentration in mature grain. Differences between cvs were more obvious towards the end of grain development (32 to 38 DPA) and were only statistically significant at 38 DPA (p-value < 0.01, Tukey’s test). At grain maturity (38 DPA), two main genotype groups were identified based on divergent (1,3;1,4)-β-glucan content, namely cvs Dew and Imidis and cvs Egmont and Gull. In addition, a potential decrease in (1,3;1,4)-β-glucan concentration was observed at 38 DPA compared with the previous grain development time point in 2-row cvs Dew [4.13% (w/w) ± 0.25] and Imidis [4.26% (w/w) ± 0.14]. Conversely, grain (1,3;1,4)-β-glucan concentration increased from 32 to 38 DPA in cvs Egmont [6.09% (w/w) ± 0.19] and Gull [6.15% (w/w) ± 0.18].
Mean (1,3;1,4)-β-glucan concentration for cvs Dew, Imidis, Egmont, Gull and Morex during grain development stages: 15, 24–26, 32 and 38 days post anthesis (DPA). Bars indicate standard errors from three biological replicates (n = 3). Letters above each bar indicate significant differences within each developmental stage determined by one-way ANOVA followed by Tukey’s test.
Transcript levels of HvCslF6, HvCslF9 and HvGlbI show genotype-dependent and temporal variation during grain development
HvCslF6, HvCslF9 and HvGlbI transcript abundance were quantified at six stages of grain development: 3–5, 8–10, 15, 24–26, 32 and 38 DPA. As described above, cvs Dew, Imidis, Egmont and Gull show differences in mature grain (1,3;1,4)-β-glucan content, varying from 4.13–6.15% (w/w) (Fig. 1). Variation in HvCslF6 expression levels was observed between genotypes with similar grain (1,3;1,4)-β-glucan concentration (either high or low) at the same time point (Fig. 2a). HvCslF6 transcript abundance peaked at 8 DPA in cvs Egmont (high) and Dew (low). The latter showed a prolonged HvCslF6 expression until 15 DPA. At this time point, higher HvCslF6 expression levels were found in cvs Imidis (low) and Dew (low) compared to Gull (high; p-value 0.023, Tukey’s test). During mid to late grain development stages (from 15 until 38 DPA), a significant decrease in HvCslF6 transcript abundance was observed across cvs Imidis (p-value 0.002, Tukey’s test) and Dew (p-value 0.011, Tukey’s test), both with lower grain (1,3;1,4)-β-glucan content compared with cvs Egmont and Gull. Conversely, HvCslF6 expression levels in cv Egmont (high) showed a moderate increase from 24–26 until 38 DPA (p-value 0.015, Tukey’s test). This expression trend was not observed in cv Gull although it contains a similarly high concentration of (1,3;1,4)-β-glucan in mature grain compared to cv Egmont.
Transcript levels of HvCslF6, HvCslF9 and HvGlbI in developing grain of cvs Dew, Imidis, Egmont, Gull and Morex at 3–5, 8–10, 15, 24–26, 32 and 38 days post anthesis (DPA). Transcript levels are expressed as arbitrary units for a) HvCslF6, b) HvCslF9 and c) HvGlbI. Expression data was averaged from three biological replicates per genotype and normalized to α-tubulin, GAPDH and HSP70 housekeeping genes. qRT-PCR reactions were performed in triplicate. Bars indicate standard errors (n = 3). Letters above each bar indicate significant differences within each developmental stage determined by one-way ANOVA followed by Tukey’s test.
While HvCslF6 transcript was found at high levels during mid grain development stages (8–10 and 15 DPA), a genotype-dependent response was observed throughout grain development in the present study. In contrast, transcript abundance of HvCslF9 in all genotypes was extremely low and variable compared to HvCslF6 (Fig. 2b). HvCslF9 transcripts were mostly detected during early grain development (3–5 DPA) (cvs Dew, Imidis and Egmont) except in cv Gull where HvCslF9 expression peaked at 8–10 DPA (p-value 0.037 Tukey’s test). From 15 to 38 DPA, HvCslF9 transcript abundance remained undetectable for most genotypes except for cv Dew which showed a relative increase towards late stages of grain development, although levels were very low compared to early grain development (3–5 DPA).
The transcript profile of HvGlbI, which encodes (1,3;1,4)-glucanase isoenzyme EI, was also determined in this experiment (Fig. 2c). The expression of HvGlbI at 15–26 DPA coincides with the differentiation stage of barley grain development. HvGlbI transcript abundance was higher in the low grain (1,3;1,4)-β-glucan cvs Dew (at 15 DPA, p-value 0.016, Tukey’s test) and Imidis (at 3–5 DPA, p-value 0.011, Tukey’s test) compared to cvs Egmont and Gull, both with high grain (1,3;1,4)-β-glucan through grain development. For the two cvs with lower levels of grain (1,3;1,4)-β-glucan, Imidis and Dew, we observed a temporal difference in the peak of HvGlbI expression, at 3–5 DPA and 15 DPA respectively. HvGlbI expression was undetectable towards late grain development stages (32 and 38 DPA).
HvCslF6 has low levels of genetic variation in coding and non-coding regions within a diverse collection of barley
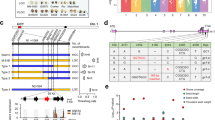
An exome capture sequencing dataset comprising 1,336 barley accessions (including wild, landrace, and elite barley) was used to survey HvCslF6 for sequence variation. Three polymorphisms (MAF < 5% cut off applied) were found within the HvCslF6 coding region: two synonymous SNPs (first and third exon) and a non-synonymous SNPs on the third exon A590T, also described by Taketa et al.29. 10 SNPs were identified in non-coding regions of HvCslF6, seven were located within the first intron and three towards the end of the second intron (Fig. 3). Based on the predicted HvCslF6 gene structure, detected SNPs within introns are not expected to affect splice junctions. Overall, the 13 SNPs found within HvCslF6 had a low frequency across the genotypes analysed (Table 2); alternate alleles were present in approximately 19% of the barley accessions included in this study except for two SNPs located in the first and second introns of HvCslF6 (46.3% and 39.9%, respectively).
HvCslF6 gene structure and relative position of the 13 SNPs identified across 1,336 barley accessions from the exome capture dataset. HvCslF6 coding regions are shown in black boxes and transmembrane domains in stripe rectangles. Triangles represent SNPs (orange within CDS and blue within introns) and the grey box indicates the location of BdTHX1 binding site53.
Limited sequence variation in a −3,000 bp HvCslF6 putative promoter region
We identified 12 SNPs across the 3,000 bp promoter region of 35 barley genotypes described in Table S2. Two of these SNPs were located in the proximal promoter (defined as −500 bp upstream the start codon), which is generally known to contain key cis-acting regulatory elements in plant species38, while the remaining polymorphisms were detected at least −500 bp upstream of the HvCslF6 start codon (Fig. 4). No correlation was found between the 12 SNPs identified within the region upstream of the HvCslF6 start codon and the natural variation in grain (1,3;1,4)-β-glucan content in the germplasm analysed (a subset of 25 elite barley cvs split in two groups with contrasting grain (1,3;1,4)-β-glucan content, 6.3% and 3.0% (w/w) respectively, Table S2). The HvCslF6 promoter sequences of Hordeum vulgare subsp. spontaneum ‘Caesarea’ and progeny lines, OSU105 and OSU127 contained six SNPs (five of them located in the distal and one in the proximal promoter region) that were unique compared with the other genotypes analysed (Table 3). Two of these were present in a set of 12 polymorphisms previously reported in six Australian barley lines15. Both single nucleotide substitutions were detected in the distal promoter.
HvCslF6 promoter structure representation containing 11 SNPs identified across a subset of 35 barley accessions. Previously described SNPs15are indicated by red triangles whereas unique SNPs identified in the current study are shown in grey. Consensus TATA box motif [TATAWAWN70] is represented by a white box. Distal, proximal and core promoter regions71 are shown as shaded boxes.
Discussion
Quantification of (1,3;1,4)-β-glucan content during late endosperm differentiation (15 DPA) until grain maturity (38 DPA) revealed two main groups with divergent grain (1,3;1,4)-β-glucan content. (1,3;1,4)-β-Glucan quantification was not assessed during early grain development stages (3–5 and 8–10 DPA) based on previous data that suggested levels increase later during grain development1,39 as well as to avoid potential errors in determining small concentrations of (1,3;1,4)-β-glucan. Based on HvCslF6 CDS variation, the only non-synonymous SNP (A590T) identified in the exome capture dataset is unlikely to explain differences in grain (1,3;1,4)-β-glucan content5,29,30. This suggests that another aspect of HvCslF6 biology, possibly transcript abundance, or the transcript abundance of other genes potentially contributes to (1,3;1,4)-β-glucan content. Previous QTL studies across different barley populations37,40,41,42,43 identified several genomic regions (1 H, 2 H, 3 H, 5 H and 7 H mainly) collocating with genes putatively involved in the synthesis, remodelling or degradation of (1,3;1,4)-β-glucan which could contribute to the variation in grain (1,3;1,4)-β-glucan content. A QTL interval on 1 H identified in these studies contained HvCslF9, a member of the same gene family as HvCslF6, and HvGlbI which is capable of hydrolysing (1,3;1,4)-β-glucan. Therefore, we quantified wholegrain transcript abundance of HvCslF6, HvCslF9, and HvGlbI in addition to (1,3;1,4)-β-glucan content. The use of wholegrain samples rather than isolated endosperm was important since this reflects the end use of barley grain in malting and feed applications. Despite this, previous studies4,13 on isolated endosperm tissue are reflective of our wholegrain development series for the targeted genes.
The dose-dependent role of HvCslF6 in (1,3;1,4)-β-glucan biosynthesis has previously been demonstrated in barley, where endosperm-specific over-expression led to a considerable increase in (1,3;1,4)-β-glucan concentration12. In addition, the functional characterization of HvCslF6 orthologs confirmed a similar role in wheat44 and rice45. In the present study, HvCslF6 and HvGlbI mRNA abundance was examined in four elite barley cvs with contrasting grain (1,3;1,4)-β-glucan to understand if the combination of transcript abundance of genes encoding biosynthetic and hydrolytic enzymes could explain mature grain (1,3;1,4)-β-glucan content. Although transcript levels and profile varied between the cvs during grain development, the combined variation did correlate with mature grain (1,3;1,4)-β-glucan levels for some cvs, but not for others. It has been observed previously that just considering the expression profile of HvCslF6 does not describe grain (1,3;1,4)-β-glucan content in Australian cvs Sloop and Himalaya13, and more recently in cvs CDC Bold, Beka, Logan, Harrington and breeding lines ‘TR251’, ‘TR306’15. However, qRT-PCR showed that for some cultivars (Dew and Egmont) at least a balance between biosynthetic and hydrolytic transcript abundance, and hence activities, likely impacts final (1,3;1,4)-β-glucan content. For others (Imidis and Gull) the lack of a direct relationship between transcript abundance of HvCslF6 and HvGlbI, and grain (1,3;1,4)-β-glucan content raises several possibilities. One, that there may be other genes contributing to final grain (1,3;1,4)-β-glucan content, and two, that wholegrain transcript abundance may not necessarily correlate with protein amount and enzyme activity. Although the lack of polymorphisms in the HvCslF6 coding sequence (see below) argues against any cultivar-specific modification to enzyme activity, it is possible that in some cultivars, polymorphisms in non-coding regions could impact interactions with upstream transcription factors and this could contribute to variation in transcript abundance and grain (1,3;1,4)-β-glucan.
Based on the current grain development series, 2-row genotypes (cvs Dew, Imidis, Egmont, Gull) had highest levels of HvCslF6 expression at 8–10 DPA and 15 DPA during grain development. Similarly Nemeth et al.44 observed that the wheat CslF6 ortholog, TaCslF6, was highly expressed during mid-endosperm development in cv Hereward, corresponding to the grain filling stage, showing maximum expression levels from 8–15 DPA. In a more recent study, the average TaCslF6 mRNA levels across 10 wheat cvs were abundant at 21 and 28 DPA although different expression patterns were found within this subset of wheat genotypes, showing significant differences in TaCslF6 transcript abundance at 21, 28 and 40 DPA46. These expression profiles are consistent with a function of (1,3;1,4)-β-glucan as a destination for energy storage in the form of metabolizable glucose during grain filling and germination47,48. As mentioned in Wong et al.15, differences in HvCslF6 expression levels during late grain development might impact (1,3;1,4)-β-glucan accumulation in mature barley grain. In our experiment cv Egmont [6.09% (w/w) ± 0.19] showed an increase in HvCslF6 expression at 38 DPA and no HvGlbI expression at this time point in contrast to low grain (1,3;1,4)-β-glucan cvs Dew [4.13% (w/w) ± 0.25] and Imidis [4.26% (w/w) ± 0.14]. Additionally, cv Egmont showed a 14.10% increase in (1,3;1,4)-β-glucan content from 32 DPA and 38 DPA coupled with an increase in HvCslF6 expression, but a lack of HvGlbI.
In the cvs included in the present study the transcriptional profile of HvGlbI has a peak in expression at 15 DPA and 24–26 DPA, suggesting a potential role for this enzyme in determining (1,3;1,4)-β-glucan content during the later stages of grain development. A particularly high level of HvGlbI expression was observed at 15 DPA in cv Dew, which is characterized by a relatively low (1,3;1,4)-β-glucan content in mature grain compared to cvs Egmont and Gull (both with higher (1,3;1,4)-β-glucan content). In contrast, for other genotypes we observed concurrent high or low HvCslF6 and HvGlbI transcript, which could initially appear to be counterintuitive. Concurrent synthase and hydrolase activity may contribute to re-modelling of the nascent polysaccharide, to hydrolysis of the polysaccharide off the synthase, or provide a rapidly degradable source of glucose in the developing grain due to its unbranched structure compared to starch20,47,49. Additionally, potential expression differences in (1,3;1,4)-β-endoglucanase HvGlbII, other putative β-glucan glucohydrolases (HvExoI and HvExoII) or β-glucosidases that were not investigated in this study may also contribute to variation in grain (1,3;1,4)-β-glucan content. Recently, Betts et al.28 reported expression differences of (1,3;1,4)-β-endoglucanases (HvGlbI and HvGlbII) in simulated malting conditions across malting and feed barley cvs. HvGlbI expression was three-fold higher than HvGlbII whose expression is known to be restricted to the aleurone under GA regulation26. However, since a direct relationship between transcript abundance and activity levels cannot be assumed, the quantification of (1,3;1,4)-β-glucanase activity in future studies could provide useful data towards understanding variation during grain development in the cultivars investigated here. Alternatively, this could indicate that the relationship between HvGlbI expression, isoenzyme EI activity, and consequently (1,3;1,4)-β-glucan content is nonlinear. It is possible that HvGlbI acts in combination with other remodelling and degrading enzyme partners to explain differences in grain (1,3;1,4)-β-glucan content.
Based on the grain development series used in the current study, the expression of HvCslF9 was predominantly detected from 3–5 DPA, except for a later expression peak in cv Gull (8–10 DPA). Burton et al.13 also observed a peak of expression for HvCslF9 for cv Sloop at 8 DAP, while at other time points in both genotypes included in their study HvCslF9 transcripts were absent or at extremely low levels. Similarly, in the current study during mid and late grain development, and coinciding with endosperm differentiation and maturation, HvCslF9 expression was almost undetectable across all genotypes. While other independent HvCslF9 expression studies support a peak expression of this gene during endosperm differentiation stages13, HvCslF9 over-expression did not increase (1,3;1,4)-β-glucan content in transgenic barley grain12, with similar results obtained in wheat addition lines50. The fact that the genomic location of HvCslF9 and HvGlbI [1 H, 48.1 cM and 1 H, 54.4 cM, respectively51] co-locates with association peaks based on two independent QTL and genome-wide association studies (GWAS) on grain (1,3;1,4)-β-glucan content5,37 might suggest that variation in (1,3;1,4)-β-glucan is due to HvGlbI (1,3;1,4)-β-glucanase activity and not HvCslF9 biosynthetic activity.
In the current study, a survey of HvCslF6 sequence variation in 1,336 exome-captured barley accessions identified three SNPs within the HvCslF6 coding sequence, two synonymous and one non-synonymous SNP, A590T which doesn’t explain differences in grain (1,3;1,4)-β-glucan content5,15,29,30. Based on the model of HvCSLF6 described by Schwerdt et al.52, the A590T substitution is proximal to a 55 amino acid insert that is specific to CSLF6 proteins of the grasses and absent in other CSLF proteins. While several amino acids under positive selection (non-synonymous to synonymous nucleotide substitution rate greater than 1) were described flanking the 55 amino acid insert and at other predicted transmembrane domains, low levels of sequence variation were found in CSLF6 unlike other CSLF proteins in grasses. The role of the CSLF6-specific amino acid insert, far from the active site, is not entirely clear however it contains charged aa residues and a conserved Cys residue which might facilitate the interaction with other protein/s12,52.
In the same exome-captured dataset, 10 SNPs were detected in non-coding regions of HvCslF6, mostly within the first intron, confirming that natural variation within this gene is rare. Recent work in Brachypodium distachyon identified a trihelix family transcription factor (BdTHX1) which binds to a GT-motif in the second intron of BdCslF653. The same motif is found in the first intron of HvCslF6 and another putative GT-motif is present in the second intron. However, the 10 SNPs within HvCslF6 introns identified in this study did not affect the THX1 binding site or GT-motifs described by Fan et al.53.
Similar to the intronic regions of HvCslF6, low levels of variation were identified in the putative HvCslF6 promoter sequence. In a −3,000 bp region upstream of the HvCslF6 start codon, DNA sequence alignments identified only 12 SNPs across 35 barley accessions representing natural (1,3;1,4)-β-glucan variation. Polymorphisms were mostly found in the HvCslF6 distal promoter, in which two SNPs were found in common with a set of six Australian cultivars previously analysed for sequence variation in 5’ and 3’ UTR regions15. These data indicate that there is remarkably narrow variation within HvCslF6 promoter and enzyme coding sequences. None of these SNPs appeared to associate with variation in (1,3;1,4)-β-glucan content. Although genetic diversity will likely be lower when considering elite cultivars, Russell et al.54 observed 745,815 SNPs in high confidence exons across 20,729 high confidence gene models, representing an average of 35.9 SNPs per gene model in a collection of 267 wild and landrace barleys. The low sequence diversity in HvCslF6 identified in our current study and by others15,30,31 appears to highlight atypical conservation of the HvCslF6 nucleotide sequence, possibly due to its indispensable role in (1,3;1,4)-β-glucan synthesis and the importance of this polysaccharide in the grasses, or a common origin of the gene55. This role has been confirmed via the characterization of barley chemically-induced mutants that completely lack grain (1,3;1,4)-β-glucan and exhibit poor agronomic traits29,56, or partially functional HvCslF6 mutants with less severe phenotypic effects31. Thus far, evidence suggests that the basis for differences in grain (1,3;1,4)-β-glucan is not linked to any polymorphism within HvCslF6 and hence other regulatory sequences or proteins, acting independently or in combination, are likely to be involved.
Mature grain characteristics, including size and shape, were not obviously correlated with the amount of grain (1,3;1,4)-β-glucan or expression differences in genes that produce enzymes that synthesize or hydrolyze (1,3;1,4)-β-glucan. Hence, natural diversity for barley grain size seems unlikely to affect (1,3;1,4)-β-glucan concentration. This is despite recent work on the barley lys3a shrunken endosperm mutant showing that defects in hordein synthesis could potentially affect HvCslF6 transcript abundance57,58. Shrunken endosperm barley mutants typically exhibit defects in starch biosynthesis and embryo development, but the (1,3;1,4)-β-glucan content was not quantified59. Since (1,3;1,4)-β-glucan has been proposed to act as an alternative source of stored glucose60, knowing the (1,3;1,4)-β-glucan content of shrunken endosperm mutants may help to explain the role of this polysaccharide and the regulation of HvCslF6. Furthermore, by comparing Brachypodium and barley grain during development Trafford et al.61 suggest that starch synthesis influences endosperm cell enlargement and as a consequence endosperm size. Therefore, although the relationship between starch and grain size is well understood, perhaps a wider screen of mutant germplasm62 might reveal a link between grain size and shape, and (1,3;1,4)-β-glucan content.
Material and Methods
Plant material for grain development series
Four elite 2-row spring cvs: Dew, Imidis, Egmont and Gull were used in this experiment. Based on previous work5, these cvs represent phenotypic extremes of grain (1,3;1,4)-β-glucan concentration (2.1–6.7% w/w). Three biological replicates across six grain development stages: 3–5, 8–10, 15, 24–26, 32 and 38 days post anthesis (DPA) were collected from material grown in glasshouse conditions of 16 h light/8 h dark, until maturity at The James Hutton Institute, United Kingdom (Feb–May 2015). In the present grain development series, DPA were considered equivalent to days after pollination (DAP), which has been used in previous studies4,13,15. For each genotype, six developing grains from the central part of the spike were collected at each developmental stage and snap frozen in liquid nitrogen for further mRNA and (1,3;1,4)-β-glucan quantification analyses.
Isolation of mRNA and cDNA synthesis
For mRNA isolation, we collected three biological replicates from the time points described above, and for each time point included three technical replicates. Samples were ground into a fine powder (100 mg) in liquid nitrogen and mixed with 1 mL TRIzol (Thermo Fisher Scientific, Waltham, USA). Manufacturer’s instructions were followed with several modifications: (1) After phase separation, supernatant was transferred to a fresh tube and mixed with 0.25 mL isopropanol followed by 0.25 mL 0.8 M sodium citrate/1.2 M sodium chloride (per 1 mL TRIzol used, both) to help removal of polysaccharides. Samples were incubated for 10 min, at room temperature. (2) After an ethanol wash (1 mL 75% ethanol per 1 ml of TRIzol used), RNA pellets were re-suspended in 400 µL RNase-free water on ice. (2) For RNA purification, 400 µL chloroform/isoamyl alcohol (Sigma Aldrich, St. Louis, USA) was added, mixed well and centrifuged (16,000 × g, 4 °C) for 10 min. The top phase (~300 µL) was removed to a clean 1.5 mL Eppendorf tube on ice and 100 µL RNase-free water was added to the original tubes, mixed well and centrifuged (same conditions) for 5 min. The top phase (~100 µL) was removed and added to the 300 µL already collected in tubes, remaining on ice. (3) For RNA precipitation, samples were mixed with 1 mL absolute ethanol and 40 µL 3 M sodium acetate and stored at −80 °C overnight. The next day, samples were centrifuged (16,000 × g, 4 °C) for 10 min and supernatants were removed. RNA pellets were washed in 1 mL 70% ethanol and re-suspended in 100 µL RNase-free water. cDNA synthesis (1 µg total RNA) was performed using the RNA to cDNA EcoDry™ Premix (Takara, Kyoto, Japan) according to the manufacturer’s instructions.
qRT-PCR
Quantitative real-time PCR (qRT-PCR) was performed in a StepOne Real-Time PCR machine (Thermo Fisher Scientific, Waltham, USA) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, USA). Three replicate qRT-PCR reactions were performed for each cDNA sample including three negative controls using RNAse-free water. Gene specific primers and qRT-PCR reaction conditions were used as described in previous studies12,13. Absolute mRNA quantification of HvCslF6, HvCslF9 and HvGlbI was performed using three housekeeping genes for normalization: α-tubulin, (α-tub) glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and heat shock protein (HSP70) (Table S3). Multiple control gene normalization was performed as described in Vandesompele et al.63. Normalized mRNA copies were calculated based on standard concentrations. Each gene standard (101–107 mRNA copies/µL) for HvCslF6, HvCslF9, HvGlbI, α-tubulin, GAPDH and HSP70 was generated by HPLC at The University of Adelaide13.
Quantification of (1,3;1,4)-β-glucan in developing grain
Grain (1,3;1,4)-β-glucan levels were determined by a modified version of the Megazyme β-Glucan (Mixed Linkage) Assay Kit (Megazyme Int., Wicklow, Ireland)12,64 which permits the analysis of small samples (15 mg). Each biological replicate consisted of three barley grain per genotype and time point. Barley grain samples were weighed before and after a 72-h incubation at 65 °C to calculate dry mass and milled in a Powerlyser tissue homogenizer (MO BIO, CA, USA). Three independent barley flour samples (3 × 15 mg samples) were obtained and averaged for final grain (1,3;1,4)-β-glucan quantification, calculated as % of dry weight (w/w). Two technical replicates were performed on all samples using the Megazyme kit apart from a standardized barley flour control [4.05%–4.15% w/w of (1,3;1,4)-β-glucan, from Megazyme kit] included in each batch. Differences in (1,3;1,4)-β-glucan content for each developmental stage were determined by one-way ANOVA followed by Tukey’s honest significant difference (HSD) test using GenStat v1965.
Phenotypic assessment of grain characteristics
A collection of 153 2-row elite barley lines was used for grain phenotypic measurements (Table S2). A sample of approximately 100–150 bulked seeds per genotype was used to measure seed weight, roundness, length, width, thickness and overall seed area using a SeedCount SC4 (Seed Count Australasia, Condell Park, Australia) at the University of Adelaide Barley Breeding Program/Laboratory, following manufacturer’s instructions. These data where combined with previously published grain (1,3;1,4)-β-glucan data5.
HvCslF6 promoter resequencing
In total, 35 barley genotypes were used to survey sequence variation within a −3,000 bp HvCslF6 upstream region including: 25 elite barley cvs with divergent grain (1,3;1,4)-β-glucan content [2.1–6.7% w/w5], four ‘Bowman’ Near Isogenic Lines (NILs): BW840, BW837, BW638 and BW83562 with introgressions spanning the region where HvCslF6 maps to chromosome 7 H, two Recombinant Chromosome Substitution Lines (RCSLs), OSU105 and OSU12766, and their corresponding parental lines (Table S2). RCSL genotypes were chosen based on their introgressions from cv Caesarea (Hordeum vulgare subsp. spontaneum), which cover the genomic location of HvCslF6. Genomic DNA was isolated from seedlings using the Qiagen DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. PCR amplification of a −3,000 bp HvCslF6 upstream region was achieved by dividing the fragment into four overlapping PCR reactions (Table S4). PCR cycle details are as follows: 98 °C for 2 min; 5 cycles of 98 °C for 15 s, 60 °C for 20 s, 72 °C for 1 min with a touchdown on the primer annealing step of −1 °C/cycle; then 35 cycles of 98 °C for 15 s, 56 °C for 20 s, 72 °C for 1 min; followed by 72 °C for 2 min. PCR amplicons were purified using ExoStar™ (GE Healthcare UK Ltd., Buckinghamshire, UK) following the manufacturer’s instructions. Sanger Sequencing was performed using an ABI3730 DNA Analyzer (Applied Biosystems Inc., Foster City, USA) at The James Hutton Institute. Resulting sequences were aligned to cv Morex HvCslF6 reference sequence retrieved from the barley genome explorer, Barlex51 and analysed with Geneious V.967 to identify polymorphisms.
Exome capture dataset
The genomic position of HvCslF6 (HORVU7Hr1G070010.3) on the physical map was retrieved using the Barlex database51, available at: http://www.barlex.barleysequence.org. Sequence variation of HvCslF6 was analysed in 1,336 exome sequenced barley accessions (Unpublished, The James Hutton Institute). This dataset comprises a georeferenced collection of exotic barley alleles including 340 landraces (Hordeum vulgare ssp. vulgare), the Spanish core collection68, 288 wild lines (Hordeum vulgare ssp. spontaneum) of which 80 correspond to the Barley1K collection from Israel69 and 2- and 6-row collections of contemporary European barley cultivars evaluated in previous projects (WHEALBI and CLIMBAR) at The James Hutton Institute. Polymorphisms identified in barley accessions with suspected heterozygosity and minimum allele frequency (MAF) of ≤5% (cut-off) were removed from the analysis as described in Russell et al.54.
References
Fincher, G. B. Morphology and chemical composition of barley endosperm cell walls. J. Inst. Brew. 81, 116–122 (1975).
Young, T. E. & Gallie, D. R. Programmed cell death during endosperm development. Plant Mol. Biol. 44, 283–301 (2000).
Sreenivasulu, N. et al. Barley grain development: Toward an integrative view. In International Review of Cell and Molecular Biology (ed. Jeon, K. W.) 49–89, https://doi.org/10.1016/S1937-6448(10)81002-0 (Academic Press, 2010).
Wilson, S. M. et al. Pattern of deposition of cell wall polysaccharides and transcript abundance of related cell wall synthesis genes during differentiation in barley endosperm. Plant Physiol. 159, 655–670 (2012).
Houston, K. et al. A genome wide association scan for (1,3;1,4)-β-glucan content in the grain of contemporary 2-row spring and winter barleys. BMC Genomics 15, 1–15 (2014).
Izydorczyk, M. S., Storsley, J., Labossiere, D., MacGregor, A. W. & Rossnagel, B. G. Variation in total and soluble β-glucan content in hulless barley: effects of thermal, physical, and enzymic treatments. J. Agric. Food Chem. 48, 982–989 (2000).
Henry, R. J. & Brown, A. H. D. Variation in the carbohydrate composition of wild barley (Hordeum spontaneum) grain. Plant Breed. 98, 97–103 (1987).
Burton, R. A. & Fincher, G. B. Current challenges in cell wall biology in the cereals and grasses. Front. Plant Sci. 3, 1–6 (2012).
Philippe, S., Saulnier, L. & Guillon, F. Arabinoxylan and (1→3),(1→4)-β-glucan deposition in cell walls during wheat endosperm development. Planta 224, 449–461 (2006).
Guillon, F. et al. Brachypodium distachyon grain: Characterization of endosperm cell walls. J. Exp. Bot. 62, 1001–1015 (2011).
Guillon, F. et al. A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J. Exp. Bot. 63, 739–755 (2012).
Burton, R. A. et al. Over-expression of specific HvCslF Cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol. J. 9, 117–135 (2011).
Burton, R. A. et al. The genetics and transcriptional profiles of the Cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 146, 1821–1833 (2008).
Pellny, T. K. et al. Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-Seq transcriptome. Plant Physiol. 158, 612–627 (2012).
Wong, S. C. et al. Differential expression of the HvCslF6 gene late in grain development may explain quantitative differences in (1,3;1,4)-β-glucan concentration in barley. Mol. Breed. 35 (2015).
Dimitroff, G. et al. (1,3;1,4)-β-Glucan biosynthesis by the CSLF6 enzyme: Position and flexibility of catalytic residues influence product fine structure. Biochemistry 55, 2054–2061 (2016).
Jobling, S. A. Membrane pore architecture of the CSLF6 protein controls (1-3,1-4)-β-glucan structure. Sci. Adv. 1, e1500069 (2015).
Izydorczyk, M. S. & Dexter, J. E. Barley β-glucans and arabinoxylans: Molecular structure, physicochemical properties, and uses in food products-a Review. Food Res. Int. 41, 850–868 (2008).
Little, A. et al. A novel (1,4)-β-linked glucoxylan is synthesized by members of the Cellulose synthase-like F gene family in land plants. ACS Cent. Sci. 5, 73–84 (2019).
Farrokhi, N. et al. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 4, 145–167 (2006).
Fincher, G. B. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 149, 27–37 (2009).
Hrmova, M., Farkas, V., Lahnstein, J. & Fincher, G. B. A barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-D-glucans. J. Biol. Chem. 282, 12951–12962 (2007).
Fincher, G. B. et al. Primary structure of the (1→3,1→4)-β-D-glucan 4-glucohydrolase from barley aleurone. Proc. Natl. Acad. Sci. USA 83, 2081–2085 (1986).
Slakeski, N. et al. Structure and tissue-specific regulation of genes encoding barley (1→3, 1→4)-β-glucan endohydrolases. MGG Mol. Gen. Genet. 224, 437–449 (1990).
Stuart, I. M., Loi, L. & Fincher, G. B. Varietal and environmental variations in (1→3,1→4)-β-glucan levels and (1→3,1→4)-β-glucanase potential in barley: Relationships to malting quality. J. Cereal Sci. 7, 61–71 (1988).
Slakeski, N. & Fincher, G. B. Developmental regulation of (1→3,1→4)-β-glucanase gene expression in barley. Plant Physiol. 99, 1226–1231 (1992).
Mundy, J. & Fincher, B. Effects of gibberellic acid and abscisic acid on levels of aleurone. FEBS Lett. 198, 349–352 (1986).
Betts, N. S. et al. Morphology, carbohydrate distribution, gene expression, and enzymatic activities related to cell wall hydrolysis in four barley varieties during simulated malting. Front. Plant Sci. 8, 1–15 (2017).
Taketa, S. et al. Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-D-glucan biosynthesis. J. Exp. Bot. 63, 381–392 (2012).
Cory, A. T., Båga, M., Anyia, A., Rossnagel, B. G. & Chibbar, R. N. Genetic markers for CslF6 gene associated with (1,3;1,4)-β-glucan concentration in barley grain. J. Cereal Sci. 56, 332–339 (2012).
Hu, G., Burton, C., Hong, Z. & Jackson, E. A mutation of the Cellulose-synthase-like (CslF6) gene in barley (Hordeum vulgare L.) partially affects the β-glucan content in grains. J. Cereal Sci. 59, 189–195 (2014).
Ames, N., Storsley, J. & Thandapilly, S. J. Functionality of beta-glucan from oat and barley and its relation with human health. In Cereal Grain-based Functional Foods: Carbohydrate and Phytochemical Components (eds Beta, T. & Camire, M. E.) 147–166, https://doi.org/10.1039/9781788012799 (Royal Society of Chemistry, 2018).
Cosola, C. et al. Beta-glucans supplementation associates with reduction in P-cresyl sulfate levels and improved endothelial vascular reactivity in healthy individuals. PLoS One 12, 1–16 (2017).
Keenan, J. M. et al. The effects of concentrated barley β-glucan on blood lipids in a population of hypercholesterolaemic men and women. Br. J. Nutr. 97, 1162–1168 (2007).
Gupta, M., Abu-Ghannam, N. & Gallaghar, E. Barley for brewing: Characteristic changes during malting, brewing and applications of its by-products. Compr. Rev. Food Sci. Food Saf. 9, 318–328 (2010).
Wang, J., Zhang, G., Chen, J. & Wu, F. The changes of β-glucan content and β-glucanase activity in barley before and after malting and their relationships to malt qualities. Food Chem. 86, 223–228 (2004).
Han, F. et al. Mapping of β-glucan content and β-glucanase activity loci in barley grain and malt. Theor. Appl. Genet. 91, 921–927 (1995).
Shahmuradov, I. A., Solovyev, V. V. & Gammerman, A. J. Plant promoter prediction with confidence estimation. Nucleic Acids Res. 33, 1069–1076 (2005).
Wilson, S. M. et al. Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta 224, 655–667 (2006).
Li, J., Båga, M., Rossnagel, B. G., Legge, W. G. & Chibbar, R. N. Identification of quantitative trait loci for β-glucan concentration in barley grain. J. Cereal Sci. 48, 647–655 (2008).
Islamovic, E. et al. Genetic dissection of grain beta-glucan and amylose content in barley (Hordeum vulgare L.). Mol. Breed. 31, 15–25 (2013).
Molina-Cano, J. L. et al. QTL analysis of a cross between European and North American malting barleys reveals a putative candidate gene for β-glucan content on chromosome 1H. Mol. Breed. 19, 275–284 (2007).
Igartua, E., Hayes, P. M., Thomas, W. T. B., Meyer, R. & Mather, D. E. Genetic control of quantitative grain and malt quality traits in barley. J. Crop Prod. 5, 131–164 (2002).
Nemeth, C. et al. Down-regulation of the CslF6 gene results in decreased (1,3;1,4)-β-d-glucan in endosperm of wheat. Plant Physiol. 152, 1209–1218 (2010).
Vega-Sanchez, M. E. et al. Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69 (2012).
Marcotuli, I., Pasqualina, C., Antonio, B. & Agata, G. Expression analysis of Cellulose synthase-like genes in durum wheat. Sci. Rep. 8, 1–9 (2018).
Burton, R. A. & Fincher, G. B. (1,3;1,4)-β-D-glucans in cell walls of the poaceae, lower plants, and fungi: A tale of two linkages. Mol. Plant 2, 873–882 (2009).
Gibeaut, D. M., Pauly, M., Bacic, A. & Fincher, G. B. Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221, 729–738 (2005).
Buckeridge, M. S., Rayon, C., Urbanowicz, B., Tiné, M. A. S. & Carpita, N. C. Mixed linkage (1→3),(1→4)-β-D-glucans of grasses. Cereal Chem. 81, 115–127 (2004).
Cseh, A. et al. Expression of HvCslF9 and HvCslF6 barley genes in the genetic background of wheat and their influence on the wheat β-glucan content. Ann. Appl. Biol. 163, 142–150 (2013).
Colmsee, C. et al. BARLEX - The barley draft genome explorer. Mol. Plant 8, 964–966 (2015).
Schwerdt, J. G. et al. Evolutionary dynamics of the Cellulose synthase gene superfamily in grasses. Plant Physiol. 168, 968–983 (2015).
Fan, M. et al. A trihelix family transcription factor is associated with key genes in mixed-linkage glucan accumulation. Plant Physiol. 178, 1207–1221 (2018).
Russell, J. et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet. 48, 1024–1030 (2016).
Little, A. et al. Revised phylogeny of the Cellulose synthase gene superfamily: insights into cell wall evolution. Plant Physiol. 177, 1124–1141 (2018).
Tonooka, T., Aoki, E., Yoshioka, T. & Taketa, S. A novel mutant gene for (1-3, 1-4)-β-D-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breed. Sci. 59, 47–54 (2009).
Moehs, C. P. et al. Development of decreased-gluten wheat enabled by determination of the genetic basis of lys3a barley. Plant Physiol. 179, 1692–1703 (2019).
Christensen, U. & Scheller, H. V. Regulation of (1,3;1,4)-β-d-glucan synthesis in developing endosperm of barley lys mutants. J. Cereal Sci. 55, 69–76 (2012).
Cook, F. et al. Barley lys3 mutants are unique amongst shrunken-endosperm mutants in having abnormally large embryos. J. Cereal Sci. 82, 16–24 (2018).
Roulin, S., Buchala, A. J. & Fincher, G. B. Induction of (1→3,1→4)-β-D-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta 215, 51–59 (2002).
Trafford, K. et al. Grain development in Brachypodium and other grasses: Possible interactions between cell expansion, starch deposition, and cell-wall synthesis. J. Exp. Bot. 64, 5033–5047 (2013).
Druka, A. et al. Genetic dissection of barley morphology and development. Plant Physiol. 155, 617–627 (2011).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 34–1 (2002).
McCleary, B. V. & Codd, R. Measurement of (1→3),(1→4)-β-D-glucan in barley and oats: A streamlined enzymic procedure. J. Sci. Food Agric. 55, 303–312 (1991).
VSN International. Genstat for Windows 19th Edition. Available at, https://www.vsni.co.uk/ (2017).
Matus, I. et al. Development and characterization of recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Genome 46, 1010–1023 (2003).
Kearse, M. et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Igartua, E. et al. The Spanish barley core collection. Genet. Resour. Crop Evol. 45, 475–481 (1998).
Hübner, S. et al. Strong correlation of wild barley (Hordeum spontaneum) population structure with temperature and precipitation variation. Mol. Ecol. 18, 1523–1536 (2009).
Juo, Z. S. et al. How proteins recognize the TATA box. J. Mol. Biol. 261, 239–254 (1996).
Pedersen, A. G., Baldi, P., Chauvin, Y. & Brunak, S. The biology of eukaryotic promoter prediction - A review. Comput. Chem. 23, 191–207 (1999).
Acknowledgements
We thank the Genome Technology team at The James Hutton Institute (UK), for technical assistance during RNA isolation from mature barley grain and amplicon Sanger sequencing. The work was supported by the Australian Research Council, the BBSRC, and Rural & Environment Science & Analytical Services Division of the Scottish Government.
Author information
Authors and Affiliations
Contributions
K.H., M.R.T., R.A.B., J.R. and R.W. conceived this work. G.G.-G. and K.H. designed the grain development series experiment. G.G.-G. performed the sampling, laboratory work and analysed the data. J.R. curated the exome capture dataset. M.K.A. collected grain phenotypic measurements. The manuscript was drafted by G.-G.G., K.H. and M.R.T. and reviewed by G.B.F., R.A.B. and R.W. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Garcia-Gimenez, G., Russell, J., Aubert, M.K. et al. Barley grain (1,3;1,4)-β-glucan content: effects of transcript and sequence variation in genes encoding the corresponding synthase and endohydrolase enzymes. Sci Rep 9, 17250 (2019). https://doi.org/10.1038/s41598-019-53798-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-53798-8
This article is cited by
-
Characterization of barley (Horduem vulgare) lys3 mutants identifies genes under the regulation of the prolamin-box binding transcription factor and elucidates its role in endosperm promoter methylation during grain development
Molecular Genetics and Genomics (2024)
-
HvBGlu3, a GH1 β-glucosidase enzyme gene, negatively influences β-glucan content in barley grains
Theoretical and Applied Genetics (2024)
-
Distinct roles of carbohydrate-binding modules in multidomain β-1,3–1,4-glucanase on polysaccharide degradation
Applied Microbiology and Biotechnology (2023)
-
Identification of genetic loci and candidate genes related to β-glucan content in barley grain by genome-wide association study in International Barley Core Selected Collection
Molecular Breeding (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.