Abstract
In this study, a deep learning-based method for developing an automated diagnostic support system that detects periodontal bone loss in the panoramic dental radiographs is proposed. The presented method called DeNTNet not only detects lesions but also provides the corresponding teeth numbers of the lesion according to dental federation notation. DeNTNet applies deep convolutional neural networks(CNNs) using transfer learning and clinical prior knowledge to overcome the morphological variation of the lesions and imbalanced training dataset. With 12,179 panoramic dental radiographs annotated by experienced dental clinicians, DeNTNet was trained, validated, and tested using 11,189, 190, and 800 panoramic dental radiographs, respectively. Each experimental model was subjected to comparative study to demonstrate the validity of each phase of the proposed method. When compared to the dental clinicians, DeNTNet achieved the F1 score of 0.75 on the test set, whereas the average performance of dental clinicians was 0.69.
Similar content being viewed by others
Introduction
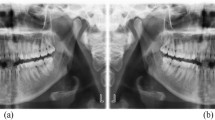
Periodontal disease caused by dental bacterial infection is one of the most common human diseases affecting gums and the support structure of the teeth. Periodontitis is an inflammatory disease which can result in periodontal bone loss (PBL) and ultimately leads to loosening or loss of teeth if not diagnosed and treated properly1. Therefore, early detection and management of PBL plays a crucial role in improving the clinical outcome of periodontal disease. To acquire valuable information for the diagnosis and treatment planning of PBL, radiological exams, including bitewing, periapical, and panoramic radiographs, have been widely used in clinical practices. While intra-oral images such as bitewing and periapical radiographs have been routinely taken for the diagnosis of PBL, extra-oral panoramic radiographs which capture the entire mouth have been widely used for their advantages over intra-oral images such as lower radiation exposure, better patient comfort, faster and easier procedure, and wider field of view2,3. However, detecting and diagnosing PBL in panoramic dental radiographs is considered a difficult task with a low intra and inter-examiner agreement rate due to their complex structures and low resolution4 as shown in Fig. 1.
Deep convolutional neural networks (DCNNs) have recently been actively adopted in medical image analysis with successful applications to computer-aided detection (CADe) and diagnosis (CADx)5,6,7,8. While the majority of these studies focus on analyzing images from radiology, pathology, dermatology, and ophthalmology, studies dealing with various dental imaging modalities using DCNNs have also been conducted recently9. In addition to the aforementioned works which focus on caries and plaque, some studies also deal with periodontal disease10,11. Lee et al.10 used intra-oral images for the detection of periodontally compromised teeth, whereas Krois et al.11 proposed a DCNN-based method for the detection of PBL in panoramic dental radiographs. The proposed method in Krois et al.11 however has some limitations: their DCNN is trained using manually cropped teeth patches, the dataset used in their study is small, and their DCNN architecture is shallow.
In this study, we develop a fully automated method to detect PBL in panoramic dental radiographs. By exploiting transfer learning and lesion correlation prior information, the proposed DCNN named DeNTNet is able to accurately detect PBL, outperforming human experts when tested on data consisting of all teeth types. To be more clinically applicable, DeNTNet is trained to predict the existence of PBL for each tooth and is thus capable of providing the teeth numberings of predicted lesions. To the best of our knowledge, this is the first study to propose a multi-phase deep learning framework to detect PBL in whole panoramic dental radiographs with teeth numbering.
Materials and Methods
This study was approved by the institutional review board of Korea University Anam Hospital(IRB No.2016AN0267) with a waiver of informed consent. The dataset collection and experiments were performed in accordance with the approved ethical guidelines and regulations.
Data collection and annotation
A total of 12,179 panoramic dental radiographs were retrospectively collected from Korea University of Anam Hospital after removing identifiable patient information during the period between \({1}^{{\rm{st}}}\) Jan. 2014 and \(1{4}^{{\rm{th}}}\) Feb. 2016. The radiographs were taken with devices from multiple vendors: DENTRI (HDXWILL, South Korea), Hyper-XCM (Asahi, Japan), CS 9300 (Carestream Dental, USA), Papaya (Genoray, South Korea), and PHT-30LFO (Vatech, South Korea). Only one radiograph per patient is included by excluding all follow-up exams taken during data collection, and the patients’ gender and age distribution is described in the appendix. Since the intra and inter-examiner agreement of PBL is considered to be low4,11, five dental clinicians who are experienced dental hygienists with 5, 9, 16, 17, 19 years of practice for assessing dental radiographs independently masked the PBL lesions and recorded the corresponding tooth number for annotation. During the course of data collection, the annotation quality was continuously monitored by a separate dental expert who has been in clinical dental practice for over 15 years as a board-certified oral and maxillofacial surgeon. For teeth numbering, the Federation Dentaire Internationale (FDI) teeth numbering system (ISO-3950) was used12 and there are 32 teeth in total, as shown in Fig. 2. To construct the reference dataset, the five teeth-level annotations were aggregated using the following rule:
where \(i\) is the data index (\(N=12,179\)), \(j\) is the tooth index (\(T=32\)), and \(l\) is the annotator index (\(L=5\)). Here we set \({C}_{R}=3\) which corresponds to majority voting. We randomly split the dataset into training (\({N}_{tr}=11,198\)), validation (\({N}_{val}=190\)), and test (\({N}_{tst}=800\)) sets.
Overall framework
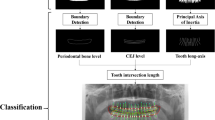
The overall framework of the proposed method consists of multiple stages, as illustrated in Fig. 2. At the first stage, we trained a region of interest (ROI) segmentation network to extract the teeth related region. In the next stage, a lesion segmentation network was trained to predict the PBL lesions. By exploiting the encoder part of this lesion segmentation network as a pre-trained model, we trained a classification network that predicts the existence of PBL in each tooth. To further improve performance, we also train a separate classification network, that predicts the existence of PBL lesions specifically for the premolar and molar teeth types. The two classification networks are ensembled to make the final PBL lesion prediction. The detailed procedures are described in the following sections, and the contribution of each stage to the final PBL detection performance is also analyzed in the ablation study.
Overall procedure for training DeNTNet. (a) ROI segmentation network used to extract teeth regions; (b) PBL lesion segmentation network as a pre-trained model; (c) Tooth-level PBL classification network with transferred weight; (d)Tooth-level PBL classification network for premolar (PM) and molar (M) teeth with transferred weight.
Region of interest segmentation
As can be seen in Fig. 1, there is a wide variability in the field of view and contextual information in the panoramic dental radiographs, depending on the patients and devices. This variability impedes the detection of PBL lesions, which are only relevant in the teeth regions in the radiograph. To accommodate this variability, we trained a segmentation network which automatically extracts the ROI. The ROI segmentation model \({f}^{R}\) has an U-shaped architecture employed by Ronneberger et al.5 which consist of an encoder part \({f}_{E}^{R}\) and a decoder part \({f}_{D}^{R}\). The detailed architecture of our ROI segmentation model can be found in the appendix.
To train and validate the ROI segmentation network, we randomly selected 440 panoramic dental radiographs from the entire training dataset (\(N\)=11,189). The ROI segmentation network was trained using 400 randomly sampled radiographs until its performance validated on the remaining 40 radiographs converged. The ROIs were annotated as binary polygon masks by an experienced dental clinician, starting from the right temporomandibular joint (TMJ) connected in order of the right mandible, left mandible, and left TMJ as shown in Fig. 2(a). The radiographs and their corresponding masks were resized to resolution \(512\times 1024\).
We used binary cross-entropy with \({L}_{2}\) regularization defined below as the loss function \({L}_{R}\) for the ROI segmentation network
where \(n\) denotes the number of pixels in the radiograph indexed by \(i\), \({y}_{i}^{R}\) is the binary label for pixel \(i\), \({\hat{y}}_{i}^{R}\) is the predicted label for pixel \(i\), and \(\lambda \) is a regularization parameter which we set to \(1{0}^{-4}\). The ROI segmentation network was trained for 50 epochs using the Adam optimizer13 and the initial learning rate was set to \(1{0}^{-4}\) obtained from hyperparameter tuning.
After training, the predicted ROI masks were post-processed with a morphology operation filling the holes in the segmented mask using binary_fill_hole function in the SciPy package14. The convex hull of the resulting masks was computed using the OpenCV15convexHull function and the regions in the corresponding bounding box were extracted and resized to \(512\times 1024\). Finally, the extracted image excluding the black background area outside of the predicted mask in the radiograph was normalized using \(z\)-score. The validation dice coefficient increased from 0.95 to 0.98 after post-processing.
Pre-training for transfer learning
While the goal of the proposed framework is to provide tooth level prediction, there are a limited number of positive PBL examples for each tooth(the number can be found in the appendix). This lack of data and class imbalance is even more severe for the molar teeth, which is problematic for training deep neural networks, where an abundant training dataset is necessary to learn complex and diverse patterns such as PBL in radiographs. We work around this issue by employing transfer learning, which utilizes the learned weight from one task as the initial weight of a new network, which is then trained for a different task. Transfer learning is especially useful in medical image analysis and has been widely adopted in this domain because acquiring annotated medical data is expensive and time-consuming7.
We trained a lesion segmentation network \({f}^{S}\) with the same U-shape architecture as the ROI segmentation network, which learns to extract salient features of PBL as shown in Fig. 2(b). The PBL lesion segmentation model has the same network architecture as the ROI segmentation network \({f}^{R}\) but has a different loss function. Specifically, focal loss was used to address the imbalance between positive and negative samples in the pixel level16:
where \(n\) is the number of pixels indexed by \(i\) in the ROI-extracted radiograph and \(\gamma \) denotes the focal loss parameter, which was set as 2 in our study. Here the PBL lesion segmentation masks provided by five dental clinicians were aggregated using the following rule:
where \(i\) is the index of the pixel in the ROI-extracted radiograph and \(l\) is the index of annotator (\(L=5\)). In order to enhance recall rate and facilitate learning various features of PBL, we set \({C}_{S}=1\) which means \({y}_{i}^{S}\) is the union of masks annotated by the five dental clinicians. The lesion segmentation network was trained for 50 epochs using Adam optimizer, and the initial learning rate was set to \(1{0}^{-5}\) obtained by hyperparameter tuning. The entire training dataset (\(N\) = 11,189) was used to train the lesion segmentation network until its performance on the validation dataset (\(N=190\) was maximized. Data augmentation was performed using random rotation \(\le \,10\) degrees, horizontal and vertical shift \(\le 10 \% \), and shift of brightness, sharpness, contrast \(\le 15 \% \).
Tooth-level PBL classification
In clinical dental practice, it is mandatory to report the dental disease along with the teeth numbers of the lesions. However, providing lesion segmentation masks requires additional effort to assign teeth numbers to the corresponding lesion, which is subjective and time-consuming. Therefore, to make the computer-assisted diagnostic support system more clinically applicable in dental practice, it is desirable to provide teeth number for the detected lesions. Therefore, we formulated the PBL detection problem as a multi-label classification task where the existence or absence of PBL is predicted simultaneously for each tooth.
As mentioned in the previous section, training this multi-label network directly from tooth-level annotations with randomly initialized weights requires a plethora of training examples for each tooth to learn diverse and complex representation of PBL lesions. To deal with the scarcity of positive training examples in the tooth-level annotations, we transferred the weights from the encoder part \({f}_{E}^{S}\) of the lesion segmentation network \({f}^{S}\) to the multi-label classification network \({f}^{A}\) as pre-trained weights. Two additional convolution blocks, one global average pooling layer, and two fully connected layers are attached to the end of the encoder \({f}_{E}^{A}\). Finally, the output layer consists of 32 nodes, which correspond to the binary classification output for each tooth. The detailed architecture of the tooth-level classification model can be found in the appendix. All the layers, including the encoder part initialized with the transferred weights from the lesion segmentation network, were trained for the tooth-level classification task.
Another difficulty which arises in PBL classification is that premolar and molar teeth represent a more complex morphological structure, which hinders the detection of PBL in the panoramic dental radiographs. To overcome this difficulty, another tooth-level classification network \({f}^{B}\) was trained specifically for the premolar and molar teeth. While \({f}^{A}\) and \({f}^{B}\) share the same network architecture, \({f}^{B}\) was trained with a vertical split of the ROI image as the input, generated by \({f}^{R}\), and the label-masked multi-label as the output as shown in Fig. 2(d). Here the label-masks were applied to incisors and canines by assigning zero to the corresponding labels; therefore, \({f}^{B}\) takes as input an ROI image and is trained to predict PBL in only premolar and molar teeth. The transferred weight from \({f}_{E}^{S}\) was also used to initialize \({f}^{B}\). To train the teeth-level classification model \({f}^{C}=\{\,{f}^{A},{f}^{B}\}\), focal loss with \({L}_{2}\) regularization is used for each output node computed as:
where \(N\) is the number of training examples indexed by \(i\), \(j\) is the corresponding tooth index, \(k\) is the trainable weight index, and \(\gamma \) denotes the focal loss parameter, which was set to 2. We applied the same data augmentation technique as in the pre-training for transfer learning section above. Both \({f}^{A}\) and \({f}^{B}\) were jointly trained using the entire training dataset (\(N\)=11,189) until their performance on the validation dataset (\(N\)=190) converged. Because \({f}^{B}\) takes the vertical split of ROI images as illustrated in Fig. 2, the effective number of training samples for \({f}^{b}\) is twice (\(N=22,378\)) the number of the training dataset. As stated in Equation 4, the majority vote of the annotators for each tooth was used as target labels to train the model.
At the inference phase, we aggregated the prediction of \({f}^{A}\) and \({f}^{B}\) as follows:
where \(j\) is the tooth type index and \(\{In\},\{Ca\},\{Pr\},\{Mo\}\) denotes the index set of incisor, canine, premolar, and molar teeth, respectively. \({x}_{{\rm{ROI}}}\) is a ROI image with resolution \(512\times 1024\) given by the output of \({f}^{R}\) and \({x}_{{\rm{ROI}}/2}\) is its vertical split image with resolution \(512\times 512\). \(\alpha \) is a hyperparameter which was set to \(0.1\) in our study attained by hyperparameter tuning.
Auxiliary co-occurrence loss
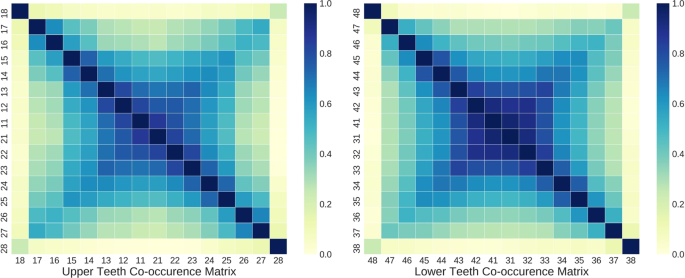
While the encoder part of the tooth-level classifier is shared to extract salient features for PBL detection, the predictions for each tooth at the output layer are made independently without considering the clinical knowledge that PBL lesions frequently co-occur across horizontally adjacent teeth. As shown in Fig. 3, there is a high co-occurrence of PBL among incisors and canines, while the co-occurrence among premolar and molar teeth is lower in both maxillary (upper jaw) and mandibular (lower jaw) teeth. Also, the third molar had noticeably low co-occurrence of PBL with other teeth except the third molar at the opposite side.
To exploit this prior information for improving the generalization performance of PBL detection, auxiliary co-occurrence loss is computed as:
where both \(j\) and \({j}^{^{\prime} }\) are the tooth index, \({y}_{j}\) is the binary label derived from Eq. (4), \({\hat{y}}_{j}\) is the network output, and \({C}_{j{j}^{^{\prime} }}\) is the element of the co-occurrence matrix \(C\in {{\mathbb{R}}}^{k\times k}\), and \(k(=16)\) is the number of teeth in the upper or lower teeth. \(Z={\max }_{j}\,{c}_{j}\) is the normalization term which guarantees all \({c}_{j}\)’s lie between 0 and 1.
The auxiliary co-occurrence loss was applied separately for upper and lower teeth based on their respective co-occurrence matrix, and the final loss function \({L}_{F}\) used to train the tooth-level classification network is defined as
where \({L}_{C}\) is focal loss defined in Eq. 5, \(\beta \) is a hyperparameter set to \(0.01\), and \({1}_{\{j\in U\}}\) and \({1}_{\{j\in L\}}\) correspond to the indicator function for the upper and lower teeth, respectively. We trained the tooth-level classification network for \(100\) epochs using Adam optimizer and the initial learning rate was set to \(1{0}^{-5}\) which was obtained by hyperparameter tuning.
Results
We compared the performance of DeNTNet on the test set consisting of 800 radiographs with the performance of each dental clinician who participated in the annotation. We also conducted an ablation study to quantitatively analyze the contribution of each training phase of DeNTNet.
PBL detection performance
The teeth-level performance of DeNTNet and five dental clinicians in PBL detection on the test dataset is summarized in Table 1. Each human expert’s performances were calculated by comparing the expert’s decision (PBL present/absent) on the target tooth with the majority voting among human experts, which served as the ground truth. The model’s performances were attained using its teeth-level classification prediction outputs with varying operating points. We measured the area under the receiver operating characteristics curve (AUROC), F1 score, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The average performance of five dental clinicians was 0.85 in AUROC, 0.69 in F1 score, 0.78 in sensitivity, 0.92 in specificity, 0.62 in PPV, and 0.96 in NPV. The baseline DeNTNet, trained directly from the original radiograph without using either transfer learning or auxiliary loss, showed comparable performance to the average of five dental clinicians. However, when the proposed multi-phase training, including ROI segmentation, pre-training for transfer learning, auxiliary loss, and ensembled classification was applied, DeNTNet outperformed dental clinicians with respect to the F1 score as shown in Fig. 4. The balanced setting, which was selected to maximize the F1 score, achieved the F1 score of 0.74, while high sensitivity and high specificity settings achieved F1 scores of 0.71 and 0.73, respectively. The high sensitivity operating point was selected such that the specificity is close to the worst dental clinician’s, whereas the high specificity operating point was selected such that the sensitivity of DeNTNet is close to the worst dental clinician’s. The detailed performance of dental clinicians and DeNTNet for each tooth and teeth type can be found in the appendix.
Performance of DeNTNet and five dental clinicians in the detection of PBL. ROC curves of both baseline (yellow curve) and improved (green curve) DeNTNet (green curve) are shown. The high sensitivity and high specificity operating point of DeNTNet is also displayed for all teeth. (All Teeth) The performance for all 32 teeth, (Incisor) For 8 upper and lower incisors, (Canine) For 4 upper and lower canines, (Premolar) For 8 upper and lower premolars, (Molar) For 12 upper and lower molars.
Ablation study
We conducted an ablation study to quantitatively assess the contribution of each step in our proposed training framework. Each additional step improved the F1 score achieved by DeNTNet, as shown in Table 2. The baseline performance improved from 0.66 to 0.68 after applying the ROI segmentation step, which removes regions in the panoramic dental radiograph unnecessary for detecting PBL, thus standardizing the classification models’ inputs. Transferring pre-trained weights from the lesion segmentation model further improved the performance by facilitating the training of the tooth-level classification model. Adopting an auxiliary co-occurrence loss also improved performance by exploiting prior clinical knowledge when training the model. Finally, ensembling the classification model \({f}^{B}\) specialized for detecting PBL in premolar and molar teeth with the multi-label classification network \({f}^{A}\) considerably improved the performance in these teeth types, which thereby improved the overall performance.
Discussion
Panoramic dental radiography is one of the most widely and frequently taken dental examinations which capture the entire mouth in a single 2D image. It is generally used as an initial evaluation of the condition of bone structure and soft tissue in the maxillofacial area. While intra-oral bitewing and periapical radiography are also widely used for diagnosing dental disease, extra-oral panoramic dental radiography has several advantages over these tooth-specific imaging methods. First of all, panoramic dental radiography provides better patient comfort and acceptance as the procedure is faster and easier for both the patient and dental staff than bitewing and periapical radiography, which require inserting a sensor into the patient’s mouth. Furthermore, it exposes less radiation and reduces infection control compared to intra-oral images. Finally and most importantly, panoramic dental radiography is a better screening examination when considering the field of view as it can provide more coverage for diagnosing periodontal bone defects, periapical lesions, and pathological jaw lesions. When combined with automated detection of dental lesions and teeth numbering, computer-assisted diagnostic support systems can substantially improve efficiency and reduce workload for clinical dental practices.
There are a few limitations in this study. The first limitation is that there is still room for improvement in PBL detection. Panoramic dental radiographs capture a wide field of view, which results in low resolution for each individual tooth. This hinders the detection of local morphological changes in PBL, and consequently, the overall sensitivity performance of both dental clinicians and DeNTNet for PBL detection is limited, as shown in Table 1. Further, although model ensembling improved the performance of PBL detection and thus outperforms human experts when tested on data including all teeth types, the DeNTNet’s performance on just the third molar (also known as wisdom teeth) is considerably lower than that attained by dental clinicians(examples of false negative cases with PBL in third molar can be found in the appendix). This is mainly because of the limited availability of third molar examples due to wide morphological variations with respect to patient demographics in this study. Due to the lack of third molar examples, we decided to use a relatively small validation dataset in order to fully expose the model to infrequently occurring examples while training instead of searching for better hyperparameters on a larger validation dataset. To fully benefit from data-driven methods such as deep learning, more annotated data must be collected, and a dedicated model for detecting PBL in the third molar may prove useful.
Second, deep learning-based methods should illuminate the rationale behind how predictions are made in order to be clinically applicable. However, most deep learning methods are considered to be a ’black-box’ because they cannot intuitively explain how predictions are made. DeNTNet is also based on DCNNs and provides predictions directly from the panoramic dental radiographs without explanations. However, to make the model at least interpretable, we applied Grad-CAM17 which is a widely used method for visualizing class activation maps (CAM)18. As shown in Fig. 5, the model accurately highlights the PBL lesions, which coincide with the annotations of the dental clinicians. When integrated into a computer-assisted diagnostic system, this visualization capability strengthens the model’s reliability to clinicians and patients.
Panoramic dental radiograph examples with dental clinicians’ annotations and DeNTNet activation maps. (Top) Original input panoramic dental radiographs; (Middle) PBL lesion masks annotated by dental clinicians; (Bottom) Class activation map highlighting the most salient region in the image for PBL prediction; The red area in the activation map corresponds to a stronger activation region; (First column) A case with vertical periodontal bone loss; (Second column) A case with horizontal periodontal bone loss; (Third column) A case with generalized severe periodontal bone loss with both vertical and horizontal bone loss.
Finally, each step in the multi-stage approach proposed critically impacts the final performance of the model. For example, when the ROI segmentation fails, in spite of DeNTNet’s very high and stable performance, the subsequent steps become unreliable. Therefore, developing a fully end-to-end approach which predicts PBL for each tooth directly from the original panoramic dental radiograph is a promising future direction when a larger dataset becomes available.
Conclusion
In this study, a fully automated method for PBL detection with tooth numbering in panoramic dental radiograph was proposed. Through the multi-step training framework, the proposed model was able to achieve a PBL detection performance superior to that of dental clinicians. We expect this approach to substantially benefit clinical practices by improving the efficiency of diagnosing PBL and reducing the workload involved in reporting tooth numbers.
Further clinical and external validation is necessary for the adoption of the proposed method in clinical practice. Expanding the targeted dental disease of the proposed framework beyond PBL also remains for future work.
Data availability
The data used in current study were collected from Korea University Anam Hospital and is available only for the granted research. However, the data can be made available if requested within data protection and regulation guideline.
References
Papapanou, P. N. et al. Periodontitis: Consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. Journal of periodontology 89, S173–S182 (2018).
Casamassimo, P. S. Radiographic considerations for special patients-modifications, adjuncts, and alternatives .
Akkaya, N., Kansu, O., Kansu, H., Cagirankaya, L. & Arslan, U. Comparing the accuracy of panoramic and intraoral radiography in the diagnosis of proximal caries. Dentomaxillofacial Radiology 35, 170–174 (2006).
Åkesson, L., Håkansson, J. & Rohlin, M. Comparison of panoramic and intraoral radiography and pocket probing for the measurement of the marginal bone level. Journal of clinical periodontology 19, 326–332 (1992).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, 234–241 (Springer, 2015).
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), 801–818 (2018).
Litjens, G. et al. A survey on deep learning in medical image analysis. Medical Image Analysis 42, 60–88 (2017).
Jung, K.-H. & Hwang, W. Deep learning for medical image analysis : Applications to computed tomography and magnetic resonance imaging. Hanyang Medical Review 37, 61–70 (2017).
Hwang, J.-J., Jung, Y.-H., Cho, B.-H. & Heo, M.-S. An overview of deep learning in the field of dentistry. Imaging Science in Dentistry 49, 1–7 (2019).
Lee, J.-H., Hyung Kim, D., Jeong, S.-N. & Choi, S.-H. An overview of deep learning in the field of dentistry. Diagnosis and prediction of periodontally compromised teeth using a deep learning-based convolutional neural network algorithm 48, 114–123 (2018).
Krois, J. et al. Deep learning for the radiographic detection of periodontal bone loss. Scientific reports 9, 8495 (2019).
Federation dentaire internationale (fdi) tooth-numbering system. American Journal of Orthodontics and Dentofacial Orthopedics 120, 465 (2001).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014).
Jones, E. et al. Open source scientific tools for Python (2001–). [Online; accessed today].
Bradski, G. The OpenCV Library. Dr. Dobb’s J. Software Tools (2000).
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE international conference on computer vision, 2980–2988 (2017).
Selvaraju, R. R. et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, 618–626 (2017).
Zhou, B., Lapedriza, A., Oliva, A. & Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2921–2929 (2016).
Acknowledgements
We thank dental clinicians for their effort of labeling the image data. This work was supported by the National Research Foundation of Korea(NRF) grant funded by the Korea government(MSIT)(No. NRF-2017R1C1B2006401).
Author information
Authors and Affiliations
Contributions
J.K., H.L., I.S.S. and K.H.J. conceived and designed the study. J.K. conducted the experiments. I.S.S collected data. J.K., H.L., I.S.S., and K.H.J. analyzed the results. J.K., K.H.J wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.K., H.L. and K.H.J are employees of VUNO Inc., Seoul, South Korea. I.S.S. declares no potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, J., Lee, HS., Song, IS. et al. DeNTNet: Deep Neural Transfer Network for the detection of periodontal bone loss using panoramic dental radiographs. Sci Rep 9, 17615 (2019). https://doi.org/10.1038/s41598-019-53758-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-53758-2
This article is cited by
-
Detection of periodontal bone loss patterns and furcation defects from panoramic radiographs using deep learning algorithm: a retrospective study
BMC Oral Health (2024)
-
Graphical user interface-based convolutional neural network models for detecting nasopalatine duct cysts using panoramic radiography
Scientific Reports (2024)
-
Deep learning for classifying the stages of periodontitis on dental images: a systematic review and meta-analysis
BMC Oral Health (2023)
-
SinusC-Net for automatic classification of surgical plans for maxillary sinus augmentation using a 3D distance-guided network
Scientific Reports (2023)
-
Classification of periodontitis stages in mandibular area from dental panoramic radiograph using Adaptive Center Line-Distance Based image processing approach
Journal of Ambient Intelligence and Humanized Computing (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.