Abstract
Antibiotic resistance in Acinetobacter baumannii is a major global health threat. New drugs with novel chemical structures are needed to overcome a myriad of resistance mechanisms in A. baumannii. In this study, we screened an open-source Pathogen Box library for anti-A. baumannii compounds. Compound MMV675968 (a diaminoquinazoline analog) was the only non-reference compound found to inhibit the growth of all four A. baumannii test strains with IC50 of 0.6–2.7 μM, IC90 of 0.7–3.9 μM, and MIC of 1.6–10 μM. We showed that MMV675968 targeted A. baumannii dihydrofolate reductase (AbDHFR) as determined by an E. coli surrogate whose growth was dependent on AbDHFR function and by an in vitro DHFR activity assay. Additionally, chemical scaffolds of DHFR inhibitors that are effective as antibiotics against A. baumannii were identified using an in vitro DHFR activity assay and A. baumannii growth inhibition. MMV675968 was the most potent among DHFR inhibitors tested in inhibiting A. baumannii growth. This study shows for the first time that MMV675968 inhibits A. baumannii growth via selective inhibition of AbDHFR and is therefore a promising scaffold for further antibiotic development against A. baumannii.
Similar content being viewed by others
Introduction
Acinetobacter baumannii, a Gram-negative opportunistic pathogen, is a leading cause of nosocomial infection worldwide1. It has developed resistance to multiple classes of antibiotics with strains resistant to all commercially available antibiotics1,2,3,4. A. baumannii possesses an array of resistance mechanisms such as downregulation of porin, mutations in drug targets and altered expression of β-lactamases, aminoglycoside-modifying enzymes, and drug efflux pumps5,6. Treatment failure is common in A. baumannii infection owing to a lack of effective alternate antibiotics against this multidrug-resistant pathogen, responsible for up to 50% mortality among infected patients1. Discovery of new drug candidates and understanding of their mechanisms of action is direly needed to tackle the problem of resistance in this pathogenic organism.
The Open-source Pathogen Box consists of 400 compounds known to be active against a number of pathogens responsible for neglected tropical diseases and have low toxicity (www.pathogenbox.org). Pathogen Box contains compounds with diverse chemical scaffolds distinct from currently available antibiotics, making the Pathogen Box library a promising source of compounds for novel antibiotics discovery. The mechanisms of action of these compounds are mostly unknown but crucial for hit-to-lead development in a drug discovery process, thereby worth investigating further7,8. In this study, we used Pathogen Box compounds to screen for those with anti-A. baumannii activity and investigated the mode of action of the most promising bioactive candidate.
Results
MMV675968 inhibits the growth of A. baumannii strains
Initially, we screened the Pathogen Box library (www.pathogenbox.org) for compounds that inhibit the growth of A. baumannii Mahidol and Naval-81 strains, an environmental and clinical strain, respectively. Using the Clinical and Laboratory Standards Institute (CLSI) microdilution method, the majority of the compounds exhibited no significant growth inhibitory activity at 10 μM (Table S1). Five compounds inhibited A. baumannii growth to less than 50% of untreated (Table 1). However, of the five compounds with inhibitory activities, auranofin, doxycycline, levofloxacin-(-)-ofloxacin, and rifampicin are reference compounds with known antibacterial activity, with doxycycline and rifampicin completely inhibited the growth of both strains tested (Table 1). Compound MMV675968, a diaminoquinazoline, completely inhibited the growth of A. baumannii Naval-81 strain and inhibited A. baumannii Mahidol strain to 25% in the primary screen (Table 1). We then used various concentrations of MMV675968 to determine growth inhibition curve against four strains of A. baumannii, namely, Mahidol, Naval-81, H72721, and 3–137 strains. IC50, IC90, and MIC values were 0.6–2.7 μM (0.22–0.97 mg/L), 0.7–3.9 μM (0.25–1.40 mg/L), and 1.6–10 μM (0.58–3.60 mg/L), respectively (Fig. 1), which are comparable to corresponding values of effective antibiotics against A. baumannii9,10,11.
Growth inhibitory curve of MMV675968. Four strains of A. baumannii, namely the Mahidol strain (Ab M, circle), the H72721 strain (Ab H72721, square), the 3-137 strain (Ab 3-137, triangle), and the Naval-81 strain (Ab N81, inverted triangle) were incubated with various concentrations of MMV675968 for 18 hours. Growth was monitored using OD600. Percent growth was calculated relative to untreated A. baumannii, which was set to 100%. The plot shows mean ± standard error of the mean (SEM) from three independent experiments.
MMV675968 selectively inhibits A. baumannii DHFR
MMV675968 inhibits DHFR of Cryptosporidium, Plasmodium, and Escherichia coli12,13,14. We hypothesized that MMV675968 might have a similar mode of action in A. baumannii. In order to test this notion, we used E. coli PA414 strain, which lacks dihydrofolate reductase (DHFR) and thymidylate synthase (TS) of thymidylate cycle required for thymidine synthesis hence making the strain a thymidine auxotroph15, as a model to test DHFR target specificity. The E. coli surrogate was co-transformed with plasmids expressing E. coli TS (EcTS) and either AbDHFR or human DHFR (hDHFR). When EcTS was expressed together with AbDHFR or hDHFR, E. coli PA414 was able to grow in the absence of thymidine (Fig. 2A), indicating that AbDHFR and hDHFR could function in E. coli and can complement for the loss of EcDHFR. The growth of the E. coli surrogates were then tested in the presence of MMV675968, trimethoprim (TMP), a bacterial DHFR inhibitor, or methotrexate (MTX), a known hDHFR inhibitor16,17. MMV675968 (10 µM) was able to inhibit the growth of the E. coli surrogate expressing AbDHFR but not that of surrogate expressing hDHFR (Fig. 2B); conversely, TMP and MTX inhibited the growth of the E. coli surrogate expressing hDHFR but not AbDHFR (Fig. 2B). These data suggest that 1) MMV675968 is more effective in inhibiting AbDHFR than TMP and MTX, 2) MMV675968 is less effective in inhibiting hDHFR than MTX and TMP, and 3) MMV675868 is likely to be more specific to AbDHFR than hDHFR since expression levels of AbDHFR and hDHFR in E. coli PA414 were comparable (Fig. S2). In addition, thymidine supplement was able to rescue growth inhibition by MMV675968 (Fig. 2C), confirming that thymidylate cycle is the target of MMV675968 in E. coli surrogate.
MMV675968 inhibits AbDHFR but not hDHFR in an E. coli model. (A) AbDHFR and hDHFR can complement for the loss of EcDHFR. Cultures of E. coli PA414 (DHFR−, TS−) carrying empty plasmids or plasmids expressing EcTS and either AbDHFR or hDHFR were spotted onto LB plate supplemented with 50 µg/ml thymidine or 0.2% arabinose. A representative picture from three independent experiments is shown. (B) MMV675968 inhibits the growth of E. coli model expressing AbDHFR not hDHFR when grown without thymidine supplement. (C) Thymidine supplement rescues growth inhibition of E. coli surrogate expressing EcTS and AbDHFR by MMV675968. E. coli PA414 expressing EcTS and either AbDHFR or hDHFR were incubated with 10 µM of DHFR inhibitors for 18 hours. Growth was monitored using OD600. Percent growth was calculated relative to untreated which was set at 100%. The graph shows mean ± SEM from three independent experiments.
In addition to testing MMV675968 inhibition of AbDHFR in an E. coli model, we also tested whether MMV675968 directly inhibited AbDHFR and hDHFR in an enzyme activity assay. DHFR activities of lysate of E. coli BL21(DE3) harboring pET15b and that overexpressing AbDHFR or hDHFR were compared. Using equal amount of total protein, DHFR activity present in the lysate of E. coli BL21(DE3) containing pET15b accounted for only 3% and 6% of that detected in the lysate of E. coli BL21(DE3) overexpressing AbDHFR and hDHFR, respectively (Fig. 3A,B). These results suggest that most of DHFR activity measured in E. coli BL21(DE3) expressing AbDHFR and hDHFR comes from overexpressed enzymes not endogenous EcDHFR. We found that MMV675968 inhibited AbDHFR and hDHFR in a dose dependent manner with IC50 values of 8.5 nM and 492.4 nM, respectively (Fig. 3C), confirming that MMV675968 prefers AbDHFR to hDHFR as target.
MMV675968 selectively inhibits AbDHFR, not hDHFR, in an in vitro DHFR activity assay. DHFR activity present in lysate of BL21(DE3) containing pET15b and that overexpressing AbDHFR (A) or hDHFR (B) were determined by measuring reduction of A340 and compared. (C) MMV675968 prefers AbDHFR to hDHFR as target. AbDHFR or hDHFR activity with various concentrations of MMV675968 was determined. DHFR activity of DMSO-treated lysate of BL21(DE3) overexpressing AbDHFR or hDHFR was set to 100%. DHFR activity in the presence of inhibitors was calculated relative to DMSO-treated. The graphs show mean ± SEM from at least three independent experiments.
Structure-function relationship reveals MMV675968 as a potent antifolate scaffold targeting A. baumannii
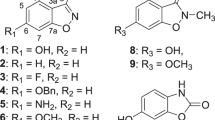
The chemical structures of DHFR inhibitors vary with different chemical scaffolds possessing different efficacy toward different targets. To determine which chemical scaffolds are the most effective antibiotic candidates against A. baumannii, MMV675968 and seven other antimicrobial DHFR inhibitors (Fig. 4A) were tested for their efficacy in inhibiting AbDHFR in vitro and in vivo and in inhibiting A. baumannii growth. AbDHFR activity in the presence of 1000 nM inhibitors were determined (Fig. 4B). At 1000 nM, PMX, cycloguanil, MMV667486, and MMV667487 showed little inhibitory activity, while MTX, TMP, PYR, and MMV675968 strongly inhibited AbDHFR activity in vitro (Fig. 4B). MTX, TMP, PYR, and MMV675968 were chosen for further IC50 determination. MTX and MMV675968 potently inhibited AbDHFR in vitro with IC50 values of 4.8 nM and 8.5 nM, respectively (Fig. 4C). PYR and TMP, both of which are diaminopyrimidine analogs, were less effective in inhibiting AbDHFR in vitro with IC50 values of 81.7 and 280.7 nM, respectively (Fig. 4C).
Structures of DHFR inhibitors and their activity in inhibiting AbDHFR in vitro, in vivo, and in inhibiting growth of A. baumannii. (A) Structures of folates and DHFR inhibitors used in this study. DHFR inhibitors are divided into four major classes: (1) diaminopteridine - methotrexate, (2) diaminodimethyltriazine - cycloguanil, MMV667486, and MMV667487, (3) diaminopyrimidine - trimethoprim and pyrimethamine, and (4) diaminoquinazoline - MMV675968. (B) In vitro AbDHFR activity in the presence of 1000 nM compounds. (C) Dose response curve of selected compounds and their anti-AbDHFR activity. (D) A. baumannii growth inhibitory activity of 10 μM of DHFR inhibitors. Four strains of A. baumannii, namely the Mahidol strain (Ab Mahidol), the H72721 strain (Ab H72721), the 3-137 strain (Ab 3–137), and the Naval-81 strain (Ab Naval-81) were incubated with 10 µM of various inhibitors. (E) AbDHFR activity of four A. baumannii strains after treatment with 10 µM for 90 minutes. Percent AbDHFR activity or percent growth was calculated compared with untreated, which was set to 100%. The graphs show mean ± SEM from at least three independent experiments.
Eight DHFR inhibitors were then tested for their A. baumannii growth inhibitory activity. MMV675968 was the only compound that completely inhibited the growth of all four A. baumannii strains (Fig. 4D). Even though MTX strongly inhibited AbDHFR in vitro (Fig. 4C), it did not inhibit the growth of any A. baumannii strains tested (Fig. 4D), implying that MTX may not be able to reach AbDHFR target within A. baumannii cells. TMP and pyrimethamine (PYR) inhibited the growth of two strains, namely, Mahidol and Naval-81 strains, but not the H72721 and 3–137 strains (Fig. 4D). All four strains share identical AbDHFR polypeptide sequence deduced from DNA sequences of amplified gene fragments (Fig. S3), indicating that differences in PYR and TMP sensitivities among the test A. baumannii strains are due to factor(s) not related to AbDHFR inhibition.
To further confirm that MMV675968 targets AbDHFR in A. baumannii, we determined whether AbDHFR activity of each A. baumannii strain decreased upon treatment with DHFR inhibitors. A. baumannii strains were treated with 10 µM TMP, PYR, or MMV675968 for 90 minutes. Normalized lysate was then tested for DHFR activity. All four A. baumannii strains showed reduced DHFR activity upon MMV675968 treatment compared with untreated cells (Fig. 4E), confirming that MMV675968 targets AbDHFR in vivo. Higher AbDHFR inhibitory activity of MMV675968 in this assay compared with other inhibitors tested is probably due to a combination of differences in cell permeability, drug efflux rate, and AbDHFR inhibitory efficacy.
Discussion
In this study, an open-source Pathogen Box was screened for bioactive compounds against A. baumannii. The advantages of using such library are the availability of pure compounds, the knowledge of chemical structures, and the access to related information such as toxicity and pharmacokinetics properties (www.pathogenbox.org). The open source format is expected to jump-start the discovery of new antibiotics urgently needed to combat drug resistant organisms. From the Pathogen Box, compound MMV675968 was discovered to be an effective inhibitor of A. baumannii growth with IC50, IC90, and MIC values comparable to known effective antibiotics against A. baumannii (Table 1, Fig. 1)9,10,11.
MMV675968 inhibits DHFR, an essential enzyme in the folate biosynthetic pathway, in Cryptosporidium, Plasmodium, and E. coli12,13,14. This study is the first to show that MMV675968 has anti-A. baumannii activity through inhibition of AbDHFR as determined by an E. coli surrogate model and an in vitro DHFR activity assay. MMV675968 inhibits AbDHFR more effectively than hDHFR, with 58 fold selectivity (Fig. 3C), consistent with biochemical data from others that this compound is 83 fold more effective in inhibiting E. coli DHFR than hDHFR12. Since structures of bacterial DHFRs and mammalian DHFRs are clustered in different phylogenetic groups18, the presence of selective bacterial DHFR inhibitors like trimethoprim is possible.
There are at least four major classes of DHFR inhibitors, namely, diaminopteridine, diaminodimethyltriazine, diaminopyrimidine, and diaminoquinazoline (Fig. 4A)19. MTX, an anti-cancer diaminopteridine, showed strong in vitro AbDHFR inhibition (Fig. 4B,C) but did not show AbDHFR inhibitory activity in E. coli surrogate (Fig. 2B) and did not have antibacterial activity against A. baumannii (Fig. 4D), suggesting that MTX may not reach AbDHFR target in both E. coli surrogate and in A. baumannii. It is possible that MTX cannot permeate E. coli and A. baumannii cells, is degraded by enzymes, and/or is pumped out via efflux pump(s) such as AcrAB/TolC efflux pump, which is shown to export MTX, rendering MTX ineffective against E. coli20.
Diaminodimethyltriazines such as cycloguanil, MMV667486, and MMV667487, which are Plasmodium DHFR inhibitors19,21,22, did not inhibit AbDHFR in vitro or show any growth inhibitory activity against A. baumannii (Fig. 4B,D), highlighting structural differences between AbDHFR and Plasmodium DHFR.
Diaminopyrimidines such as PYR and TMP are inhibitors of Plasmodium and bacterial DHFR, respectively19. While being moderately effective in inhibiting AbDHFR in vitro (Fig. 4B,C), TMP did not inhibit AbDHFR in E. coli surrogate (Fig. 2B), possibly due to compound permeability and/or compound efficacy against AbDHFR. Diaminopyrimidines inhibited the growth of only two of the four test A. baumannii strains (Fig. 4D). The reason for differential diaminopyrimidine sensitivity among A. baumannii strains was not due to AbDHFR sequence (Fig. S3) and requires further investigation. Various A. baumannii strains may differ in terms of levels of AbDHFR expression, alteration of cell permeability, and/or differences in efflux pump.
MMV675968, a diaminoquinazoline, inhibited AbDHFR 33 fold better than TMP (Fig. 4C), inhibited the growth of both TMP-sensitive and -resistant A. baumannii strains (Fig. 4D), and inhibited DHFR activity in A. baumannii (Fig. 4E). MMV675968 is therefore more effective in inhibiting AbDHFR than TMP and can bypass TMP- and PYR-resistance mechanism present in A. baumannii H72721 and the 3–137 strains. Since Acinetobacter species cannot salvage thymine or thymidine23,24,25, enzymes of thymidylate cycle become even more critical to its survival and could serve as promising drug targets. MMV675968 and its diaminoquinazoline analogs are, therefore, worth investigating further as antibiotic candidates against A. baumannii.
DHFR, an essential enzyme that converts dihydrofolate to tetrahydrofolate, is a validated drug target in bacteria and protozoan parasites. There may be concerns that AbDHFR might not be an effective drug target because resistant mutations can arise and lead to drug resistance. However, since DHFR active site needs to accommodate dihydrofolate substrate for its essential function, there should be a limit number of mutations that AbDHFR active site can tolerate. Designing a DHFR inhibitor to stay in the substrate space has proved to be beneficial in making the P218 compound bind to and inhibit both wild type and pyrimethamine-resistant mutant of Plasmodium DHFR and effectively inhibits both wild type and mutant parasites26. It is therefore theoretically possible to use AbDHFR as a drug target despite possible rise of resistant mutations.
Another strategy to prevent rapid emergence of drug resistance is the use of drug combinations, such as using DHFR inhibitors in combination with dihydropteroate synthase (DHPS) inhibitors, e.g. trimethoprim-sulfamethoxazole combination (Bactrim) and pyrimethamine-sulfadoxine combination (Fansidar). Using DHPS inhibitors in combination with MMV675968 or its analogs should help reduce the required doses and forestall resistance.
The E. coli surrogate expressing exogenous AbDHFR or hDHFR constructed in this study can be used for screening other compound libraries for selective inhibitors of AbDHFR in an antibiotic discovery research program. Model organisms lacking endogenous DHFR activity have been used to study the activity of exogenously expressed DHFR from pathogenic species and for inhibitor screening27,28,29. The advantages of such surrogate models are the ease and safety of handling model organisms rather than the pathogenic organisms of interest. However, one caveat of using model organisms for studying target enzymes is the vast differences in cell permeability between species. For example, compounds that are active against a Plasmodium enzyme expressed in yeast may not be able to penetrate Plasmodium membrane. However, since E. coli and A. baumannii are both Gram-negative bacteria, they share similar cell wall/cell membrane structure, making the results from the surrogate assay developed in this study easier to translate from one organism to the other.
In summary, we identified a diaminoquinazoline MMV675968 that could serve as a starting compound for drug development against multidrug-resistant A. baumannii. Further comparative studies to understand enzyme kinetics and structures of AbDHFR and hDHFR and studies on in vivo efficacy, toxicity, pharmacokinetics, and pharmacodynamics can provide insights into improving the starting compound into more effective and/or more selective antibiotic.
Methods
Bacteria strains and culture conditions
A. baumannii Mahidol strain was isolated from soil as described previously30, and multidrug-resistant clinical Naval-81, 3–137 (OIFC137), and H72721 isolates were from BEI Resources, NIAID, NIH, USA31,32. E. coli PA414 strain carrying null mutations in folA and thyA encoding dihydrofolate reductase (DHFR) and thymidylate synthase (TS), respectively was used as a host for expressing DHFR from other organisms and TS from E. coli15. E. coli and A. baumannii were grown in Luria Bertani (LB) broth supplemented as necessary with 50 μg/ml thymidine, 100 μg/ml ampicillin, 10 μg/ml chloramphenicol, 0.2% arabinose, or their combination and incubated with shaking at 37 °C.
Growth inhibition assay
We performed bacterial growth inhibition assay according to the Clinical and Laboratory Standards Institute (CLSI) microdilution method33. In brief, compounds were solubilized and diluted in dimethyl sulfoxide (DMSO). Approximately 5 × 104 colony forming units (CFUs) were incubated with 10 μM (unless otherwise indicated) of each compound in cation adjusted Mueller Hinton broth (CAMHB) (Beckton Dickinson, Franklin Lakes, NJ, USA) at 37 °C for 18 hours. Optical density at 600 nm (OD600) was measured using SpectraMax M5 (Molecular Devices, San Jose, CA, USA). OD600 of wells containing A. baumannii (either compound-treated or untreated) was subtracted with OD600 of blank. Percent growth of compound-treated A. baumannii of the same strain was calculated using the following formula:
Percent growth of each DMSO-treated A. baumannii strain was therefore 100%. Hits are defined as compounds that inhibit bacteria growth >50%. For IC50 and IC90 (50% and 90% inhibitory concentration) determination, A. baumannii strains were treated with various concentrations of test compound and cultured according to the microdilution method above. Minimum inhibitory concentration (MIC) is defined as the lowest concentration that completely inhibits bacteria growth as determined by unaided eye33.
Construction of expression plasmids
Abdhfr was amplified from clarified cell lysates of four strains of A. baumannii using Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA) and primers 5′ctggtgccgcgcggcagccatATGGCATGGCAAAATGTAG3′ and 5′tcgggctttgttagcagccggatccTTATTTTTTATAAGTGGCAAATTCG3′ (lower case denoting sequence homologous to pET15b plasmid), and human DHFR from pL003534 using primers 5′ctggtgccgcgcggcagccatATGGTTGGTTCGCTAAAC3′ and 5′tcgggctttgttagcagccggatccTTAATCATTCTTCTCATATACTTCAAATTTG3′. Amplicons were inserted into NdeI and BamHI digested pET15b plasmid using a Gibson Assembly kit (New England Biolabs). The thymidylate synthase gene (thyA, ts) was amplified from lysate of E. coli DH5α strain using Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA) and primers 5′ctagcggagctcggagtgaaacgATGAAACAGTATTTAGAACTGATGC3′ and 5′ gcctgcaggtcgactctagaTTAGATAGCCACCGGCGC3′ (lower case denoting sequence homologous to pBAD33 plasmid). Amplicon was inserted into SacI and XbaI sites of pBAD33 using the Gibson Assembly kit (New England Biolabs, Ipswich, MA, USA). Cloned plasmids were sequenced by 1st Base DNA sequencing service (Singapore) and nucleotide sequences were deposited at GenBank, accession numbers: MH152688, MH152689, MH152690, and MH152691.
Agar plate growth assay
E. coli PA414 strain carrying various combination of expression plasmids was grown overnight at 37 °C with shaking in LB broth supplemented with 50 μg/ml thymidine, 10 μg/ml chloramphenicol, and 100 μg/ml ampicillin. Cells from overnight cultures were pelleted by centrifugation, washed with fresh medium and the OD600 adjusted to 4.0. Ten microliters of undiluted, 10-fold and 100-fold serially diluted cells were spotted onto LB agar plates supplemented with necessary antibiotics and 50 μg/ml thymidine or 0.2% arabinose and incubated at 37 °C overnight.
Overexpression of AbDHFR and hDHFR in E. coli BL21(DE3)
Overnight cultures of E. coli BL21(DE3) carrying pET15b, pET15b-AbDHFR, and pET15b-hDHFR were diluted into fresh LB supplemented with 100 µg/ml ampicillin and grown with shaking at 37 °C for 3 hours. After addition IPTG to final concentration of 40 mM, the cultures were grown overnight with shaking at 16 °C. Bacteria cells were harvested by centrifugation and lysed by sonication. Protein concentration of clarified bacteria lysate was determined using Bradford assay (Biorad, Hercules, CA, USA) according to manufacturer’s protocol.
DHFR activity assay
The assay was performed as previously described26. Briefly, 0.2 µg of total protein from lysate of E. coli containing pET15b or overexpressing AbDHFR or 5 µg of total protein from lysate of E. coli containing pET15b or overexpressing hDHFR was added to reaction mixture containing 50 mM TES, pH 7.0, 75 mM β-mercaptoethanol, 1 mM EDTA, 1 mg/ml BSA, 0.1 mM NADPH, 0.1 mM dihydrofolate, and various concentrations of inhibitors to initiate the enzymatic reaction. Absorbance at 340 nm (A340) was monitored over 100 seconds. DHFR activity was measured as changes in A340 per minute.
Ex vivo AbDHFR activity assay
To determine whether AbDHFR activity in A. baumannii cells is affected by DHFR inhibitors, overnight cultures of A. baumannii strains were harvested, OD600-adjusted to 5.6 OD/ml and treated with 10 µM inhibitors for 90 minutes. To determine DHFR activity in treated A. baumannii, cells were harvested, washed once with and resuspended in DHFR buffer (20 mM potassium phosphate buffer, pH 7.0, 0.1 mM EDTA, 10 mM DTT, 50 mM KCl, 20% glycerol). A. baumannii suspension was lysed by sonication, normalized for protein content, and used to initiate DHFR activity assay. DHFR activity in untreated A. baumannii was set to 100% activity. DHFR activity of inhibitor-treated A. baumannii was calculated relative to untreated control.
References
Munoz-Price, L. S. & Weinstein, R. A. Acinetobacter Infection. N. Engl. J. Med. 358, 1271–1281 (2008).
Lu, C. L. et al. Antimicrobial susceptibilities of commonly encountered bacterial isolates to fosfomycin determined by agar dilution and disk diffusion methods. Antimicrob. Agents Chemother. 55, 4295–4301 (2011).
Leite, G. C. et al. Antimicrobial Combinations against Pan-Resistant Acinetobacter baumannii Isolates with Different Resistance Mechanisms. PLoS One 11, e0151270 (2016).
Gales, A. C., Jones, R. N. & Sader, H. S. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: Report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin. Microbiol. Infect. 12, 315–321 (2006).
Bonomo, R. A. & Szabo, D. Mechanism of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43, 49–56 (2006).
Peleg, A. Y., Adams, J. & Paterson, D. L. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 2065–2069 (2007).
Bleicher, K. H., Böhm, H. J., Müller, K. & Alanine, A. I. Hit and lead generation: Beyond high-throughput screening. Nat. Rev. Drug Discov. 2, 369–378 (2003).
Congreve, M., Murray, C. W. & Blundell, T. L. Keynote review: Structural biology and drug discovery. Drug Discov. Today 10, 895–907 (2005).
Cheol Park, G. et al. In Vitro Interactions of Antibiotic Combinations of Colistin, Tigecycline, and Doripenem Against Extensively Drug-Resistant and Multidrug-Resistant Acinetobacter baumannii. Ann Lab Med 36, 124–130 (2016).
Siricilla, S. et al. A New Combination of a Pleuromutilin Derivative and Doxycycline for Treatment of Multidrug-Resistant Acinetobacter baumannii HHS Public Access. J Med Chem. April 13, 2869–2878 (2017).
The Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing CLSI supplement M100S. Clinical and Laboratory Standards Institute, Wayne, PA. (2016).
Nelson, R. G. & Rosowsky, A. Dicyclic and tricyclic diaminopyrimidine derivatives as potent inhibitors of Cryptosporidium parvum dihydrofolate reductase: Structure-activity and structure-selectivity correlations. Antimicrob. Agents Chemother. 45, 3293–3303 (2001).
Lau, H., Ferlan, J. T., Hertle Brophy, V., Rosowsky, A. & Sibley, C. H. Efficacies of Lipophilic Inhibitors of Dihydrofolate Reductase against Parasitic Protozoa. 45, 187–195 (2001).
Popov, V. M. et al. Analysis of complexes of inhibitors with Cryptosporidium hominis DHFR leads to a new trimethoprim derivative. Bioorganic Med. Chem. Lett. 16, 4366–4370 (2006).
Ahrweiler, P. M. & Frieden, C. Construction of a fol mutant strain of Escherichia coli for use in dihydrofolate reductase mutagenesis experiments. J. Bacteriol. 170, 3301–4 (1988).
Goodsell, D. S. The molecular perspective: methotrexate. Oncologist 4, 340–1 (1999).
Rajagopalan, P. T. R. et al. Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics. Proc. Natl. Acad. Sci. USA 99, 13481–6 (2002).
Hecht, D., Tran, J. & Fogel, G. B. Structural-based analysis of dihydrofolate reductase evolution. Mol. Phylogenet. Evol. 61, 212–230 (2011).
Lele, A. C., Mishra, D. A., Kamil, T. K., Bhakta, S. & Degani, M. S. Repositioning of DHFR Inhibitors. Curr. Top. Med. Chem. 16, 2125–43 (2016).
Kopytek, S. J., Dyer, J. C., Knapp, G. S. & Hu, J. C. Resistance to methotrexate due to AcrAB-dependent export from Escherichia coli. Antimicrob. Agents Chemother. 44, 3210–2 (2000).
Foote, S. J., Galatis, D. & Cowman, A. F. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc. Natl. Acad. Sci. USA 87, 3014–7 (1990).
Aroonsri, A. et al. Identifying antimalarial compounds targeting dihydrofolate reductase-thymidylate synthase (DHFR-TS) by chemogenomic profiling. Int. J. Parasitol. 46, 527–535 (2016).
Ovrebo, S. & Kleppe, K. Pyrimidine metabolism in Acinetobacter calcoaceticus. J. Bacteriol. 116, 331–6 (1973).
Rajamani, S. et al. Bioengineering of bacterial pathogens for noninvasive imaging and in vivo evaluation of therapeutics OPEN. Sci. Reports 8, 12618 (2018).
Towner, K. The Genus Acinetobacter. In The Prokaryotes 746–758, https://doi.org/10.1007/0-387-30746-X_25 (Springer New York, 2006).
Yuthavong, Y. et al. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target.
Talawanich, Y., Kamchonwongpaisan, S., Sirawaraporn, W. & Yuthavong, Y. Use of bacterial surrogates as a tool to explore antimalarial drug interaction: Synergism between inhibitors of malarial dihydrofolate reductase and dihydropteroate synthase. Acta Trop. 149, 64–69 (2015).
Tirakarn, S. et al. Cloning and heterologous expression of Plasmodium ovale dihydrofolate reductase-thymidylate synthase gene. Parasitol. Int. 61, 324–332 (2012).
Wooden, J. M., Hartwell, L. H., Vasquez, B. & Sibley, C. H. Analysis in yeast of antimalaria drugs that target the dihydrofolate reductase of Plasmodium falciparum. Mol. Biochem. Parasitol. 85, 25–40 (1997).
Thotsaporn, K., Sucharitakul, J., Wongratana, J., Suadee, C. & Chaiyen, P. Cloning and expression of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii: Evidence of the divergence of enzymes in the class of two-protein component aromatic hydroxylases. Biochim. Biophys. Acta - Gene Struct. Expr. 1680, 60–66 (2004).
Tien, H. C. et al. Multi-drug resistant Acinetobacter infections in critically injured Canadian forces soldiers. BMC Infect. Dis. 7, 1–6 (2007).
Chan, A. P. et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 16 (2015).
The Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. (2018).
Braks, J. A. M., Franke-Fayard, B., Kroeze, H., Janse, C. J. & Waters, A. P. Development and application of a positive-negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res. 34, e39 (2006).
Acknowledgements
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. UL was supported by a grant from National Center for Genetic Engineering and Biotechnology (BIOTEC) (P16-52034), SY by a RI research grant (P16-51873) from Food Biotechnology Research Unit, BIOTEC, and PC by the Thailand Research Fund (RTA5980001) and Vidyasirimedhi Institute of Science and Technology. Pathogen Box and specific compounds from Pathogen Box and Malaria Box were obtained from Medicines for Malaria Venture (www.pathogenbox.org). The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository Resources, NIAID, NIH: Plasmid pL0035, for Transfection in Plasmodium berghei, MRA-850, contributed by Andrew P. Waters; Acinetobacter baumannii strain Naval-81, NR-17786; Acinetobacter baumannii strain H72721, NR-9667; and Acinetobacter baumannii strain 3-137 (OIFC137), NR-17777. The authors thank Dr. Philip J. Shaw and Prof. Prapon Wilairat for critical proofreading of the manuscript.
Author information
Authors and Affiliations
Contributions
W.S., S.Y., P.C. and U.L. designed the study. W.S., S.Y., L.B., E.N. and S.P. performed the experiments and analyzed the data. W.S., S.Y., P.C. and U.L. wrote the manuscript. All authors approved of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Songsungthong, W., Yongkiettrakul, S., Bohan, L.E. et al. Diaminoquinazoline MMV675968 from Pathogen Box inhibits Acinetobacter baumannii growth through targeting of dihydrofolate reductase. Sci Rep 9, 15625 (2019). https://doi.org/10.1038/s41598-019-52176-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52176-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.