Abstract
Caveolin-1 is a protein (encoded by the CAV1 gene) supposedly harboring a protective effect against fibrosis. CAV1 rs4730751 single nucleotide polymorphism (SNP) AA genotype was initially associated with lower graft survival compared to non-AA. However, subsequent studies could not find the same effect. CAV1 rs4730751 SNP was investigated on 918 kidney donors. Multivariate Cox-model analyses were performed to evaluate risk factors for graft loss. Longitudinal changes on long-term estimated glomerular filtration rate (eGFRs) were evaluated with a linear mixed model. Histopathological findings from protocolled biopsies after 3 months post transplantation were also analyzed. Donor CAV1 rs4730751 genotyping proportions were 7.1% for AA, 41.6% for AC and 51.3% for CC. The AA genotype, compared to non-AA, was not associated with lower graft survival censored or not for death (multivariate analysis: HR = 1.23 [0.74–2.05] and HR = 1.27 [0.84–1.92]). Linear mixed model on long-term eGFRs revealed also no significant difference according to the genotype, yet we observed a trend. AA genotype was also not associated with a higher degree of fibrosis index on protocolled kidney biopsies at 3 months. To conclude, donor CAV1 rs4730751 SNP may impact on kidney transplantation outcomes, but this study could not confirm this hypothesis.
Similar content being viewed by others
Introduction
Genomic studies are unraveling the genetic architecture of complex diseases and traits, and evidence is emerging that genetic information might be clinically relevant in some scenarios. In the field of kidney transplantation, several genetic polymorphisms such as those affecting APOL11, ABCB12,3,4, CYP3A45,6,7 or CYP3A58,9,10, have been shown to influence allograft outcome. Moreover, the importance of the donor’s genetic make-up was largely overlooked. Caveolin-1 has recently gained interest with the discovery of one CAV1 Single Nucleotide Polymorphism (SNP) associated with allograft failure11. Caveolin-1 is the primary structural component of caveolae, involved in endocytosis and cell signaling12. It is ubiquitously expressed, especially in the kidney, from glomerular to epithelial cells13. As the lipid-raft caveolae contribute to TGFβ receptor degradation pathway, and thus decrease TGFβ signaling14, Caveolin-1 exerts a protective effect on fibrosis15, a pathological feature occurring post-transplantation16. Moore and colleagues were the first team which identified a significant association between CAV1 rs4730751 SNP and a higher risk of allograft failure (donor AA versus AC and CC: HR = 1.77 [1.08–2.90])11. Analysis of kidney biopsies from grafts that had failed revealed a higher degree of fibrosis in the group of patients harboring an AA-genotype graft. Interestingly, the rs4730751 SNP is an intronic variant that has not been found to be in linkage disequilibrium with other exonic variants likely to alter Caveolin-1 protein function11. Thus, the precise roles of this SNP and its functional consequences have not been uncovered so far.
This seminal study has led to the evaluation of CAV1 SNPs involvement in various diseases, such as chronic kidney diseases17, pancreas transplantation18, Anti-Neutrophilic Cytoplasmic Autoantibody (ANCA) vasculitis19 or cancers20,21. However, the enthusiasm has been somewhat tempered by the controversies that have risen about the real impact of CAV1 SNPs in the field of kidney transplantation. Indeed, Ma and colleagues found opposite results, as the screening of 16 CAV1 SNPs (including rs4730751) in 1233 kidney transplants could not reproduce Moore’s observations22. Recently, graft survival was also not associated with CAV1 rs4730751 SNP either from donors or recipients in two other cohorts23,24.
Hence, considering these uncertainties, we carried out a study in a large-scaled cohort in order to evaluate the impact of donor CAV1 rs4730751 SNP on kidney transplantation outcomes, using a combined analysis of graft survivals, long-term estimated Glomerular Filtration rates (eGFRs) and histopathological data from systematic kidney biopsies.
Results
Study population and baseline characteristics
From the 1st of January 2000 to the 31st of December 2016, 918 donors for kidney transplantation were genotyped for the CAV1 rs4730751 SNP. Alleles A and C were in equilibrium according to the Hardy-Weinberg law (respectively p = 0.27 and q = 0.73). CAV1 rs4730751 AA, AC, and CC genotypes were observed in respectively 7.1% (n = 65), 41.6% (n = 382), and 51.3% (n = 471) of donors. All donors and recipients’ demographical characteristics are summarized in Table 1. There was no difference between AA and non-AA donors, or between their respective recipients. Median follow-up was 47.7 months (23.7–119.1).
Graft outcomes: survival estimates and Cox models
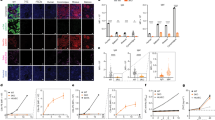
Using the Log-Rank test (Fig. 1), no long-term difference was observed for graft survival - censored for death (GS-DC), nor for graft survival - non-censored for death (GS-DNC), between AA and non-AA genotype (p = 0.64 and p = 0.64 respectively). Using a Cox model for multivariate analysis (Table 2), an AA genotype was not significantly associated in uni- or multivariate analyses with GS-DC or GS-DNC (multivariate analysis: HR = 1.23 [0.74–2.05] and HR = 1.27 [0.84–1.92] respectively).
The significant risk factors of GS-DC in multivariate analysis were donor age (HR per 10 years = 1.41 [1.25–1.60]) and graft rejection occurrence (HR = 3.17 [2.24–4.49]). A first transplantation was found to be protective (HR = 0.62 [0.44–0.86]). Considering GS-DNC, in addition to the above-mentioned risk and protective factors, the donor sex (male) was also found to be a risk factor (HR = 1.34 [1.06–1.70]).
As a secondary analysis, we tested if carrying an A allele was significantly associated with a higher risk of graft failure. CC versus non-CC donors and recipients were similar (Supplemental Table 1). Carrying an A allele was also not associated with a higher risk of graft failure in uni- or multivariate analysis: GS-DC HR = 0.97 [0.77–1.21]; GS-DNC HR = 0.91 [0.69–1.20] (Supplemental Figs 1 and 2; Supplemental Table 2).
Post-transplantation outcomes: eGFR variations
Given the normal distribution of eGFR, this variable was not transformed. According to the Bayesian Information Criterion (BIC), the best model relation between eGFR and time was linear. Using then a linear mixed model (Fig. 2), longitudinal changes of eGFR according to the Modification of Diet in Renal Disease (MDRD) formula over time were compared between AA (n = 59) and non-AA donors (n = 764) (Supplemental Table 3). We used 4785 available values of eGFR for 823 patients with a median follow up of 4 years [1.98–7.96]. The model was adjusted on significant variables in univariate analysis, i.e. recipient sex, age, time on dialysis, Body Mass Index (BMI), previous transplantation, cold ischemia time and donor age. There was no significant difference according to CAV1 genotype on long-term eGFRs (fixed effect of AA genotype at 3 months post transplantation eGFR: 2.95 mL/min/1.73 m² [−0.87–6.77, p = 0.13] and fixed effect of AA genotype on slope: −0.62 mL/min/1.73 m² per year [−1.33–0.13, p = 0.10]). When comparing CC donors (n = 421) versus non-CC donors (n = 402), there was also no difference on long-term eGFRs (fixed effect of CC genotype at 3 months post-transplantation eGFR: 0.21 mL/min/1.73 m² [−1.79–2.21, p = 0.83] and fixed effect of CC genotype on slope: −0.13 mL/min/1.73 m² per year [−0.52–0.26, p = 0.51]) (Supplemental Fig. 3).
Linear mixed model for long-term estimated glomerular filtration rate comparison between CAV1 rs4730751 single nucleotide polymorphism AA versus non-AA. n = 4785 samples. Fixed effect of AA phenotype at 3 months post transplantation eGFR: 2.95 mL/min/1.73 m² [−0.87–6.77, p = 0.13] and fixed effect of AA genotype on slope: −0.62 mL/min/1.73 m² per year [−1.33–0.13, p = 0.10].
Post-transplantation outcomes: proteinuria
As a surrogate marker, we also tested if AA genotype impacted on proteinuria after transplantation. There were 7936 available values related to 695 patients with a median follow-up of 2.2 years [0.72–8.00]). Using a linear mixed model (Supplemental Table 4), longitudinal changes of urine protein/creatinine ratio were compared between AA donors (n = 47) and non-AA donors (n = 648). There was no significant difference on both baseline effect and slope effect according to CAV1 genotype for long-term urine protein/creatinine ratio. (Supplemental Fig. 4).
Post-transplantation outcomes: 3-months kidney biopsies
As protocolled kidney biopsies at 3 months post-transplant were systematically analyzed according to Banff classification since 200725, only data for the last 394 recipients were available. Those included 25 CAV1 AA and 369 non-AA donors. Over the tested parameters, only the proportion of globally scarred glomeruli was significantly different between AA and non-AA (4.80% versus 7.70%, p = 0.034). Of note, there was no significant difference between AA and non-AA for Banff lesions scores especially as regards markers of chronic injury, such as mesangial matrix expansion (mm score), glomerular double contours (cg score), interstitial fibrosis (ci score), or tubular atrophy (ct score). There was also no significant difference for IFTA score (Table 3).
We also evaluated the impact of carrying an A allele (n = 208 CC and n = 186 non-CC) on kidney biopsies graded according to Banff criteria, and could not find any significant difference between the two groups (Supplemental Table 5) in particular for fibrosis.
Discussion
The objective of our study was to evaluate the donor CAV1 rs4730751 SNP involvement on kidney allograft outcomes. It is currently the only study which investigates precisely the association between CAV1 rs4730751 SNP genotype and its related phenotype, through large scale clinical data, biological outcomes, and histopathological analyses. First of all, we could not find any significant impact of this polymorphism on graft survival. Secondly, one of the major strengths of our study was that we performed a longitudinal analysis of eGFR values using a mixed model, as a surrogate marker of graft survival. Again, eGFR trajectories did not differ significantly according to CAV1 genotype. Nevertheless, we observed a trend of a higher slope decrease for AA donors, without statistical significance, which may be due to a lack of power. Finally, while using a standardized analysis with Banff classification, we could not find any significant difference on kidney fibrosis in 3-months post-transplant protocolled kidney biopsies.
Considering the previous attempts to test CAV1 rs4730751 SNP impact on kidney allograft survival, Moore et al. showed for the first time a deleterious effect of this SNP11, when tested on two independent cohorts, the Birmingham cohort (n = 785) and the Belfast cohort (n = 679). In both cohorts, rs4730751 SNP was associated with a higher risk of graft loss (multivariate analysis AA versus non-AA: HR = 1.77 [1.08–2.90] and HR = 1.56 [1.07–2.27]). Even with a small prevalence of the AA genotype in both cohorts (respectively 7.3% and 7.1%), the risk conferred by the described association justified the authors to recommend further investigations on CAV1 rs4730751 SNP. Indeed, the impact of caveolin-1 involvement in post transplantation outcomes could be supported with several mouse models. Knock-out mice for CAV1 exhibited higher degree of kidney interstitial fibrosis than control mice, after unilateral ureteral obstruction challenge26,27. In a pro-fibrotic environment, with sustained TGFβ stimulation, caveolin-1 deficiency may be associated with an accelerated fibrotic process and impaired outcomes, as caveloae are supposed to internalize TGFβ receptor13. Moreover, caveolin-1 deficiency may also be associated with a different inflammatory response and a pro-fibrotic polarization of M2 macrophages after unilateral ureteral obstruction challenge in knock-out CAV1 mice compared to wild type27. Unfortunately, concerning CAV1 rs4730751 SNP, no in vitro or in vivo data are available regarding caveolin-1 functionality in patients harboring this particular intronic SNP. The study from Moore et al. presents several differences compared to ours which may explain the difference of results. First, fibrogenesis is a multifactorial process16, in which CAV1 is known to play a role. Indeed, CAV1 rs4730751 SNP is supposed to be involved in an accelerated fibrosis process due to impaired abilities of protection against pro-fibrotic injuries. After kidney transplantation, several events cause pro-fibrotic kidney damages, such as cold and warm ischemia times, rejection, infections like Polyomavirus-associated nephropathy, and immunosuppressive agents, especially calcineurin inhibitors28. Thus, there may be differences between our cohorts regarding pro-fibrotic injuries, which would explain the difference of results. Second, Moore et al. validated the CAV1 rs4730751 SNP impact on two independent cohorts, including both living and brain-deceased donors, compared to our single cohort, including only brain-deceased donors. Finally, this study provided data which may be more accurate to assess long-term results, as the independent cohorts had a median follow-up of respectively 81 (54–113) and 69 (24–124) months, compared to 47.7 months (23.7–119.1) in our single cohort. Moore et al. provided also data on long term indication biopsies for allograft dysfunction11 and showed that the main cause of graft failure in the AA donor cohort was interstitial fibrosis (13 graft losses among 22 AA donors), mostly related to chronic cellular rejection (n = 6/13). On the contrary, we presented data on 3-month protocolled post-transplantation kidney biopsies, which may be too early to assess differences considering kidney fibrosis. Also, even if we could not find any effect of this SNP in our cohort, we cannot exclude that its effect could not be observed because of the differences mentioned above. After Moore and coworkers study, other teams studied the impact of CAV1 rs4730751 SNP on kidney transplantation outcomes. Ma et al. presented data on CAV1 rs4730751 SNP from 1233 kidney transplantations from Afro-American (n = 675) and European American donors (n = 558)22. The prevalence for AA donors was 4.00% in Afro-American donors and 7.84% in European American donors. As in our study, AA genotype was not associated with a higher risk of graft failure, but the authors were able to show interactions between other CAV1 SNPs and APOL1 SNPs, in particular the CAV1 rs6466583 SNP. We cannot exclude in our study that interactions with other SNPs affecting CAV1 sequence are underestimated. Furthermore, in our cohort ethnicity could not be collected for both donors and recipients, due to French ethical issue29. This could be an underlying confounder, especially considering the impact of donor APOL1 genotype on allograft outcomes1. To compare Ma et al. study with Moore and coworkers work, the differences on survival could be due to the same reasons than ours, i.e. difference of pro-fibrotic injuries. Moreover, as in our study, the study from Ma et al. also had a lower median follow-up of 34.3 months (13.8–57.9) compared to Moore et al.11. It could have been interesting to provide surrogate markers, in particular long term eGFR, which would support the lack of impact of CAV1 rs4730751 SNP in their cohort. In the same way, Van der Hauwaert et al. could not find any impact on graft survival in a smaller cohort (475 kidney donors, AA genotype prevalence: 7.6%)24, however AA genotype exhibited a significant decrease in eGFR. As in the present study, information concerning ethnicity was missing which may interfere with the results. As Van der Hauwaert et al.24, our results may suggest an impact on eGFR decrease as there was a trend of a higher slope decrease, using a linear mixed-model, for AA donors compared to non AA. Considering the low frequency of the allele, the absence of significance may be due to a lack of power. Furthermore, even if we could not find any difference on interstitial fibrosis-tubular atrophy in 3-months kidney biopsies score, we observed a significant higher percentage of globally scarred glomeruli in AA donors compared to non AA donors. This may be in line with the results of the previous studies on CAV1 genotype11,24, where AA donors related recipients, who had experienced a delayed graft functioning, had lower eGFRs at 3-months than non-AA donors related recipients, suggesting a decreased ability of renal recovery. In this study, AA genotype was also associated with several markers of fibrosis, with a higher risk of chronic allograft dysfunction24, interstitial fibrosis24 or vascular fibrosis11. AA genotype was also associated with a higher proportion of recipient showing a significant proteinuria at 5 years post-transplantation24. Unfortunately we could not reproduce these results with a more powerful linear mixed model.
We only focused on donor CAV1 rs4730751 SNP effect, since evidence exists that the donor genetic background may impact on kidney transplantation outcomes30. Yet, considering that one of the major risk factor for graft loss in our population was graft rejection, which may be related also to the recipient genetic background, the interrelationship between donor and recipient genetic background could be relevant to investigate. Sluczanowska-Glabowska et al. provided data on the impact of allograft recipient CAV1 rs4730751 SNP in a cohort of 270 kidney recipients. The prevalence of AA genotype was 7.4%23. There was no difference between AA versus non AA recipients, regarding long-term serum creatinine levels, or fibrosis index on kidney biopsies. They could also not find any difference between AA and non-AA recipients considering survivals. However, given the AA sample size, these results are probably subject to a lack of power.
Considering all these elements, assessing the real impact of CAV1 rs4730751 SNP on kidney transplantation outcomes still remains a matter of debate. The first seminal study had a robust design and managed to observe an effect of this SNP on two independent large-sized cohorts11, whereas four other studies22,23,24, including ours, could not reproduce these results on strong outcomes such as graft survival. AA genotype may be associated with graft survival and an accelerated decrease of eGFR, but converging proof still remains to be produced. There may also be other specific fields of research in kidney transplantation, such as caveolin-1 genotype involvement in BK virus nephropathy, as the kidney tubular cells way of infection is thought to be caveolae-endocytosis mediated31. Targeting specific recipients, carrying pro-fibrotic risk factors, could also be one relevant field of research in the future.
Patients and Methods
Ethical statement
This observational retrospective study was performed according to the Declaration of Helsinki and the Declaration of Istanbul. No organs were procured from prisoners. As the French Biomedical Agency regulates the allocation system in France, every organ was allocated by the Agency and transplanted in Lille, France (Centre Hospitalier Régional, Lille). The protocol was certified to be in accordance with French laws by the local Institutional Review Board (Centre Hospitalier Régional, Lille), since French health authorities waived for consent requirement from deceased donors. DNA collection was registered to the French Ministry of Research under the number DC-2008-642.
Patients
918 recipients of kidney allograft from brain-deceased donors were consecutively included from the 1st of January 2000 to the 31st of December 2016. Recipients under 18 years-old or receiving combined grafts were excluded from the study.
Data collection
All data were collected from the CRISTAL database (French National Biomedical Agency registry) and from the recipient personal files. General demographic parameters and well-characterized risk factors of allograft failure were extracted from the database: donor age, sex, weight, height, BMI, cause of death, time of cold ischemia; and recipient age, sex, weight, height, BMI, cause of end-stage renal disease (ESRD), rank of transplantation, type of ESRD treatment (hemodialysis, peritoneal dialysis, or no treatment), number of HLA mismatches (on HLA A, B, DR), and longitudinal assessment of serum creatinine as well as eGFRs (MDRD).
Immunosuppressive therapy
The immunosuppressive regimen followed our standard care procedure. All of the patients received an induction therapy consisting in basiliximab or thymoglobulin. Maintenance immunosuppression associated for every recipient tacrolimus, mycophenolate mofetil and steroids. Tacrolimus first doses were 0.15 mg/kg/d, and secondly adapted to tacrolimus trough level (Tac-T0). From D0 to D15, Tac-T0 targets were between 10 and 15 ng/mL. After D15, targets were between 6 and 8 ng/mL. Daily doses of mycophenolate mofetil were 750 mg twice a day. Early steroid withdrawal (day 7) was performed for non-sensitized recipients of a first renal graft.
DNA samples and genotyping
Deceased donor DNA was extracted from lymphocytes used for the pre-transplantation cross match test. Genotyping of the CAV1 rs4730751 SNP was performed with TaqMan allelic discrimination assays (C-29772987_10 assay) on an ABIPrism 7900HT (Life Technologies), in a 96-well plate, according to the manufacturer’s instructions. For quality control, all runs included duplicates of a null sample and of samples with known genotypes. After PCR, end-point fluorescence was measured and genotype calling was carried out using the allelic discrimination analysis module.
Histopathologic diagnosis
From 2007, 3-month protocol biopsies after transplantation, systematically analyzed according to Banff classification30, were performed in our center for each kidney recipients. Tissue was embedded in paraffin, cut in 3–4 μm thick sections and stained with hematoxylin and eosin, periodic acid-Schiff, Masson’s trichrome and silver methenamine. All biopsies had adequate cortical tissue and were evaluated according to Banff criteria30. Fibrosis was evaluated by the IFTA score, depending on the percentage of the total kidney cortical area suffering from tubular atrophy and interstitial fibrosis (<25% = 1; 25–50% = 2; >50% = 3)25. Retrospectively, histopathological data were available for 394 patients.
Statistical analysis
First, a descriptive analysis of the patients’ characteristics according to CAV1 genotype was conducted to identify possible differences. Continuous variables are expressed as means ± standard deviations or medians (interquartile ranges), as appropriate. Categorical variables are presented as absolute numbers and percentages. Comparisons were made using the Student’s T test for quantitative variables and the Chi2 test for qualitative variables.
Second, the median overall survival and median follow-up times were estimated using Kaplan-Meier and inverse Kaplan-Meier methods, respectively. Between-group comparisons were performed using the log-rank test. Univariate followed by multivariable Cox analyses were performed to identify independent predictors of the death-censored graft survival and of the death-uncensored graft survival in two separate models.
The proportional hazard assumption was checked by log-minus-log survival curves plotting and by the scaled Schoenfeld residuals test for all covariates. When the log-linearity assumption was not met for continuous covariates, the variable was categorized in order to minimize the Bayesian information criterion (BIC).
Multivariate Cox model types were built by including all the covariates that were associated in univariate analyses, using a p < 0.20 threshold for selection, and suppressing redundant covariates. Characteristics which are known to impact long term survival were also maintained in the final multivariate models regardless of the univariate significance (i.e. recipient and donor age, sex, and BMI, cold ischemia time, donor cause of death, cause of ESRD, HLA mismatch and previous transplantation).
Third, a linear mixed model estimated by Restricted Maximum Likelihood was used to compare longitudinal changes in eGFR and urine protein/creatinine ratio according to CAV1 genotype over time, beginning at 3 months post transplantation. The CAV1 genotype was treated as a fixed effect associated to two random effects for baseline value and slope. If the dependent variable was not normally distributed, we considered a relevant transformation. We then chose the best fit model for eGFR variations over time on the basis of BIC values.
Univariable analyses were conducted using three effects for each variable: on baseline value, slope (interaction with time) and CAV1 genotype. Among these parameters, those which were not significant (p > 0.20) were removed. If the association on the slope was significant, the corresponding association on baseline value was also considered. Finally, the selected significant variables were further analyzed in a multivariate linear mixed model to determine those acting independently (backward selection procedure, p < 0.05). The normal distribution of random effect on intercept, random effect on slope, residuals and homoscedasticity assumption were graphically assessed. Finally, chronic allograft injury parameters assessed on the 3 months post-transplantation systematic kidney biopsies were compared between groups of AA and non-AA genotype patients.
All analyses were performed using the 3.5.1 version of the R software with “nlme” and “survival” packages.
References
Freedman, B. I., Locke, J. E., Reeves-Daniel, A. M. & Julian, B. A. Apolipoprotein L1 Gene Effects on Kidney Transplantation. Semin. Nephrol. 37, 530–537 (2017).
Palanisamy, A., Reeves-Daniel, A. M. & Freedman, B. I. The impact of APOL1, CAV1, and ABCB1 gene variants on outcomes in kidney transplantation: donor and recipient effects. Pediatr. Nephrol. Berl. Ger. 29, 1485–1492 (2014).
Sun, B. et al. Influence of CYP3A and ABCB1 polymorphisms on cyclosporine concentrations in renal transplant recipients. Pharmacogenomics 18, 1503–1513 (2017).
Bloch, J. et al. Donor ABCB1 genetic polymorphisms influence epithelial-to-mesenchyme transition in tacrolimus-treated kidney recipients. Pharmacogenomics 15, 2011–2024 (2014).
Andreu, F. et al. A New CYP3A5*3 and CYP3A4*22 Cluster Influencing Tacrolimus Target Concentrations: A Population Approach. Clin. Pharmacokinet. 56, 963–975 (2017).
Elens, L. & Haufroid, V. Genotype-based tacrolimus dosing guidelines: with or without CYP3A4*22? Pharmacogenomics 18, 1473–1480 (2017).
Wang, C., Lu, K.-P., Chang, Z., Guo, M.-L. & Qiao, H.-L. Association of CYP3A4*1B genotype with Cyclosporin A pharmacokinetics in renal transplant recipients: A meta-analysis. Gene 664, 44–49 (2018).
Barry, A. & Levine, M. A systematic review of the effect of CYP3A5 genotype on the apparent oral clearance of tacrolimus in renal transplant recipients. Ther. Drug Monit. 32, 708–714 (2010).
Glowacki, F. et al. Influence of Cytochrome P450 3A5 (CYP3A5) Genetic Polymorphism on the Pharmacokinetics of the Prolonged-Release, Once-Daily Formulation of Tacrolimus in Stable Renal Transplant Recipients. Clin. Pharmacokinet. 50, 451–459 (2011).
Rojas, L. et al. Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: a systematic review and meta-analysis of observational studies. Pharmacogenomics J. 15, 38–48 (2015).
Moore, J. et al. Association of caveolin-1 gene polymorphism with kidney transplant fibrosis and allograft failure. JAMA 303, 1282–1287 (2010).
Chidlow, J. H. & Sessa, W. C. Caveolae, caveolins, and cavins: complex control of cellular signalling and inflammation. Cardiovasc. Res. 86, 219–225 (2010).
Lee, E. K., Lee, Y. S., Han, I.-O. & Park, S. H. Expression of Caveolin-1 reduces cellular responses to TGF-beta1 through down-regulating the expression of TGF-beta type II receptor gene in NIH3T3 fibroblast cells. Biochem. Biophys. Res. Commun. 359, 385–390 (2007).
Moriyama, T. et al. The significance of caveolae in the glomeruli in glomerular disease. J. Clin. Pathol. 64, 504–509 (2011).
Shihata, W. A., Putra, M. R. A. & Chin-Dusting, J. P. F. Is There a Potential Therapeutic Role for Caveolin-1 in Fibrosis. Front. Pharmacol. 8, 567 (2017).
Vanhove, T., Goldschmeding, R. & Kuypers, D. Kidney Fibrosis: Origins and Interventions. Transplantation 101, 713–726 (2017).
Chand, S. et al. Caveolin-1 single-nucleotide polymorphism and arterial stiffness in non-dialysis chronic kidney disease. Nephrol. Dial. Transplant. 31, 1140–1144 (2016).
Hamilton, A. et al. Genetic Variation in Caveolin-1 Correlates With Long-Term Pancreas Transplant Function: Caveolin-1 in Pancreas Transplant Function. Am. J. Transplant. 15, 1392–1399 (2015).
Chand, S. et al. Caveolin-1 single nucleotide polymorphism in antineutrophil cytoplasmic antibody associated vasculitis. PloS One 8, e69022 (2013).
Wang, S., Zhang, C., Liu, Y., Xu, C. & Chen, Z. Functional polymorphisms of caveolin-1 variants as potential biomarkers of esophageal squamous cell carcinoma. Biomarkers 19, 652–659 (2014).
Zhao, R. et al. Genetic Variants in Caveolin-1 and RhoA/ROCK1 Are Associated with Clear Cell Renal Cell Carcinoma Risk in a Chinese Population. PLOS ONE 10, e0128771 (2015).
Ma, J. et al. Deceased donor multidrug resistance protein 1 and caveolin 1 gene variants may influence allograft survival in kidney transplantation. Kidney Int. 88, 584–592 (2015).
Słuczanowska-Głąbowska, S. et al. Caveolin-1 rs4730751 gene polymorphism in kidney allograft recipients. J. Appl. Biomed. 16, 133–137 (2018).
Van der Hauwaert, C. et al. Donor caveolin 1 (CAV1) genetic polymorphism influences graft function after renal transplantation. Fibrogenesis Tissue Repair 8, 8 (2015).
Loupy, A. et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am. J. Transplant. 17, 28–41 (2017).
Park, H. C. et al. Postobstructive regeneration of kidney is derailed when surge in renal stem cells during course of unilateral ureteral obstruction is halted. Am. J. Physiol.-Ren. Physiol. 298, F357–F364 (2010).
Chand, S., Hazeldine, J., Smith, S. W. & Borrows, R. Genetic Deletion of the Lipid Raft Protein Caveolin-1 Leads to Worsening Renal Fibrosis. J. Clin. Nephrol. Ren. Care 4, (2018).
Nankivell, B. J., P’Ng, C. H., O’Connell, P. J. & Chapman, J. R. Calcineurin Inhibitor Nephrotoxicity Through the Lens of Longitudinal Histology: Comparison of Cyclosporine and Tacrolimus Eras. Transplantation 100, 1723–1731 (2016).
INSEE. Ethnic-based statistics. (2016).
Chand, S., McKnight, A. J. & Borrows, R. Genetic polymorphisms and kidney transplant outcomes. Curr. Opin. Nephrol. Hypertens. 23, 605–610 (2014).
Moriyama, T., Marquez, J. P., Wakatsuki, T. & Sorokin, A. Caveolar Endocytosis Is Critical for BK Virus Infection of Human Renal Proximal Tubular Epithelial Cells. J. Virol. 81, 8552–8562 (2007).
Acknowledgements
We would like to thank Dr. Paul Chamley (University of Lille, Department of Nephrology), as an English native speaker, for his comments on the main body text. We also would like to thank Mr. Sébastien Gomis (CHU Lille, Data-Manager) for his help concerning the data-mining. This study was supported in part by a following grant from Santelys Association. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Study concept & design: M.M., R.L., A.H., C.V., C.C., F.G. Acquisition of data and data management: M.M., R.L., A.H., C.H., J.G., B.H., N.P. Drafting of the manuscript: M.M., R.L., A.H., B.L., J.G., M.F., C.C., F.G. Critical Revision: M.M., R.L., A.H., C.V., B.L., J.G., M.F., G.S., B.H., N.P., F.B., F.P., M.H., C.C., F.G. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maanaoui, M., Lenain, R., Hamroun, A. et al. Caveolin-1 rs4730751 single-nucleotide polymorphism may not influence kidney transplant allograft survival. Sci Rep 9, 15541 (2019). https://doi.org/10.1038/s41598-019-52079-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52079-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.