Abstract
3D imaging approaches based on X-ray microcomputed tomography (microCT) have become increasingly accessible with advancements in methods, instruments and expertise. The synergy of material and life sciences has impacted biomedical research by proposing new tools for investigation. However, data sharing remains challenging as microCT files are usually in the range of gigabytes and require specific and expensive software for rendering and interpretation. Here, we provide an advanced method for visualisation and interpretation of microCT data with small file formats, readable on all operating systems, using freely available Portable Document Format (PDF) software. Our method is based on the conversion of volumetric data into interactive 3D PDF, allowing rotation, movement, magnification and setting modifications of objects, thus providing an intuitive approach to analyse structures in a 3D context. We describe the complete pipeline from data acquisition, data processing and compression, to 3D PDF formatting on an example of craniofacial anatomical morphology in the mouse embryo. Our procedure is widely applicable in biological research and can be used as a framework to analyse volumetric data from any research field relying on 3D rendering and CT-biomedical imaging.
Similar content being viewed by others
Introduction
One of the formidable phenomena in developmental biology is how the shape diversity observed among living organisms is defined and controlled during development and growth. Embryonic patterning is a highly dynamic process implicating multiple molecular mechanisms and cell interactions at the basis of organ formation. In the human embryo, defects in such cellular processes can affect the developmental program leading to congenital disorders. Congenital defects have an incidence of 3% in the human population1 and they are causal for up to one-quarter of all reported neonatal deaths2. Thus, contextual visualisation of embryonic development is critical to elucidate the origins of malformations.
Multi-disciplinary collectives composed of clinical doctors, biologists, engineers and imaging experts are currently pushing forward the understanding of biological questions using three dimensional (3D) approaches. While the analysis of histological sections remains a mainstay in developmental biology, the reconstruction of 3D volumes from 2D slices has provided important information to understand morphogenesis in mouse and human embryos3,4,5,6. Non-destructive technologies such as 3D imaging by confocal microscopy and light sheet methods7,8,9, optical projection tomography (OPT)10,11,12 and micro-computed tomography (microCT)13,14,15,16 constitute emerging powerful methods to analyse the topography of developing structures in 3D context, to gain insight into the pathogenesis of congenital disorders. However, one major challenge of 3D approaches is sharing of complex datasets effectively and intuitively between colleagues from different fields for discussion, and ultimately presenting them in a publication format. Generally, 3D datasets need special software for visualisation or are reduced to 2D pictures in which important information might be lost.
The study of the developing head constitutes a good example of the need for 3D approaches given its complex anatomy. The craniofacial region is built of diverse embryonic cell types giving rise to hard and soft tissues including skeletal, muscular and nervous components17. The final shape of the face strongly depends on the geometry of the skeletal elements and their interaction with adjacent soft tissues such as muscles and the nervous system15,16. Numerous congenital craniofacial abnormalities affect the form and function of the face and explanations of these malformations still await the fundamental understanding of the underlying failure of morphogenesis18. The microCT approach followed by 3D reconstruction has enabled high-quality information of the complex morphological aspects of head and face development15,16.
To facilitate assimilation and visualisation of microCT datasets, interactive 3D Portable Document Format (PDF) has been used in different field including in developmental biology6,19,20, in human physiology and anatomy4,21,22,23, in entomology24 and marine biology25,26. However, a major limitation of previously published methods is the requirement of advanced programming skills and/or installation of further prepaid software packages21. Most of these approaches depend on the use of Adobe Acrobat Pro Extended software4,6,19,23,24,25,26,27,28 or JavaScript programming language20,27.
Here, we provide an alternative, user-friendly way to create interactive 3D PDF files from microCT datasets taking the complexity of mouse craniofacial anatomy as a model example. Our innovative and efficient pipeline comprises microCT data acquisition, segmentation and final establishment of a 3D PDF using a combination of free and pre-paid software. The resulting file can be viewed with standard PDF viewers and offers an interactive interface for microCT data sharing, analysis and presentation.
Pipeline for the Creation of Interactive 3D PDF
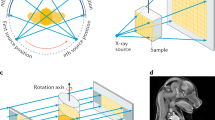
To create an interactive 3D PDF from microCT data, the critical steps are: (i) chemical contrasting of biological samples if soft tissues are to be visualized, (ii) data acquisition and (iii) CT virtual reconstruction (Fig. 1). This is followed by data processing, the most critical step consisting of segmentation, surface extraction, adjustment of segmented 3D models, and conversion into a final interactive PDF file. Here, we provide a detailed description of each step of the pipeline (see also Supplementary Material 1).
X-Ray Microtomography Measurement
MicroCT is an established technology for imaging mineralized tissues in animal specimens. However, its use in comparative morphology has been limited by the low intrinsic X-ray contrast of non-mineralised soft tissues. To overcome this problem, methods have been developed to increase tissue contrast, including chemical treatment with contrasting agents29,30, phase-contrast imaging31,32,33,34,35,36 or dual-energy computed tomography (DECT)37.
Chemical contrasting treatments have mostly been applied for the characterization of musculoskeletal tissues on small fixed samples. For in vivo CT imaging including clinical diagnoses, non-toxic iodine-based contrast agents (e.g. iohexol or hexabrix) are used for the analysis of the cardiovascular system and cavities, but not for direct analysis of muscle tissues38,39. Clinical CT imaging of the musculoskeletal system is commonly performed without a contrasting agent, thereby limiting the analysis to discrete soft organs40. Therefore, on small fixed samples, multiple contrasting protocols have been used, each with its own advantages and limitations41,42. The stainings based on iodine, osmium or the toxic phosphotungstic acid (PTA) are the most commonly used29,30. For the pipeline proposed here on the study of craniofacial mouse development, we propose the use of PTA contrasting treatment that permits high contrast imaging of a wide variety of soft and mineralised tissues composing the mouse head (Fig. 2). However, microCT data obtained with other contrasting agents or in their absence, such as clinical data or data from non-biological specimens, will be equally ameneable for the subsequent steps of the 3D PDF pipeline.
Tomographic measurements and segmentation of craniofacial structures in a mouse embryo 15 days post-fertilisation. Colour planes on 3D models (left panels) indicate the position of the raw tomographic slices (middle panels) and some segmented structures (right panels) including the central nervous system (purple), the lens (dark blue), the dental placodes (light blue) and extraocular muscles (red).
Data Processing
Following the acquisition of high-contrast tomographic slices, the segmentation of complex craniofacial structures constitutes a major challenge. Fully-automatic approaches have been tried and were generally unsuccessful43. Thus, the manual approach is usually the only method available to achieve precise and accurate segmentations44. However, some semi-automatic methods such as local segmentation or interpolation between manually segmented slices can be used45,46. For verification of the accuracy of manually segmented structures, Fig. 2 shows both original and segmented tomographic slices. This approach permitted the segmentation of soft tissues such as eyeballs, extraocular muscles and the central nervous system, as well as of hard tissues including the chondrocranium, bones and future teeth (dental placodes) (Fig. 3).
For 3D volume rendering, it is then more convenient to use 3D mesh formats rather than a stack of 2D images. 3D mesh formats represent a series of 2D polygons (typically triangles or quadrangles) linked together to recreate the surface of a 3D object28. It encodes the 3D model’s geometry, colours, textures, etc. For further work, the segmented masks need to be transferred to the meshes. Most software can transfer the mask into a wide range of equivalent formats. However, only some of the formats contain information about the colour (e.g. OBJ, VRML and STEP) which is important for the visualisation of several structures in one model. Other formats represent only the mesh (e.g. STL and Matlab m-file).
Another problem to be solved is the size of the model (i.e. the number of faces in the mesh). When transferring the segmented mask into the mesh, an unnecessarily large number of faces can be created. Comparison of a different number of faces and corresponding size can be found in Fig. 4. Surprisingly, reducing by four times the number of faces does not significantly compromise details in the 3D models. Simple shapes such as the eyeball do not show notable errors besides the slight deformation of the sphere. However, further simplification of complex shapes, such as the chondrocranium and extraocular muscles (EOM), reduces the model quality and some details are lost (Fig. 4, red arrows). Thus, depending on the structures analysed and the resolution needed, different face reductions should be tested and validated before further analysis. Therefore, the compression of the data should be customised according to the complexity of the model and the resolution of tomographic slices.
Among the 3D printing software, some free software enable colour modifications or mesh simplification. Here, we made use of the Meshlab47 and Blender48 packages for this purpose. Notably, the latter allows unifying different sub-models into one mesh preserving the individuality of each object (see Supplementary Material 1 for more details).
Creation of an Interactive 3D PDF
Once the models are colour-labelled and simplified, they are then converted into an interactive file. We exploited 3D PDF Maker Standalone49 taking advantage of the pre-prepared model that provides an interactive window in the PDF file by clicking on Add 3D button. Before embedding the model into an interactive file, the user can prepare an ordinary PDF file using standard software (e.g. Microsoft Office, Apache OpenOffice etc.). This file is used as a template showing, for example, individual structures or predefined views. These areas (i.e. images or signs) are taken into life in 3D PDF Maker by assigning whatever area in the page to specific view/structure (see for more details Supplementary Materials 1 and 2).
The model document presented here (Supplementary Material 3) consists of four predefined views (ventral, dorsal, lateral and medial) and a possibility of showing individual craniofacial structures (brain, cartilage, bone and teeth). It helps to manipulate the 3D model and can be set according to the areas/views of interest that are important to show. For example, for better visualisation, some structures can be set to the semi-transparent mode, or structures can be switched on/off individually in the model tree. The interactive 3D PDF also allows the user to rotate, turn and pan the model, change the pre-defined 3D-rendering, change the lights and background colour or add a virtual cross-section on the model.
Discussion and Conclusion
Comparative morphological studies in the field of developmental biology have been challenging, but contrast-enhanced X-ray computed tomography has brought new possibilities of high-resolution 3D visualisation15,16,50. In this study, we present a detailed, user-friendly protocol for the surface rendering of craniofacial structures including soft tissues (muscles, eyeballs, central nervous system) as well as of hard tissues (cartilages, bones and teeth), with a step-by-step procedure from sample collection to the creation of an interactive 3D PDF. The final PDF is readable on all operating systems with the free standard AdobeReader®/AcrobatReaderDC (Adobe Inc., California USA).
Our 3D reconstruction methodology has already shown its utility and strength for visualisation and phenotypic analysis of complex structures such as the neck musculoskeletal system50 and nasal capsules of control and mutant mouse embryos16. Therefore, a user-friendly method for creating such files will be of great utility for biologists. In addition, our procedure can be used as a framework to analyse volumetric data for any field of research that relies on 3D rendering, e.g. for visualisation of volumetric information of geological samples by laser-induced breakdown spectroscopy51.
The use of interactive 3D PDF files has a great potential for data sharing, communication and publications21, but is still sparsely used. We believe that our work will inspire researchers working with 3D imaging to present their data in such a user-friendly format.
Methods
Use of experimental animals
All animal work was approved and permitted by the Local Ethical Committee on Animal Experiments (North Stockholm Animal Ethics Committee) and conducted according to The Swedish Animal Agency’s Provisions and Guidelines for Animal Experimentation recommendations. Mice were sacrificed with isoflurane (Baxter KDG9623) overdose or cervical dislocation, and embryos were collected into ice-cold PBS. Subsequently, tissue samples were fixed into freshly prepared 4% paraformaldehyde (PFA) in PBS solution for 24 hours at +4 °C with slow rotation and washed in PBS.
Tissue contrasting
Staining protocol has been adapted and modified from the original protocol developed by Brian Metscher laboratory33,34. After fixation, the samples were dehydrated in increasing concentration of ethanol series (30%, 50%, 70%, 90%), one day in each concentration to minimise the shrinkage of tissues. For tissue contrasting, samples were then transferred into 1% PTA (phosphotungstic acid) in 90% methanol for three weeks. The PTA-methanol solution was changed every two days. Subsequently, the samples were rehydrated by ethanol series (90%, 70%, 50% and 30%) and shipped to the CT-laboratory for scanning.
X-ray microCT measurements
After fixation and contrasting treatments, samples were fully rehydrated in distilled water, embedded in 1.0% agarose gel and placed in a polypropylene tube to avoid movement artefacts during tomography scanning. The polypropylene tube was fixed on a plastic rod by a silicone gun. The rod was put in the centre of the rotation stage axis. The microCT scanning was performed using the laboratory system GE Phoenix v|tome|x L 240 (GE Sensing & Inspection Technologies GmbH, Germany) with a 180 kV/15 W maximum power nanofocus X-ray tube and flat panel dynamic 41|100 with 4000 × 4000 px and a pixel size of 100 × 100 μm. The exposure time was 600 ms in each of the 2200 projections acquired over a total angle of 360°. Three projections were acquired and averaged in every position to reduce the noise in the tomographic data. Thus, the scanning time was 73 minutes. The acceleration voltage and current of the X-ray tube were 60 kV and 200 μA, respectively. The beam was filtered by a 0.2 mm-thick aluminium filter. The linear voxel resolution of the measurement was set to 4.2 μm in all dimensions. The tomographic reconstruction was conducted using the software GE phoenix datos|x 2.0 (GE Sensing & Inspection Technologies GmbH, Germany).
Data processing and analysis
Segmentation of craniofacial structures was performed semi-manually in Avizo (Thermo Fisher Scientific, USA). We used some automatic tools using a region growing method and thresholding on 2D slices. Segmentations were done on one of three to five slices, and the mask was interpolated. Subsequently, the segmented models were smoothed in VG Studio MAX 3.2 (Volume Graphics GmbH, Germany) according to45. The segmented mask was then exported into a mesh (*.OBJ format) and was colour-labelled and adjusted in Meshlab47 and Blender software48. The colour-coded models were transferred into a pre-prepared PDF file in 3D PDF Maker Standalone49. The detailed manual can be found in Supplementary Material 1. The use of the interactive 3D PDF is available in Supplementary Material 2.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
World Health Organization. Congenital anomalies, Fact sheet No. 370. https://www.who.int/news-room/fact-sheets/detail/congenital-anomalies (2016).
World Health Organization. Birth defects: report by the Secretariat, Executive board 125th session EB125/7. http://www.who.int/iris/handle/10665/2271 (2009).
Weninger, W. et al. High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol 211, 213–221, https://doi.org/10.1007/s00429-005-0073-x (2006).
De Bakker, B. et al. An interactive three-dimensional digital atlas and quantitative database of human development. Science 354, 1019–1028, https://doi.org/10.1126/science.aag0053 (2016).
De Bakker, B., De Jong, K., Hagoort, J., Oostra, R. & Moorman, A. Towards a 3-dimensional atlas of the developing human embryo: The Amsterdam experience. Reproductive Toxicol 34, 225–236, https://doi.org/10.1016/j.reprotox.2012.05.087 (2012).
De Boer, B., Van den Berg, G., De Boer, P., Moorman, A. & Ruijter, J. Growth of the developing mouse heart: An interactive qualitative and quantitative 3D atlas. Dev Biol 368, 203–213, https://doi.org/10.1016/j.ydbio.2012.05.001 (2012).
Belle, M. et al. Tridimensional Visualization and Analysis of Early Human Development. Cell 169, 161–173, https://doi.org/10.1016/j.cell.2017.03.008 (2017).
Renier, N. et al. iDISCO: A Simple, Rapid Method to Immunolabel Large Tissue Samples for Volume Imaging. Cell 159, 896–910, https://doi.org/10.1016/j.cell.2014.10.010 (2014).
Ragazzi et al. G. Fluorescence confocal microscopy for pathologists. Mod Pathol 27, 460–471, https://doi.org/10.1038/modpathol.2013.158 (2014).
Dickinson, M. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514, https://doi.org/10.1038/nature19356 (2016).
Sharpe, J. Optical Projection Tomography as a Tool for 3D Microscopy and Gene Expression Studies. Science 296, 541–545, https://doi.org/10.1126/science.1068206 (2002).
Sharpe, J. Optical projection tomography as a new tool for studying embryo anatomy. J Anat 202, 175–181, https://doi.org/10.1046/j.1469-7580.2003.00155.x (2003).
Wong, M., Dorr, A., Walls, J., Lerch, J. & Henkelman, R. A novel 3D mouse embryo atlas based on micro-CT. Dev 139, 3248–3256, https://doi.org/10.1242/dev.082016 (2012).
Hsu, C. et al. Three-dimensional microCT imaging of mouse development from early post-implantation to early postnatal stages. Dev Biol 419, 229–236, https://doi.org/10.1016/j.ydbio.2016.09.011 (2016).
Kaucka, M. et al. Oriented clonal cell dynamics enables accurate growth and shaping of vertebrate cartilage. elife 6, e25902, https://doi.org/10.7554/eLife.25902 (2017).
Kaucka, M. et al. Signals from the brain and olfactory epithelium control shaping of the mammalian nasal capsule cartilage. eLife 7, e34465, https://doi.org/10.7554/eLife.34465 (2018).
Noden, D. & Trainor, P. Relations and interactions between cranial mesoderm and neural crest populations. J Anat 207, 575–601, https://doi.org/10.1111/j.1469-7580.2005.00473.x (2005).
Buchanan, E., Xue, A. & Hollier, L. Craniofacial Syndromes. Plast Reconstr Surg 134, 128e–153e, https://doi.org/10.1097/PRS.0000000000000308 (2014).
De Boer, B. et al. The interactive presentation of 3D information obtained from reconstructed datasets and 3D placement of single histological sections with the 3D portable document format. Dev 138, 159–167, https://doi.org/10.1242/dev.051086 (2010).
De Laurier, A. et al. The Mouse Limb Anatomy Atlas: An interactive 3D tool for studying embryonic limb patterning. BMC Dev Biol 8, 1–7, https://doi.org/10.1186/1471-213x-8-83 (2008).
Newe, A. & Becker, L. Three-Dimensional Portable Document Format (3D PDF) in Clinical Communication and Biomedical Sciences: Systematic Review of Applications, Tools, and Protocols. JMIR Med Inform 6, e10295, https://doi.org/10.2196/10295 (2018).
Danz, J. & Katsaros, C. Three-dimensional portable document format: A simple way to present 3-dimensional data in an electronic publication. Am J Orthod Dentofac Orthop 140, 274–276, https://doi.org/10.1016/j.ajodo.2011.04.010 (2011).
Valera-Melé, M. et al. A Novel and Freely Available Interactive 3d Model of the Internal Carotid Artery. J Med Syst 42, 6, https://doi.org/10.1007/s10916-018-0919-4 (2018).
Van de Kamp, T. et al. Three-Dimensional Reconstructions Come to Life – Interactive 3D PDF Animations in Functional Morphology. PLoS ONE 9, e102355, https://doi.org/10.1371/journal.pone.0102355 (2014).
Ruthensteiner, B. & Heß, M. Embedding 3D models of biological specimens in PDF publications. Microsc Res Techn 71, 778–786, https://doi.org/10.1002/jemt.20618 (2008).
Ruthensteiner, B., Baeumler, N. & Barnes, D. Interactive 3D volume rendering in biomedical publications. Micron 41, 886.e1–886.e17, https://doi.org/10.1016/j.micron.2010.03.010 (2010).
Menn, J. & Seliger, G. Increasing Knowledge and Skills for Assembly Processes through Interactive 3D-PDFs. Procedia CIRP 48, 454–459, https://doi.org/10.1016/j.procir.2016.02.093 (2016).
Semple, T., Peakall, R. & Tatarnic, N. A comprehensive and user-friendly framework for 3D-data visualisation in invertebrates and other organisms. J Morphol 280, 223–231, https://doi.org/10.1002/jmor.20938 (2019).
Metscher, B. D. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol 9, 11, https://doi.org/10.1186/1472-6793-9-11 (2009).
Metscher, B. D. MicroCT for developmental biology: A versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn. 238, 632–640, https://doi.org/10.1002/dvdy.21857 (2009).
Wilkins, S., Gureyev, T., Gao, D., Pogany, A. & Stevenson, A. Phase-contrast imaging using polychromatic hard X-rays. Nature 384, 335–338, https://doi.org/10.1038/384335a0 (1996).
Baran, P. et al. High-Resolution X-Ray Phase-Contrast 3-D Imaging of Breast Tissue Specimens as a Possible Adjunct to Histopathology. IEEE Trans Med Imaging 37, 2642–2650, https://doi.org/10.1109/TMI.2018.2845905 (2018).
Wagner, W. et al. Towards synchrotron phase-contrast lung imaging in patients – a proof-of-concept study on porcine lungs in a human-scale chest phantom. J Synchrotron Rad 25, 1827–1832, https://doi.org/10.1107/S1600577518013401 (2018).
Momose, A., Takeda, T., Itaj, Y. & Hirano, K. Phase−contrast X−ray computed tomography for observing biological soft tissues. Nat. Med. 2 (1996).
Saccomano, M. et al. Synchrotron inline phase contrast µCT enables detailed virtual histology of embedded soft‐tissue samples with and without staining. J. Synchrotron Radiat. 25, https://doi.org/10.1107/S1600577518005489 (2018).
Larsson, D. H., Vågberg, W., Yaroshenko, A., Yildirim, A. Ö. & Hertz, H. M. High-resolution short-exposure small-animal laboratory x-ray phase-contrast tomography. Sci. Rep. 6, https://doi.org/10.1038/srep39074 (2016).
Yang, M. et al. Theoretical variance analysis of single- and dual-energy computed tomography methods for calculating proton stopping power ratios of biological tissues. Phys Med Biol 55, 1343–1362, https://doi.org/10.1088/0031-9155/55/5/006 (2010).
Yan, D., Zhang, Z., Luo, Q., Yang, X. & Chen, C. A Novel Mouse Segmentation Method Based on Dynamic Contrast Enhanced Micro-CT Images. PLoS ONE 12, https://doi.org/10.1371/journal.pone.0169424 (2017).
Lusic, H. & Grinstaff, M. W. X-Ray Computed Tomography Contrast Agents. Chem Rev 3, 113, https://doi.org/10.1021/cr200358s (2013).
Heude, E., Rivals, I., Couly, G. & Levi, G. Masticatory muscle defects in hemifacial microsomia: A new embryological concept. Am J Med Genet Part A 155, 1991–1995, https://doi.org/10.1002/ajmg.a.34095 (2011).
de Bournonville, S., Vangrunderbeeck, S. & Kerckhofs, G. Contrast-Enhanced MicroCT for Virtual 3D Anatomical Pathology of Biological Tissues: A Literature Review. Contrast Media Mol. Imaging 2019, 1–9, https://doi.org/10.1155/2019/8617406 (2019).
Zikmund, T. et al. J. High-contrast differentiation resolution 3D imaging of rodent brain by X-ray computed microtomography. J Instrum 13, C02039–C02039, https://doi.org/10.1088/1748-0221/13/02/C02039 (2018).
Weinhardt, V. et al. Quantitative morphometric analysis of adult teleost fish by X-ray computed tomography. Sci Rep 8, 16531, https://doi.org/10.1038/s41598-018-34848-z (2018).
Boccardi, M. et al. Survey of Protocols for the Manual Segmentation of the Hippocampus: Preparatory Steps Towards a Joint EADC-ADNI Harmonized Protocol. J Alzheimers Dis 26, 61–75, https://doi.org/10.3233/JAD-2011-0004 (2011).
Tesařová, M. et al. Use of micro computed-tomography and 3D printing for reverse engineering of mouse embryo nasal capsule. J Instrum 11, C03006–C03006, https://doi.org/10.1088/1748-0221/11/03/C03006 (2016).
Tesařová, M. et al. A quantitative analysis of 3D-cell distribution in regenerating muscle-skeletal system with synchrotron X-ray computed microtomography. Sci Rep 8, 14145, https://doi.org/10.1038/s41598-018-32459-2 (2018).
Cignoni, P. et al. MeshLab: an Open-Source Mesh Processing Tool. http://vcg.isti.cnr.it/Publications/2008/CCCDGR08/MeshLabEGIT.final.pdf (2008).
Hess, R. The essential Blender: guide to 3D creation with the open source suite Blender (ed. Roosendaal, T.) (No Starch Press, 2007).
3D PDF Maker [software] www.3dpdfmaker.com (2019).
Heude, E. et al. Unique morphogenetic signatures define mammalian neck muscles and associated connective tissues. eLife 7, e40179, https://doi.org/10.7554/eLife.40179 (2018).
Prochazka, D. et al. Joint utilization of double-pulse laser-induced breakdown spectroscopy and X-ray computed tomography for volumetric information of geological samples. J Anal Atom Spectrom 33, 1993–1999, https://doi.org/10.1039/c8ja00232k (2018).
Acknowledgements
This research was carried out under the project CEITEC 2020 (LQ1601) with financial support from the Ministry of Education, Youth and Sports of the Czech Republic under the National Sustainability Programme II, CEITEC Nano Research Infrastructure (MEYS CR, 2016–2019) and) and Ceitec Nano+ project, CZ.02.01/0.0./.0.0./16_013/0001728 under the program OP RDE. We acknowledge support by grant AFM Telethon 21853. J.K. and M.T. acknowledge the support of the Brno University of Technology through grant FSI-S-17-4506. M.T. was financially supported by grant CEITEC VUT-J-19-5764 and by the Brno City Municipality as a Brno Ph.D. Talent Scholarship Holder.
Author information
Authors and Affiliations
Contributions
M.T., E.H. and G.C. worked on the data analysis and methodology. M.K. prepared the samples and stained them for CT measurement. M.T. and T.Z. performed CT measurements. I.A., S.T. and J.K. designed the experiments. M.T., T.Z. and J.K. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tesařová, M., Heude, E., Comai, G. et al. An interactive and intuitive visualisation method for X-ray computed tomography data of biological samples in 3D Portable Document Format. Sci Rep 9, 14896 (2019). https://doi.org/10.1038/s41598-019-51180-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51180-2
This article is cited by
-
Contrast enhanced X-ray computed tomography imaging of amyloid plaques in Alzheimer disease rat model on lab based micro CT system
Scientific Reports (2021)
-
A combinatorial method to visualize the neuronal network in the mouse spinal cord: combination of a modified Golgi-Cox method and synchrotron radiation micro-computed tomography
Histochemistry and Cell Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.