Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system has facilitated dramatic progress in the field of genome engineering. Whilst microinjection of the Cas9 protein and a single guide RNA (sgRNA) into mouse zygotes is a widespread method for producing genetically engineered mice, in vitro and in vivo electroporation (which are much more convenient strategies) have recently been developed. However, it remains unknown whether these electroporation methods are able to manipulate genomes at the chromosome level. In the present study, we used these techniques to introduce chromosomal inversions of several megabases (Mb) in length in mouse zygotes. Using in vitro electroporation, we successfully introduced a 7.67 Mb inversion, which is longer than any previously reported inversion produced using microinjection-based methods. Additionally, using in vivo electroporation, we also introduced a long chromosomal inversion by targeting an allele in F1 hybrid mice. To our knowledge, the present study is the first report of target-specific chromosomal inversions in mammalian zygotes using electroporation.
Similar content being viewed by others
Introduction
Chromosomal rearrangements are the cause of many hereditary disorders and play a role in the pathogenesis of diseases such as cancer1,2. To study these molecular pathologies, in vivo models of chromosomal rearrangement are indispensable, but producing them requires substantial effort and prudent technique3,4,5,6. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) system has enabled genome editing of specific sites and has been applied for the generation of gene knockouts and knockins in rats, mice and other organisms7. Recently, chromosomal rearrangements, including large deletions, inversions and duplications, were also performed via microinjection of Cas9 and single guide RNAs (sgRNAs) into mouse zygotes8,9. However, this method still requires specialised equipment and highly skilled personnel. To overcome this limitation, various groups have reported successful genome editing via the in vitro electroporation of Cas9 and a sgRNA into mouse zygotes10,11,12,13, which greatly simplifies their delivery and significantly improves embryo/pup survival compared to microinjection-based methods11,12. More recently, Ohtsuka et al. developed a novel genome editing method called improved genome editing via oviductal nucleic acid delivery (i-GONAD)14,15, which directly delivers Cas9 and a sgRNA into rodent zygotes through the oviducts14,15. This method does not require the isolation, culture, transfer or other in vitro handling of embryos. Whereas this approach has been applied for making knockout and knockin rodents, it is unknown whether it is able to generate more drastic chromosomal rearrangements, such as inversions. In the present study, we present an editing method to generate chromosomal rearrangements of substantial size using in vitro and in vivo electroporation in mouse zygotes. The inverted region in our study was 7.67 megabases (Mb), which is approximately 1.5 times longer than a previously reported inversion produced in mouse zygotes8. Moreover, we demonstrate a method for targeting a selected allele in F1 hybrid mice using in vivo electroporation. This method resulted in successful genomic rearrangements, generating large-scale inversions and recessive lethal deletion alleles at specific sites. Our study shows that the application of the CRISPR/Cas9 system via electroporation in vitro and in vivo would be useful for the study of genetic diseases with large-scale chromosomal rearrangements.
Results and Discussion
Chromosomal inversion via in vitro electroporation
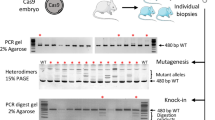
We examined whether in vitro electroporation-mediated CRISPR/Cas9 could generate large-scale genome editing, such as megabase-sized chromosomal inversion. The CRISPR/Cas9 mixture was electroporated into mouse zygotes as described in the Methods section (Fig. 1a, Supplementary Fig. 1a,b). We first designed a 7.67 Mb inversion between Adamts20 and the Krt18 neighbourhood locus (K18N-locus) on the right arm of chromosome 15 (Fig. 1b), which includes the largest region of the type II keratin gene cluster; we targeted this region because disruption of Adamts20 would show easily identifiable phenotypes16. In the present study, we used the cloning-free CRISPR/Cas system, consisting of Cas9 protein, chemically synthesised CRISPR RNA (crRNA), and trans-activating crRNA (tracrRNA). In addition, we designed two single-stranded oligodeoxynucleotides (ssODNs) that joined the chromosomal breakpoints together; each ssODN had a 100 bp sequence that was homologous to each junction point so that chromosomal inversion would be induced by the homology-directed repair (HDR) process to occur between the targeted regions and the homologous ssODNs (Supplementary Fig. 2).
Chromosomal engineering of mouse zygotes via in vitro electroporation. (a) Experimental procedures for induction of chromosomal inversion using in vitro electroporation. (b) Schematic of the chromosomal rearrangements created by an inversion between Adamts20 and the K18N-locus in chromosome 15. (c) Alignment of sequences corresponding to the Adamts20 and the K18N-locus genomic breakpoint junctions. (d) Schematic of an inversion event and the associated FISH signal pattern. Wild-type (WT) shows a separation of the green and red signals. Inversion (Inv) shows the co-localization of the signals. (e) Summary of the experimental efficiency of chromosomal inversions introduced via in vitro electroporation.
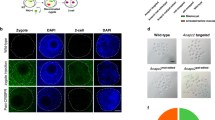
We electroporated CRISPR/Cas9 ribonucleoproteins (RNPs) targeting Adamts20, the K18N-locus, and the ssODNs into in vitro fertilised C57BL/6JJmsSlc zygotes. Embryos were allowed to develop to the two-cell stage, and then they were transferred into pseudopregnant Slc:ICR mice. Six of the 12 Slc:ICR foster mothers bore pups through caesarean section, some of which showed the white-spotted phenotype (Supplementary Fig. 3). We screened for F0 mice using PCR amplification and DNA sequencing of both junction points, and we detected chromosomal inversion in one of the 10 F0 mice (10%) (Fig. 1c,e). However, in one possible inversion mouse line, named #9, only one junction point was detected, similar to what has been previously reported8; this result suggests that one or both of the primer binding sites may have been deleted (Fig. 1c). To confirm the inversion of line #9, we performed fluorescence in situ hybridization (FISH). Whereas the normal chromosome showed discrete green and red signals from the Adamts20 and K18N loci, respectively, overlapping signals indicative of the inverted Adamts20-K18N locus were observed in the chromosome of line #9, suggesting that line #9 contained the 7.67 Mb inversion (Fig. 1d). Previous studies have demonstrated that recombination between the wild-type and chromosomal inversion line does not occur within these inversion events17,18,19 (Supplementary Fig. 4a). To determine whether crossovers were suppressed in mouse line #9, heterozygous #9 (C57BL/6JJmsSlc background) and C57BL/6JJmsSlc females were mated with C3H/HeJYokSlc males, and the F1 heterozygotes were further backcrossed to C3H/HeJYokSlc mice. Among the 18 mice harbouring heterozygous inversions that were examined, there was no recombination detected within this inversion. In contrast, the mice with homozygous wild-type chromosomes did not show these suppressive effects, supporting the idea that inversion-suppressed recombination was present in line #9 (Supplementary Fig. 4b,c). These results demonstrate that our electroporation method introduced a 7.67 Mb chromosomal inversion in the mouse zygotes, which is 1.5 times longer than the inversion produced in a previous study using microinjection-based methods8. Moreover, the efficiency of the inversion produced using electroporation was comparable to that achieved using the microinjection method8,9. However, survival rates were low: only 37.4% (89/238) at the two-cell stage. This may be due to the embryonic lethality of homologous inversion in these loci, although further investigation is needed.
Chromosomal inversion via in vivo electroporation
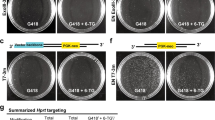
We attempted to generate a chromosomal inversion via a previously developed in vivo electroporation technique called improved genome editing via oviductal nucleic acid delivery (i-GONAD)14,15 (Fig. 2a, Supplementary Fig. 1a,c,d). This method can bypass the following three steps: (1) zygote isolation, (2) microinjection, and (3) zygote transfer14. We attempted to introduce a 4.54 Mb inversion between the Pafah1b1 neighbourhood locus (PN-locus) and the Gm30470 neighbourhood locus (G30470N-locus) in the middle of chromosome 11 (Fig. 2b); we selected this site because the disruption of neither of the two target regions would produce a lethal phenotype. We injected the CRISPR/Cas9 RNPs targeting the PN-locus and G30470N-locus and the ssODNs into the oviduct lumen of five superovulated pregnant C57BL/6NCrSlc females at day 0.7 of pregnancy (corresponding to the late one-cell stage) and electroporated the oviduct in vivo. Only one female became pregnant and two F0 pups were obtained through caesarean section, and one pup had a chromosomal inversion (#11) (Fig. 2c,d), with one complete copy of the ssODN at one breakpoint. However, another breakpoint was unmapped, suggesting that mouse line #11 may have had an inversion with a deletion of the primer binding sites, similar to what was observed in line #9 (Fig. 2c). Whilst the efficiency is low, these results suggest that in vivo electroporation could be used to introduce a megabase-sized chromosomal inversion.
Chromosomal engineering of mouse zygotes via in vivo electroporation. (a) Experimental procedures for induction of chromosomal inversion using in vivo electroporation. (b) Schematic of the chromosomal rearrangements created by an inversion between the PN-locus and G30470-locus in chromosome 11. (c) Alignment of the sequences corresponding to the PN-locus and the G30470-locus genomic breakpoint junctions. (d) Summary of the experimental efficiency of chromosomal inversion via in vivo electroporation.
Allele-specific inversion in F1 hybrid mice via in vivo electroporation
Whereas a previous study of in vivo electroporation genome editing in mouse zygotes reported an average litter size of approximately 514, only two pups were obtained in our experiment.
As the efficiency of the CRISPR/Cas system is very high, isolation of chromosomal rearrangement in mouse zygotes is sometimes difficult using this system because of the frequent deletion of both copies of the target regions, which results in lethality20. Hence, the low efficiency in our in vivo experiments may result in part from the biallelic megabase-sized deletion. To exclude this possibility, we developed a unique in vivo electroporation method wherein one allele was selectively edited in F1 hybrid (B6C3F1) mice (Fig. 3a). Using the Mouse Phenome Database (https://phenome.jax.org/), we selected two SNPs; both existed in the protospacer adjacent motif (PAM) sequences of the C57BL/6NCrSlc genome, but they are absent in the PAM sequences of the C3H/HeJYokeSlc genome. We performed in vivo electroporation on three C57BL/6NCrSlc females that had been mated to C3H/HeJYokSlc males, and we used two sgRNAs that selectively targeted the C57BL/6NCrSlc genome. We obtained 7 F0 pups through caesarean section and successfully obtained one mouse in which an inversion at both junction points was detected (Fig. 3b,c). Additionally, we examined 11 blastocysts from the pregnant female mice three days after the in vivo electroporation procedure and found that one embryo contained the inversion at both break points (Supplementary Fig. 5). To improve the low inversion efficiency, we tried the experiment again with approximately double the concentration of donor ssODN (1000 ng/µL) but could not find any breakpoints in 11 pups (Fig. 3c). In contrast, we reduced the concentration of ssODN to 250 ng/μL and analysed the results at the blastocyst stage. As shown in Supplementary Fig. 5, we detected one breakpoint in two of 16 embryos (12.5%), but we did not detect two breakpoints in any of them. No pups were delivered in the absence of ssODN (Fig. 3c).
Chromosomal engineering of mouse zygotes via targeting the selected allele in F1 hybrid mice using in vivo electroporation. (a) Schematic of the chromosomal rearrangements in F1 hybrid mice. The rearrangements were created via an inversion between the PN-locus and G30470-locus in only the C57BL/6NCrSlc allele. (b) Alignment of sequences corresponding to the PN-locus and G30470-locus genomic breakpoint junctions. (c) Summary of the experimental efficiency of chromosomal inversion via in vivo electroporation.
Together, our results indicate that genome editing via ssODN-dependent in vivo electroporation could elicit chromosomal inversion in mouse embryos, although further improvement of the method is required to increase the efficiency.
Upon closer inspection of the repaired regions in the inversions of thirteen mice (#1, #9, #11, #18, #23, #24, #27, #28, #29, #33, #32, #47, #48) obtained using the ssODNs, we found that only two mice (#11, #48) contained the complete sequence of the ssODN at one breakpoint (Fig. 2c, Supplementary Fig. 5b). Three mice (#1, #18, #32) had indels at both breakpoints (Figs 1c, 3b, Supplementary Fig. 5b), and seven (#9, #23, #24, #27, #28, #29, #30, #47) had indels at one breakpoint (Fig. 1c, Supplementary Fig. 5b). Interestingly, we observed that the upstream inversion junctions in seven mice had deletions near a 3-bp microhomology sequence of ‘CTA’ (#18, #23, #24, #27, #28, #29, #32) (Fig. 3b, Supplementary Fig. 5b). Consistent with a previous report, this observation suggests that a breakpoint flanked by microhomology sequences can generate inversion events via microhomology-mediated repair21. Thus, of the thirteen breakpoints examined, two appeared to be repaired by HDR, whereas eleven were likely to be repaired by microhomology-mediated end joining (MMEJ) or nonhomologous end joining (NHEJ).
Allele-specific recessive lethal deletion in F1 hybrid mice via in vivo electroporation
Next, we examined whether this strategy could detect a recessive lethal deletion (Fig. 4a). We attempted to delete an essential gene, Rad51, the loss of which in both alleles seemed to produce a lethal embryonic phenotype22,23. We electroporated the genome editing CRISPR/Cas9 mixture into the oviduct of C57BL/6NCrSlc females that had been mated to C3H/HeJYokSlc males, and we used C57BL/6NCrSlc females mated to C57BL/6NCrSlc males as a control. In the C57BL/6NCrSlc/C3H/HeJYokSlc F1 hybrid strains, we obtained two F0 pups and found that one of them had a large deletion in the target locus (Fig. 4b,c). It should be noted that the control C57BL/6NCrSlc/C57BL/6NCrSlc strain failed to deliver their pups, suggesting that these embryos died because of the deletion of both Rad51 gene copies (Fig. 4c). In fact, the deletion line #20 generated exhibited a recessive lethal embryonic phenotype (Supplementary Fig. 6). Together, our results suggest that this unique method may be successfully applied for the generation of target-specific chromosomal rearrangements, such as large-scale inversions and recessive lethal deletions.
Lethal gene deletion in mouse zygotes via targeting the selected allele in F1 hybrid mice using in vivo electroporation. (a) Schematic of Rad51 gene deletion in F1 hybrid mice, which was generated in only the C57BL/6NCrSlc allele. (b) Alignment of sequences corresponding to the Rad51 intron 1 and intron 2 genomic breakpoint junctions. (c) Summary of the experimental efficiency of chromosomal deletion via in vivo electroporation.
Conclusions
To our knowledge, this is the first study to successfully introduce chromosomal inversions in mouse zygotes using electroporation-based methods. Moreover, in vivo allele-specific genome editing in F1 hybrid mice has allowed us to efficiently introduce chromosomal inversions and recessive lethal deletions. Together, our methods provide a simple and efficient approach for engineering chromosomal rearrangements and contribute to the study of disease caused by chromosomal rearrangements.
Methods
Animal strains
C57BL/6JJmsSlc, C57BL/6NCrSlc, C3H/HeJYokSlc, and Slc:ICR mice (8–12 weeks old) were obtained from Japan SLC (Shizuoka, Japan). The animals were kept at a constant temperature (22 °C ± 2 °C) and humidity (50% ± 10%), with a 12 h light/12 h dark cycle. All animal experiments were approved by the Institutional Animal Care and Use Committee of Chubu University (Permit Numbers #2910066, #2910067 at Chubu University) and were conducted in accordance with institutional guidelines.
CRISPR RNP and ssODN preparation
The CRISPR guide RNAs were designed using CHOPCHOP (http://chopchop.cbu.uib.no/)24 (Supplementary Table 1). To avoid possible off-target events, we selected gRNAs that are as specific for the target sequence as possible. No mutations were detected using the T7 endonuclease I (T7E1) (New England BioLabs, MA, USA) assay at four off-target sites that differed by one or two base-pairs from the on-target sites (Supplementary Fig. 7). The T7E1 assay was performed as previously described25. The CRISPR RNP consists of Alt-R S.p. Cas9 Nuclease 3NLS (Integrated DNA Technologies, IL, USA) and a crRNA:tracrRNA duplex, which included the custom guide RNA (crRNA) and a universal structural RNA (tracrRNA) (Integrated DNA Technologies). The crRNA and tracrRNA were heated to 95 °C for 5 min and slowly cooled to room temperature. This crRNA:tracrRNA duplex and the Alt-R S.p. Cas9 Nuclease 3NLS were incubated at room temperature for 10 min to form the RNP complex.
The ssODNs were manufactured by Eurofins Genomics (Tokyo, Japan) and were designed to join two DNA sequences so that the junction would be positioned at the centre of the predicted cleavage sites, which were located within 3 bp of the PAM sequences (Supplementary Table 2).
In vitro electroporation
C57BL/6JJmsSlc female mice were injected intraperitoneally with 7.5 IU PMSG (ASKA Animal Health., Tokyo, Japan), followed by 7.5 IU of hCG (ASKA Animal Health., Tokyo, Japan) 48 h later. Thirteen hours after the hCG injection, superovulated female mice were euthanised via cervical dislocation, and unfertilised oocytes isolated from the female mice were subjected to in vitro fertilisation (IVF) with freshly isolated spermatozoa from euthanised C57BL/6JJmsSlc male mice, as previously described26. To generate chromosomes with an inversion, the following concentrations of CRISPR reagents were used: 200 ng/μL Cas9 protein, 100 ng/μL upstream crRNA/tracrRNA, 100 ng/μL downstream crRNA/tracrRNA, 100 ng/μL upstream ssODN and 100 ng/μL downstream ssODN diluted in Opti-MEM (Thermo Fisher Scientific, MA, USA). The in vitro electroporation procedures were performed as previously described10. Briefly, the embryos were cultured in KSOM medium, washed with Opti-MEM, and then placed in an electrode cuvette (CUY505P5 [NEPA GENE., Chiba, Japan]) with CRISPR solutions (47 μL total volume), followed by electroporation using a NEPA21 (NEPA GENE). The following parameters were used for electroporation: poring pulse (voltage: 225 V; pulse length: 2.0 msec; pulse interval: 50 msec; number of pulses: 4; decay rate: 40%; polarity: +), transfer pulse (voltage: 20 V; pulse length: 50 msec; pulse interval: 50 msec; number of pulses: 5; decay rate: 40%; polarity: ±). After electroporation, the embryos were cultured to the two-cell stage in KSOM medium (Kyudo, Saga, Japan). Before embryo transfer, pseudopregnant Slc:ICR mice were anaesthetised with a mixture of three drugs: medetomidine (0.75 mg/kg), midazolam (4 mg/kg), and butorphanol (5 mg/kg). Following the embryo transfer, we placed the oviducts back in their original location and sutured the incisions. After the operation, atipamezole hydrochloride (0.3 mg/kg) was intraperitoneally injected to reverse the effects of the medetomidine.
In vivo electroporation
For superovulation, C57BL/6NCrSlc female mice were injected intraperitoneally with 2 IU PMSG, and then they were injected with 5 IU hCG 48 h later, as previously described27. They were subsequently mated to C57BL/6NCrSlc or C3H/HeJYokSlc males. The presence of copulation plugs was confirmed via visual inspection the next morning, and plug-positive mice were subjected to in vivo electroporation experiments. To generate chromosomes with an inversion, the following concentrations of CRISPR solutions were used: 540 ng/μL Cas9 protein, 33 μM upstream and downstream crRNA/tracrRNA, 250, 495 or 1000 ng/μL upstream and downstream ssODN, and 0.05% Fast Green FCF (Wako) marker diluted in Opti-MEM (Thermo Fisher Scientific). To generate a chromosomal deletion, the following concentrations of CRISPR solutions were used: 500 ng/μL Cas9 protein, 30 μM upstream crRNA/tracrRNA, and 30 μM downstream crRNA/tracrRNA diluted in Opti-MEM (Thermo Fisher Scientific). Before in vivo electroporation, the females were anaesthetised with a mixture of three drugs: medetomidine (0.75 mg/kg), midazolam (4 mg/kg) and butorphanol (5 mg/kg). The CRISPR mixture (1 μL) was injected into the oviductal lumen upstream of the ampulla with a glass micropipette, which was made using a vertical capillary puller (NARISHIGE, Tokyo, Japan). After injecting CRISPR solutions, the oviduct regions were grasped by tweezer electrodes (CUY652P2.5 × 4 [NEPA GENE]), and electroporation was performed as previously described14 using a NEPA21 (NEPA GENE). The following parameters were used for electroporation: poring pulse (voltage: 50 V; pulse length: 5.0 msec; pulse interval: 50 msec; number of pulses: 3; decay rate: 10%; polarity: ±), transfer pulse (voltage: 10 V; pulse length: 50 msec; pulse interval: 50 msec; number of pulses: 3; decay rate: 40%; polarity: ±). After electroporation, we placed the oviducts back in their original location and sutured the incisions. After the operation, atipamezole hydrochloride (0.75 mg/kg) was intraperitoneally injected to reverse the effects of medetomidine.
Analysis of the chromosome-engineered mice
To screen for chromosomal rearrangements, genomic DNA was isolated from the tails or ears of founder mice using lysis buffer (100 mM NaCl, 200 mM sucrose, 10 mM EDTA, 300 mM Tris (pH 8.0), and 1% SDS), and the DNA was examined via PCR amplification (Supplementary Figs 6a, 8 and Supplementary Table 3). PCR products were cloned into the pTAC-1 vector (Biodynamics, Tokyo, Japan), and sequences of individual clones were determined via Sanger sequencing (Eurofins Genomics).
DNA extraction from mouse blastocysts
Crude DNA derived from each blastocyst was prepared to be a PCR template according to the method described28. Briefly, we euthanised electroporated females on embryonic day 3, flushed the uterine horn with 0.5 mL of M2 medium, collected blastocysts in a petri dish, and transferred a single blastocyst to a 0.2 ml PCR tube using a glass micropipette. Then, 10 μL of lysis buffer (50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.1 mg/mL gelatin, 0.45% NP40, 0.45% Tween 20 and 125 μg/mL proteinase K) was added directly to each tube, and the samples were incubated at 56 °C for 30 min and then at 95 °C for 10 min. The target genomic regions were amplified by nested PCR with the primers listed in Supplementary Table 3.
Cytogenetic analysis
Chromosomal samples were prepared from bone marrow cells of mouse line #9. The mice were injected intraperitoneally with 0.1 mg/μL colcemid and euthanised approximately 60 min afterwards. Chromosome isolation and FISH were performed as described with modification29,30. Each DNA probe, Adamts20 (15qE3), K18N (15qF2), and G41405N (15qF2), was amplified from the genomic DNA of a C57BL/6NCrSlc mouse by PCR (Supplementary Table 3). Adamts20 (15qE3) was labelled with Biotin-16-dUTP (Roche, Mannheim, Germany), and K18N (15qF2) and G41405N (15qF2) were labelled with Digoxigenin-11-dUTP (Roche), respectively. DNA probes were detected with Streptavidin Alexa Fluor® 488 conjugate (Thermo Fisher Scientific) or anti-digoxigenin-Rhodamine (Roche). Finally, the chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Roche).
Test for recombination suppression
To examine whether recombination was suppressed in mouse line #9, heterozygous #9 (C57BL/6JJmsSlc background) females and C57BL/6JJmsSlc females were mated with C3H/HeJYokSlc males, and the F1 heterozygotes were further backcrossed to C3H/HeJYokSlc mice. The Arf3 mutation (rs245124066) genotype of each individual was determined using PCR-restriction fragment length polymorphism analysis. Following PCR amplification, the PCR products were digested for 4 h at 37 °C with 5 units of StuI restriction enzyme, and they were then analysed via 1.2% agarose gel electrophoresis. Digestion of the PCR products with StuI yielded fragments of 269 and 146 bp in the C57BL/6JJmsSlc mice (Arf3 (+/+)) vs. a single product of 415 bp in the C3H/HeJYokSlc mice (Arf3 (−/−)). An Arf3 (+/−) heterozygote yielded bands of 415 bp, 269 bp and 146 bp.
References
Collins, R. L. et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 18(1), 36 (2017).
Xia, L. C. et al. Identification of large rearrangements in cancer genomes with barcode linked reads. Nucleic Acids Res 46(4), e19 (2017).
Yu, Y. & Bradley, A. Engineering chromosomal rearrangements in mice. Nat Rev Genet 2(10), 780 (2001).
Brault, V., Pereira, P., Duchon, A. & Hérault, Y. Modeling chromosomes in mouse to explore the function of genes, genomic disorders, and chromosomal organization. PLoS Genet 2(7), e86 (2006).
Walrath, J. C., Hawes, J. J., Van Dyke, T. & Reilly, K. M. Genetically engineered mouse models in cancer research. Adv Cancer Res 106, 113–164 (2010).
Cheon, D. J. & Orsulic, S. Mouse models of cancer. Annu Rev Pathol Mech 6, 95–119 (2011).
Sander, J. D. & Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32(4), 347 (2014).
Kato, T. et al. Creation of mutant mice with megabase-sized deletions containing custom-designed breakpoints by means of the CRISPR/Cas9 system. Sci Rep 7(1), 59 (2017).
Birling, M. C. et al. Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Sci Rep 7, 43331 (2017).
Kaneko, T., Sakuma, T., Yamamoto, T. & Mashimo, T. Simple knockout by electroporation of engineered endonucleases into intact rat embryos. Sci Rep 4, 6382 (2014).
Qin, W. et al. Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics 200, 423–430 (2015).
Hashimoto, M. & Takemoto, T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci Rep 5, 11315 (2015).
Chen, S. et al. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. Journal of Biological Chemistry M 116, 733154 (2016).
Ohtsuka, M. et al. i-GONAD: a robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol 19(1), 25 (2018).
Kobayashi, T. et al. Successful production of genome-edited rats by the rGONAD method. BMC Biotechnol 18(1), 19 (2018).
Silver, D. et al. The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet 4(2), e1000003 (2008).
Zheng, B. et al. Engineering a mouse balancer chromosome. Nature Genet 22(4), 375 (1999).
Nishijima, I. et al. Two new balancer chromosomes on mouse chromosome 4 to facilitate functional annotation of human chromosome 1p. Genesis 36(3), 142–148 (2003).
Ye, Z. et al. Generation of a Mouse Full-length Balancer with Versatile Cassette-shuttling Selection Strategy. Int J Biol Dci 12(8), 911 (2016).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153(4), 910–918 (2013).
Li, J. et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol 7(4), 284–298 (2015).
Tsuzuki, T. et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. PNAS 93(13), 6236–6240 (1996).
Lim, D. S. & Hasty, P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol 16(12), 7133–7143 (1996).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47(1), 171–174 (2019).
Jacobi, A. et al. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods 121, 16–28 (2017).
Takeo, T. & Nakagata, N. In vitro fertilization in mice. Cold Spring Harb Protoc 2018(6), pdb-prot094524 (2018).
Sato, M., Ohtsuka, M. & Nakamura, S. Intraoviductal instillation of a solution as an effective route for manipulating preimplantation mammalian embryos in vivo. In New Insights into Theriogenology IntechOpen, https://doi.org/10.5772/intechopen.79106 (2018).
Scavizzi, F. et al. Blastocyst genotyping for quality control of mouse mutant archives: an ethical and economical approach. Transgenic Res 24(5), 921–927 (2015).
Fukushi, D. et al. Molecular cytogenetic analysis of the highly repetitive DNA in the genome of Apodemus argenteus, with comments on the phylogenetic relationships in the genus Apodemus. Cytogenet Cell Genet 92(3-4), 254–263 (2001).
Fukushi, D. et al. Scanning Near-field Optical/Atomic Force Microscopy detection of fluorescence in situ hybridization signals beyond the optical limit. Exp Cell Res 289(2), 237–244 (2003).
Acknowledgements
We are grateful to Dr. Masato Ohtsuka (Tokai Univ.), Dr. Hiromi Miura (Tokai Univ.) and Dr. Masahiro Sato (Kagoshima Univ.) for teaching the in vivo electroporation, and Mr. Yasuhiko Hayakawa (NEPAGENE) for teaching the in vitro electroporation. We also thank our laboratory members for support with animal care and experiments. This study was supported by Grant-in-Aid for Early Career Scientists (18K14616) to S.I. from Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Contributions
S.I. designed and performed the experiments and drafted the manuscript. H.N., M.J. and M.N. performed the animal experiments. D.F. carried out FISH analysis. T.I. supervised the studies and corrected the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Iwata, S., Nakadai, H., Fukushi, D. et al. Simple and large-scale chromosomal engineering of mouse zygotes via in vitro and in vivo electroporation. Sci Rep 9, 14713 (2019). https://doi.org/10.1038/s41598-019-50900-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-50900-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.