Abstract
Adult neurogenesis occurs in the dentate gyrus of the hippocampus during adulthood and contributes to sustaining the hippocampal formation. To investigate whether neurogenesis-related pathways are associated with hippocampal volume, we performed gene-set enrichment analysis using summary statistics from a large-scale genome-wide association study (N = 13,163) of hippocampal volume from the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) Consortium and two year hippocampal volume changes from baseline in cognitively normal individuals from Alzheimer’s Disease Neuroimaging Initiative Cohort (ADNI). Gene-set enrichment analysis of hippocampal volume identified 44 significantly enriched biological pathways (FDR corrected p-value < 0.05), of which 38 pathways were related to neurogenesis-related processes including neurogenesis, generation of new neurons, neuronal development, and neuronal migration and differentiation. For genes highly represented in the significantly enriched neurogenesis-related pathways, gene-based association analysis identified TESC, ACVR1, MSRB3, and DPP4 as significantly associated with hippocampal volume. Furthermore, co-expression network-based functional analysis of gene expression data in the hippocampal subfields, CA1 and CA3, from 32 normal controls showed that distinct co-expression modules were mostly enriched in neurogenesis related pathways. Our results suggest that neurogenesis-related pathways may be enriched for hippocampal volume and that hippocampal volume may serve as a potential phenotype for the investigation of human adult neurogenesis.
Similar content being viewed by others
Introduction
Neurons are generated from neural stem cells in two regions of the brain, the dentate gyrus of the hippocampus and the olfactory bulb throughout the life span. Dentate gyrus (DG) neurons are incorporated into the hippocampal network. Adult neurogenesis-related pathways include signaling transduction, epigenetic regulation, immune system, proliferation of progenitor cells and differentiation, migration, and maturation of adult neurons1,2,3. Adult neurogenesis in DG of the hippocampus is regulated by multiple intrinsic and extrinsic factors such as hormones, transcription factors, cell cycle regulators and environmental factors that control neural stem cell (NSC) proliferation, maintenance, and differentiation into mature neurons. The estimated annualized hippocampal atrophy rate is 1.41% for cognitively normal older adults and in adults, new neurons are added in each hippocampus daily via adult neurogenesis with an annual turnover of 1.75% and a modest decline during aging4,5. Combination of structural MRI and immunohistological markers for newborn neurons and neural stem/progenitor cells in neurogenesis-related brain regions in mice revealed that neurogenesis is associated with increased hippocampal gray matter volumes in mice6,7. There is hippocampal atrophy and reduction of hippocampal neurogenesis in adult rats exposed to oxygen deprivation during birth8. Recently, it has been found that cognitively normal individuals had preserved neurogenesis compared to less angiogenesis and neuroplasticity9. Environmental factors enhance transcriptional and epigenetic changes between ventral and dorsal part of the dentate gyrus that may have an effect on hippocampal volume10. Molecular pathways and genes affect the induction of neurogenic niche and neural/progenitor cell turnover to newborn neurons for the formation of the hippocampal structure during hippocampal neurogenesis.
To our knowledge, there is no study assessing the association of adult neurogenesis related pathways with hippocampal volume measured from MRI scans in living people. In this study, in order to investigate whether genetic variants associated with variation in hippocampal volume are enriched for neurogenesis-related pathways, we performed a gene set enrichment analysis using summary statistics from a large-scale human neuroimaging genetics meta-analysis from the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) Consortium (N~13,000). Neurogenesis is an important contributor to the formation of the hippocampus in mice but less is known about the relationship between human adult neurogenesis and hippocampal volume/atrophy.
Materials and Method
Enhancing neuro imaging genetics through meta-analysis (ENIGMA)
The Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) Consortium was initiated in December 2009. The research group involved in neuroimaging and genetics worked together on a range of large-scale studies that integrated data from 70 institutions worldwide. The goal of ENIGMA was to merge neuroimaging data with genomic data to identify common genetic variants that might affect brain structure. The first project of ENIGMA focused on identifying common genetic variants associated with hippocampal volume or intracranial volume (ICV)11. The aim of ENIGMA2, follow-on study of ENIGMA1, was to perform genome-wide association study (GWAS) using subcortical volumes as phenotypes12. In ENIGMA2, GWAS was conducted using mean hippocampal volume as a phenotype controlling for age, age2, sex, ancestry (the first four multidimensional scaling components), ICV, and diagnostic status, and MRI scanner (when multiple scanners were used at the same site), and genetic imputation were processed and examined by following standardized protocols freely available online (http://enigma.ini.usc.edu/protocols/imaging-protocols/). In this study, we used GWAS summary statistics in the discovery sample of 13,163 subjects of European ancestry from the ENIGMA Consortium12. 3,824 of the 13,163 participants (21%) have anxiety, Alzheimer’s disease, attention-deficit/hyperactivity disorder, bipolar disorder, epilepsy, major depressive disorder or schizophrenia, and the remaining 9,339 (79%) are cognitively normal subjects.
Alzheimer’s disease neuroimaging initiative (ADNI)
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD, and recruited from 59 sites across the U.S. and Canada. ADNI includes over 1700 subjects consisting of cognitively normal older individuals (CN), significant memory concern (SMC), mild cognitive impairment (MCI) and Alzheimer’s Disease (AD) aged 55–90 (http://www.adni-info.org/). The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Participants for this study included 367 CN, 94 SMC, 280 early MCI, 512 late MCI and 310 AD. Demographic information, APOE, clinical information, neuroimaging and GWAS genotyping data were downloaded from the ADNI data repository (http://adni.loni.usc.edu). The CN group does not have any significant memory concern or impairment of their daily activities. The SMC group has self-reported significant memory concerns quantified using the Cognitive Change Index13 and the Clinical Dementia Rating (CDR) of zero. Individuals with MCI and AD have to have memory complains. The range of Mini-Mental State Examination (MMSE) score was 24–30 for CN and MCI, and 20–26 for AD as well as objective memory loss measured by education-adjusted scores on Wechsler Memory Scale-Revised (WMS-R) Logical Memory II14. As diagnosis criteria, CDR score was used as 0 for CN, 0.5 for MCI with the memory box score being 0.5 or greater, and 0.5–1 for AD15. A composite memory score was calculated using Logical Memory and the Rey Auditory Verbal Learning Test (RAVLT), as well as memory items from the AD Assessment Scale - Cognitive (ADAS-Cog) and Mini-Mental State Examination (MMSE)16. Hippocampal volume was determined using MRI scans and FreeSurfer version 5.1 was used to extract hippocampal and total intracranial volumes (ICV)17,18,19,20. Table 1 shows selected demographic and clinical characteristics of these participants at baseline.
Genotyping data and quality control
The genotyping data of ADNI participants were collected using the Illumina Human 610-Quad, HumanOmni Express, and HumanOmni 2.5 M BeadChips. Standard quality control procedures of GWAS data for genetic markers and subjects were performed using PLINK v1.07 (pngu.mgh.harvard.edu/∼purcell/plink). Quality control procedures included excluding samples and SNPs with criteria including SNP call rate < 95%, Hardy-Weinberg equilibrium test p < 1 × 10−6, and frequency filtering (MAF < 5%), participant call rate < 95%, sex check and identity check for related individuals21,22,23,24,25. Non-Hispanic Caucasian participants were selected using HapMap 3 genotype data and the multidimensional scaling (MDS) analysis (Supplementary Fig. 1) after performing standard quality control procedures for genetic markers and subjects. For imputation of un-genotyped SNPs, MaCH (Markov Chain Haplotyping) software based on the 1000 Genomes Project as a reference panel was used26,27.
Gene-set enrichment analysis
Gene-set enrichment analysis using GWAS summary statistics was performed to identify pathways and functional gene sets with significant associations with hippocampal volume. All SNPs (n = 6,571,356) and subjects with European ancestry were included in this study. Pathway annotations were downloaded from the Molecular Signatures Database version 5.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp/). This annotation data comprised a collection of Gene Ontology (GO). GO includes 1,454 pathways and is publicly available. 825 gene sets are assigned to GO biological processes, 233 gene sets are assigned to GO cellular components, and 396 gene sets are assigned to GO molecular functions. GSA-SNP software28 uses a p-value of each SNP from GWAS summary statistics to test if a pathway-phenotype association is significantly different from all other pathway-phenotype associations. In GSA-SNP, all SNPs within each gene are considered in turn and the negative log of the p value is noted; all of these are ranked. To avoid spurious predictions, we used the SNP with the second highest negative log p value to summarize strength of association with each gene. Each pathway (gene set) was assessed by z-statistics for the identification of the enriched pathways29. Gene-set enrichment analysis was restricted to pathways containing between 10 and 200 genes. False discovery rate (FDR) with the Benjamini-Hochberg procedure was used for multiple comparison correction30. We identified as significantly enriched pathways with hippocampal volume with FDR-corrected p-value < 0.05.
Genetic association analysis
Genome-wide gene-based association analysis using GWAS p-values was performed using KGG (Knowledge-based mining system for Genome-wide Genetic studies) software. KGG uses HYST (hybrid set-based test) to determine the overall association significance in a set of SNPs at the gene level. HYST is the combination of the gene-based association test using extended Simes procedure (GATES) and the scaled chi-square test31,32. First, SNPs in each gene were divided into different LD blocks depending on pairwise LD coefficients (r2) for all SNPs. Second, for each block, a block-based p-value for association was calculated, and the key SNP was derived and marked. Next, the block-based p-values were combined accounting for LD between the key SNPs using the scaled chi-square33.
Targeted gene-based association analysis was performed using a set-based test in Plink v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/)22. SNPs with p < 0.05 for each gene were chosen. A mean test statistic for each SNP within a gene was computed to determine with which other SNPs it is in linkage disequilibrium (LD); i.e., if the correlation coefficient between them was r2 > 0.5. A quantitative trait analysis (QT) was then performed with each SNP. For each gene, the top independent SNPs (i.e., not in LD; maximum of 5) are selected if their p-values are less than 0.05. The SNP with the smallest p-value is selected first; subsequent independent SNPs are selected in order of decreasing statistical significance. From these subsets of SNPs, the statistic for each gene is calculated as the mean of these single SNP statistics34. The analysis was performed using an additive model or in other words, the additive effect of the minor allele on the phenotypic mean was estimated22,35. Covariates included age, sex, years of education, and diagnosis for composite scores for memory. An empirical p-value (20,000 permutations) was reported for each gene for multiple comparison adjustment22.
Gene expression correlation analysis
We analyzed gene expression data in the hippocampal subfields, CA1 and CA3, from 32 normal controls brain samples in the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) archives. The Illumina HumanHT-12 v3 Expression BeadChip (48,803 probes) was used to measure expression of over 25,000 annotated genes. We processed gene expression data and removed the outliers as previously described36. We excluded probes if they were present in three or fewer samples or if they do not correspond to any gene symbol annotations. Lastly we removed duplicate probes for a gene and kept only the probe with the highest expression level. After all data cleaning process, 15,037 genes remained. We performed a weighted gene correlation network analysis (WGCNA) using processed expression data to identify clusters of highly correlated genes expressed in specific brain regions (CA1 and CA3) as modules. Pearson correlations between gene pairs were calculated. This matrix was transformed into a signed adjacency matrix by using a power function. Then, topological overlap (TO) was calculated by using the components of this matrix. Genes were clustered hierarchically by the distance measure, 1-TO, and the dynamic tree algorithm determined initial module assignments37. Gene module membership between each gene and each module eigengene was calculated. We tested these modules for enrichment of neurogenesis-related pathways.
Results
Gene-set enrichment analysis using large-scale GWAS summary statistics for hippocampal volume (N = 13,163) identified 44 significantly enriched biological pathways (FDR-corrected p-value < 0.05) (Table 2) including 38 pathways related to neurogenesis (Supplementary Table S1). We classified the 38 neurogenesis-related pathways as primary (N = 19) and secondary (helper) (N = 19) based on existing knowledge and literature mining (Fig. 1). The primary neurogenesis-related pathways were related to cellular processes such as neuronal proliferation, differentiation and survival, cellular morphogenesis, axonogenesis, neuronal development, signal transduction, and cell-cell adhesion. The secondary neurogenesis-related pathways consisted of enzyme activities related to neurogenesis, metabotropic receptor activity, lipoprotein binding and extracellular matrix. Six pathways were not related to any neurogenesis-related process such as oxidoreductase activity, phagocytosis, perinuclear region of cytoplasm and cornified envelope.
Since the inhibition of neurogenesis could be relevant to hippocampal atrophy38, we also examined if neurogenesis-related pathways were enriched with hippocampal atrophy over two years from baseline in cognitively normal individuals without amyloid-β pathology based on [18F]Florbetapir PET or CSF amyloid-β measurement (N = 112) in ADNI. Seven pathways related to neurogenesis processes were significantly enriched with hippocampal atrophy (FDR-corrected p-value < 0.05) in cognitively normal adults (Supplementary Table S2). These pathways were related to cellular differentiation, cellular morphogenesis during development, neurite development, axonogenesis, cell-cell adhesion and neuron development (Table 3).
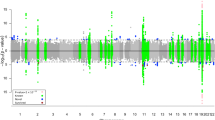
Furthermore, we performed targeted gene-based association analysis of hippocampal neurogenesis related pathway associated candidate genes using ENIGMA GWAS summary statistics31. The gene-based analysis revealed that 4 genes (MSRB3, TESC, DPP4, and ACVR1) were significantly associated with hippocampal volume (corrected p-value < 0.05; Table 4). Since hippocampal volume is correlated with memory performance, we performed an association analysis of these four genes (with 682 SNPs) with composite memory scores in ADNI. The gene-based association analysis showed that TESC is significantly associated with composite memory scores after adjusting for multiple testing (p-value = 5.7 × 10−3; Table 5). One novel SNP (rs117692586) upstream of TESC was significantly associated with composite memory scores (p-value = 4.3 × 10−4; Table 6). rs117692586-T is associated with poorer memory performance (Fig. 2).
Finally, we analyzed gene expression data in the Gene Expression Omnibus (GEO) repository to investigate if neurogenesis-related pathways were enriched in the CA1 and CA3 regions of the hippocampus in normal controls. A weighted gene correlation network analysis yielded 20 modules of co-expressed genes. These 20 modules were tested for enrichment of neurogenesis-related pathways. Six modules were found to be significantly enriched with neurogenesis-related pathways after adjusting for multiple testing. The six significantly enriched modules are all related to neurogenesis-related pathways such as neuronal proliferation and differentiation as well as cellular process (Table 7).
Discussion
Using large-scale GWAS summary statistics for hippocampal volume in 13,163 subjects of European ancestry from the ENIGMA Consortium, we performed gene-set enrichment analysis to identify 44 pathways with enrichment for hippocampal volume. These enriched pathways showed that genes associated with variation in hippocampal volume are related to neurogenesis and cellular processes including neuronal cell proliferation, differentiation and maturation as well as cell adhesion. In addition, co-expression network-based functional analysis of gene expression data in the hippocampal subfields, CA1 and CA3, from 32 normal controls showed that co-expression modules were mostly enriched in neurogenesis-related pathways.
The enriched pathways showed significant relationships between neurogenesis and hippocampal volume/atrophy. Since several studies showed neurogenesis occurs in the dentate gyrus of the hippocampus4,39, it is not surprising that hippocampal volume is significantly related to neurogenesis-related pathways. In particular, we observed significant enrichment of pathways related to cell proliferation, neuron differentiation, neuron generation, neurite development, neuronal development, cell recognition, neurogenesis and axonogenesis. The neural progenitor cells in the subgranular zone of the hippocampus differentiate and incorporate into neural network circuitry as mature neurons in the adult human brain4. In addition, these newly developed neurons enhance the formation of the hippocampus during neurogenesis and many genes are involved in these processes40,41. Moreover, our pathway enrichment analysis found that hippocampal volume is significantly related to signal transduction processes such as glutamate signaling, protein kinase signaling, and the Jun N-Terminal Kinase (JNK) cascade. Previously we identified five neurogenesis related pathways and the signal transduction pathway was one of the important pathways in adult neurogenesis processes3. During adult neurogenesis, functional granule cells in the dentate gyrus of the adult hippocampus release glutamate, project to target cells in the CA3 region, and receive glutamatergic and γ-aminobutyric acid (GABA)-ergic inputs to control their spiking activity in neuronal networks that support the formation of memory and learning42,43. Phosphoinositide 3-kinase (PI3K)/protein kinase pathways enhance neuronal differentiation and inhibit apoptosis of progenitor cells44,45. In addition, studies showed that JNK1 in the JNK cascade plays a role in neuronal differentiation and neuronal and axonal maturation46,47,48. Also, it has been shown that absence of JNK1 enhances hippocampal neurogenesis and reduces anxiety-related phenotypes in mouse models46.
Pathways related to enzyme activities such as protein tyrosine kinases, protein tyrosine phosphatases and 3’5’ cyclic nucleotide phosphodiesterases were enriched for hippocampal volume. Studies showed that three subfamilies, Tyro3, Axl and Mertk (TAM), of receptor protein tyrosine kinases play a crucial role in adult neurogenesis. TAM receptors impact proliferation and differentiation of neural stem cells to immature neurons by controlling overproduction of pro-inflammatory cytokines49. Protein tyrosine phosphatases control neural stem cell differentiation during neurogenesis50.
Our results revealed the influence of neurogenesis pathway-related genetic variation on hippocampal volume. Particularly, two genes, tescalcin (TESC) and activin receptor 1 (ACVR1), were significantly associated with hippocampal volume. In addition, TESC was significantly associated with memory performance. Previous structural neuroimaging studies showed TESC-regulating polymorphisms are significantly associated with hippocampal volume and hippocampal gray matter structure11,51. TESC cooperates with the plasma membrane Na(+)/H(+) exchanger NHE1 that catalyzes electroneutral influx of extracellular Na(+) and efflux of intracellular H(+) and establishes intracellular pH level as well as cellular hemostasis52,53. TESC was expressed in tissues such as heart and brain and plays an important role during embryonic development53. TESC plays a crucial role in controlling cell proliferation and differentiation for the formation of the hippocampal structure during brain development51. In addition, ACVR1, a member of a protein family called bone morphogenetic protein (BMP) type I receptors, regulates the hippocampal dentate gyrus stem cells during neurogenesis54. In addition, our gene co-expression analysis showed that TESC and ACVR1 were co-expressed together in the neurogenesis pathway-related module.
A limitation of the present report is that we used Gene Ontology pathways from MSigDB. For a pathway enrichment analysis design, there is no gold standard. There are several tools and strategies for pathway enrichment analysis, and alternate databases and algorithms for pathway enrichment analysis can affect the analytic results55,56. Another limitation is the lack of replication in the gene-set enrichment analysis, even though we used a large-scale GWAS result (N = 13,163). Replication in independent samples will be important. It is noteworthy that recently, Sorrell et al. reported that human hippocampal neurogenesis drops sharply in childhood to undetectable levels in adults, although some aspects are still under controversy57,58, but Boldrini et al. reported that healthy older adults display preserved neurogenesis9.
In summary, our results suggest that neurogenesis-related pathways may be enriched for hippocampal volume and that hippocampal volume may serve as a potential phenotype for the investigation of human adult neurogenesis. Genetic variation in neurogenesis pathway-related genes may have compensatory advantages or confer vulnerability to biological processes during adult neurogenesis but studies are needed to identify mechanisms by which genetic variants affect neural stem cells differentiation, proliferation, and their maturation to new neurons in human brain.
Data Availability
The data analyzed in the study are available from the ADNI website (http://adni.loni.usc.edu/) and the ENIGMA website (http://enigma.ini.usc.edu/).
References
Suh, H., Deng, W. & Gage, F. H. Signaling in adult neurogenesis. Annual review of cell and developmental biology 25, 253–275, https://doi.org/10.1146/annurev.cellbio.042308.113256 (2009).
Zhao, C., Deng, W. & Gage, F. H. Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660, https://doi.org/10.1016/j.cell.2008.01.033 (2008).
Horgusluoglu, E., Nudelman, K., Nho, K. & Saykin, A. J. Adult neurogenesis and neurodegenerative diseases: A systems biology perspective. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics 174, 93–112, https://doi.org/10.1002/ajmg.b.32429 (2017).
Spalding, K. L. et al. Dynamics of hippocampal neurogenesis in adult humans. Cell 153, 1219–1227, https://doi.org/10.1016/j.cell.2013.05.002 (2013).
Risacher, S. L. et al. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Current Alzheimer research 6, 347–361 (2009).
Biedermann, S. V. et al. The hippocampus and exercise: histological correlates of MR-detected volume changes. Brain Struct Funct 221, 1353–1363, https://doi.org/10.1007/s00429-014-0976-5 (2016).
Sierra, A., Encinas, J. M. & Maletic-Savatic, M. Adult human neurogenesis: from microscopy to magnetic resonance imaging. Frontiers in neuroscience 5, 47, https://doi.org/10.3389/fnins.2011.00047 (2011).
Takada, S. H. et al. Impact of neonatal anoxia on adult rat hippocampal volume, neurogenesis and behavior. Behavioural brain research 296, 331–338, https://doi.org/10.1016/j.bbr.2015.08.039 (2016).
Boldrini, M. et al. Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell 22, 589–599 e585, https://doi.org/10.1016/j.stem.2018.03.015 (2018).
Zhang, T. Y. et al. Environmental enrichment increases transcriptional and epigenetic differentiation between mouse dorsal and ventral dentate gyrus. Nat Commun 9, 298, https://doi.org/10.1038/s41467-017-02748-x (2018).
Stein, J. L. et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nature genetics 44, 552–561, https://doi.org/10.1038/ng.2250 (2012).
Thompson, P. M. et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 8, 153–182, https://doi.org/10.1007/s11682-013-9269-5 (2014).
Rattanabannakit, C. et al. The Cognitive Change Index as a Measure of Self and Informant Perception of Cognitive Decline: Relation to Neuropsychological Tests. J Alzheimers Dis 51, 1145–1155, https://doi.org/10.3233/JAD-150729 (2016).
Kovacevic, S., Rafii, M. S., Brewer, J. B. & Alzheimer’s Disease Neuroimaging, I. High-throughput, fully automated volumetry for prediction of MMSE and CDR decline in mild cognitive impairment. Alzheimer Dis Assoc Disord 23, 139–145, https://doi.org/10.1097/WAD.0b013e318192e745 (2009).
Petersen, R. C. et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 74, 201–209, https://doi.org/10.1212/WNL.0b013e3181cb3e25 (2010).
Crane, P. K. et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Brain Imaging Behav 6, 502–516, https://doi.org/10.1007/s11682-012-9186-z (2012).
Risacher, S. L. et al. APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern. Alzheimer’s & dementia: the journal of the Alzheimer’s Association 11, 1417–1429, https://doi.org/10.1016/j.jalz.2015.03.003 (2015).
Risacher, S. L. et al. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI). Frontiers in aging neuroscience 5, 11, https://doi.org/10.3389/fnagi.2013.00011 (2013).
Dale, A. M., Fischl, B. & Sereno, M. I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage 9, 179–194, https://doi.org/10.1006/nimg.1998.0395 (1999).
Fischl, B., Sereno, M. I. & Dale, A. M. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. NeuroImage 9, 195–207, https://doi.org/10.1006/nimg.1998.0396 (1999).
Saykin, A. J. et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement 6, 265–273, https://doi.org/10.1016/j.jalz.2010.03.013 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81, 559–575, https://doi.org/10.1086/519795 (2007).
Ramanan, V. K. et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Molecular psychiatry 19, 351–357, https://doi.org/10.1038/mp.2013.19 (2014).
Li, J. et al. Genetic Interactions Explain Variance in Cingulate Amyloid Burden: An AV-45 PET Genome-Wide Association and Interaction Study in the ADNI Cohort. BioMed research international 2015, 647389, https://doi.org/10.1155/2015/647389 (2015).
Saykin, A. J. et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement 11, 792–814, https://doi.org/10.1016/j.jalz.2015.05.009 (2015).
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature genetics 44, 955–959, https://doi.org/10.1038/ng.2354 (2012).
Nho, K. et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Molecular psychiatry 18, 781–787, https://doi.org/10.1038/mp.2013.24 (2013).
Nam, D., Kim, J., Kim, S. Y. & Kim, S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic acids research 38, W749–754, https://doi.org/10.1093/nar/gkq428 (2010).
Kim, S. Y. & Volsky, D. J. PAGE: parametric analysis of gene set enrichment. BMC bioinformatics 6, 144, https://doi.org/10.1186/1471-2105-6-144 (2005).
Benjamini, Y. & Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995).
Li, M. X., Gui, H. S., Kwan, J. S. & Sham, P. C. GATES: a rapid and powerful gene-based association test using extended Simes procedure. American journal of human genetics 88, 283–293, https://doi.org/10.1016/j.ajhg.2011.01.019 (2011).
Moskvina, V. et al. Evaluation of an approximation method for assessment of overall significance of multiple-dependent tests in a genomewide association study. Genetic epidemiology 35, 861–866, https://doi.org/10.1002/gepi.20636 (2011).
Li, M. X., Kwan, J. S. & Sham, P. C. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. American journal of human genetics 91, 478–488, https://doi.org/10.1016/j.ajhg.2012.08.004 (2012).
Horgusluoglu-Moloch, E. et al. Targeted neurogenesis pathway-based gene analysis identifies ADORA2A associated with hippocampal volume in mild cognitive impairment and Alzheimer’s disease. Neurobiol Aging 60, 92–103, https://doi.org/10.1016/j.neurobiolaging.2017.08.010 (2017).
Swaminathan, S. et al. Amyloid pathway-based candidate gene analysis of [(11)C]PiB-PET in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Brain imaging and behavior 6, 1–15, https://doi.org/10.1007/s11682-011-9136-1 (2012).
Miller, J. A., Woltjer, R. L., Goodenbour, J. M., Horvath, S. & Geschwind, D. H. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome medicine 5, 48, https://doi.org/10.1186/gm452 (2013).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559, https://doi.org/10.1186/1471-2105-9-559 (2008).
Sapolsky, R. M. D. antidepressants, and the shrinking hippocampus. Proceedings of the National Academy of Sciences of the United States of America 98, 12320–12322, https://doi.org/10.1073/pnas.231475998 (2001).
Eriksson, P. S. et al. Neurogenesis in the adult human hippocampus. Nature medicine 4, 1313–1317, https://doi.org/10.1038/3305 (1998).
Gould, E., Beylin, A., Tanapat, P., Reeves, A. & Shors, T. J. Learning enhances adult neurogenesis in the hippocampal formation. Nature neuroscience 2, 260–265, https://doi.org/10.1038/6365 (1999).
Aimone, J. B. et al. Regulation and function of adult neurogenesis: from genes to cognition. Physiological reviews 94, 991–1026, https://doi.org/10.1152/physrev.00004.2014 (2014).
Toni, N. & Schinder, A. F. Maturation and Functional Integration of New Granule Cells into the Adult. Hippocampus. Cold Spring Harbor perspectives in biology 8, a018903, https://doi.org/10.1101/cshperspect.a018903 (2015).
Williamson, L. L. & Bilbo, S. D. Chemokines and the hippocampus: a new perspective on hippocampal plasticity and vulnerability. Brain, behavior, and immunity 30, 186–194, https://doi.org/10.1016/j.bbi.2013.01.077 (2013).
Doze, V. A. & Perez, D. M. G-protein-coupled receptors in adult neurogenesis. Pharmacological reviews 64, 645–675, https://doi.org/10.1124/pr.111.004762 (2012).
Toledo, E. M., Colombres, M. & Inestrosa, N. C. Wnt signaling in neuroprotection and stem cell differentiation. Progress in neurobiology 86, 281–296, https://doi.org/10.1016/j.pneurobio.2008.08.001 (2008).
Mohammad, H. et al. JNK1 controls adult hippocampal neurogenesis and imposes cell-autonomous control of anxiety behaviour from the neurogenic niche. Molecular psychiatry. https://doi.org/10.1038/mp.2016.203 (2016).
Chang, L., Jones, Y., Ellisman, M. H., Goldstein, L. S. & Karin, M. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Developmental cell 4, 521–533 (2003).
Oliva, A. A. Jr., Atkins, C. M., Copenagle, L. & Banker, G. A. Activated c-Jun N-terminal kinase is required for axon formation. The Journal of neuroscience: the official journal of the Society for Neuroscience 26, 9462–9470, https://doi.org/10.1523/JNEUROSCI.2625-06.2006 (2006).
Ji, R., Meng, L., Li, Q. & Lu, Q. TAM receptor deficiency affects adult hippocampal neurogenesis. Metabolic brain disease 30, 633–644, https://doi.org/10.1007/s11011-014-9636-y (2015).
Kim, S. Y. et al. Profiling analysis of protein tyrosine phosphatases during neuronal differentiation. Neuroscience letters 612, 219–224, https://doi.org/10.1016/j.neulet.2015.12.027 (2016).
Dannlowski, U. et al. Multimodal imaging of a tescalcin (TESC)-regulating polymorphism (rs7294919)-specific effects on hippocampal gray matter structure. Molecular psychiatry 20, 398–404, https://doi.org/10.1038/mp.2014.39 (2015).
Baumgartner, M., Patel, H. & Barber, D. L. Na(+)/H(+) exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. American journal of physiology. Cell physiology 287, C844–850, https://doi.org/10.1152/ajpcell.00094.2004 (2004).
Bao, Y. et al. Expression and evolutionary conservation of the tescalcin gene during development. Gene expression patterns: GEP 9, 273–281, https://doi.org/10.1016/j.gep.2009.03.004 (2009).
Choe, Y., Kozlova, A., Graf, D. & Pleasure, S. J. Bone morphogenic protein signaling is a major determinant of dentate development. The Journal of neuroscience: the official journal of the Society for Neuroscience 33, 6766–6775, https://doi.org/10.1523/JNEUROSCI.0128-13.2013 (2013).
Gui, H., Li, M., Sham, P. C. & Cherny, S. S. Comparisons of seven algorithms for pathway analysis using the WTCCC Crohn’s Disease dataset. BMC research notes 4, 386, https://doi.org/10.1186/1756-0500-4-386 (2011).
Ramanan, V. K. et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain imaging and behavior 6, 634–648, https://doi.org/10.1007/s11682-012-9196-x (2012).
Sorrells, S. F. et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381, https://doi.org/10.1038/nature25975 (2018).
Horgusluoglu, E. Neurogenesis in the adult brain, gene networks, and Alzheimer’s Disease PhD thesis, Indiana University (2017).
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen. Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health. (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. ENIGMA was supported in part by a Consortium grant (U54EB020403 to PMT) from the NIH Institutes contributing to the Big Data to Knowledge (BD2K) Initiative, including the NIBIB and NCI. Additional support for data analysis was provided by NLM R01 LM012535, NIA R03 AG054936, NIA R01 AG19771, NIA P30 AG10133, NLM R01 LM011360, NSF IIS-1117335, DOD W81XWH-14-2-0151, NCAA 14132004, NIGMS P50GM115318, NCATS UL1 TR001108, NIA K01 AG049050, NCI R01 CA129769 and R35 CA197289, the Alzheimer’s Association, the Indiana Clinical and Translational Science Institute, and the IU Health-IU School of Medicine Strategic Neuroscience Research Initiative. This manuscript is based in part on the published PhD thesis of Emrin Horgusluoglu in Medical and Molecular Genetics at Indiana University School of Medicine entitled “Neurogenesis in the adult brain, gene networks, and Alzheimer’s Disease”58, under the supervision of the corresponding authors.
Author information
Authors and Affiliations
Consortia
Contributions
A.J.S., K.N. and E.H. conceived and designed the study; E.H. analyzed data; S.L.R. contributed to the extraction of AD-related endophenotypes; P.K.C. contributed to the generation of composite scores for memory and executive functioning; D.H. and P.M.T. contributed to the generation of ENIGMA GWAS data for hippocampal volume; E.H., A.J.S. and K.N. wrote the manuscript, with contributions from all of the authors; All authors critically reviewed the paper. Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A comprehensive list of consortium members appears at the end of the paper
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Horgusluoglu-Moloch, E., Risacher, S.L., Crane, P.K. et al. Genome-wide association analysis of hippocampal volume identifies enrichment of neurogenesis-related pathways. Sci Rep 9, 14498 (2019). https://doi.org/10.1038/s41598-019-50507-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-50507-3
This article is cited by
-
Distributed genetic architecture across the hippocampal formation implies common neuropathology across brain disorders
Nature Communications (2022)
-
FKBP51 modulates hippocampal size and function in post-translational regulation of Parkin
Cellular and Molecular Life Sciences (2022)
-
A peripheral lipid sensor GPR120 remotely contributes to suppression of PGD2-microglia-provoked neuroinflammation and neurodegeneration in the mouse hippocampus
Journal of Neuroinflammation (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.