Abstract
The prevalence of many chronic diseases has increased over the last decades. It has been postulated that dysbiosis driven by environmental factors such as antibiotic use is shifting the microbiome in ways that increase inflammation and the onset of chronic disease. Dysbiosis can be defined through the loss or gain of bacteria that either promote health or disease, respectively. Here we use multiple independent datasets to determine the nature of dysbiosis for a cluster of chronic diseases that includes urinary stone disease (USD), obesity, diabetes, cardiovascular disease, and kidney disease, which often exist as co-morbidities. For all disease states, individuals exhibited a statistically significant association with antibiotics in the last year compared to healthy counterparts. There was also a statistically significant association between antibiotic use and gut microbiota composition. Furthermore, each disease state was associated with a loss of microbial diversity in the gut. Three genera, Bacteroides, Prevotella, and Ruminococcus, were the most common dysbiotic taxa in terms of being enriched or depleted in disease populations and was driven in part by the diversity of operational taxonomic units (OTUs) within these genera. Results of the cross-sectional analysis suggest that antibiotic-driven loss of microbial diversity may increase the risk for chronic disease. However, longitudinal studies are needed to confirm the causative effect of diversity loss for chronic disease risk.
Similar content being viewed by others
Introduction
Many chronic diseases are on a trend of increasing prevalence. Cardiovascular disease, obesity, diabetes, urinary stone disease (USD), asthma, and inflammatory bowel disease (IBD) are all on the rise, among others1,2,3,4,5,6. These chronic inflammatory diseases often exist as co-morbidities with common physiological manifestations, which both compound the burden on patients and suggests common origins7,8,9. While a number of genetic and environmental factors contribute to the manifestation of chronic disease, an emerging hypothesis postulates that dysbiosis, an imbalance in the composition and metabolic capacity of our microbiota, increases the risk of developing chronic disease10,11,12,13. Dysbiosis can contribute to the onset of chronic disease in one of three general ways. First, pathogens and their functions can be acquired or opportunistically overgrow to promote disease, termed gain of function dysbiosis. Gain of function dysbiosis leads to infectious diseases such as cholera or streptococcal pharyngitis, but can also lead to chronic inflammation14,15,16. Second, health-protective bacteria and their functions may be lost or suppressed, which then promotes the onset of disease, termed loss of function dysbiosis. Loss of function dysbiosis has been linked to chronic diseases such as IBD, USD, obesity, and others17,18,19,20,21,22,23,24,25,26. Finally, some combination of loss and gain of function dysbiosis may be required for the onset of disease, such as with recurrent Clostridium difficile infection27.
The complexity of the microbiome presents unique challenges to our understanding of the role of dysbiosis in disease. Each individual harbors thousands of unique microbial operational taxonomic units (OTUs)28,29. Compounding this issue is the considerable inter-individual variability in the composition of the microbiome30,31. Given the hyper-variable nature of high-throughput 16S rRNA data combined with the high levels of inter-individual variability, any two random populations of individuals are likely to harbor a proportion of OTUs unique to each population, with no relevance to any phenotype. To help restrict false-discoveries associated with this issue, false-discovery rate corrections are commonly integrated into microbiome studies32. Despite the limitations of microbiome studies, many clinical studies use the differential abundance of OTUs between healthy and disease populations at a single time-point to determine if dysbiosis contributes to the disease, which can lead to erroneous results when it comes to the nature of dysbiosis and microbial taxa involved. Thus, a number of questions remain surrounding conclusions that dysbiosis contributes to the manifestation of disease. First, is the differential abundance of OTUs between two populations driven by inter-individual variability of the two populations or disease status? Second, what features of microbial taxa contribute to them being identified as dysbiotic? Finally, are there taxa consistently found to be dysbiotic across chronic disease states, indicative of common, dysbiosis-driven disease processes?
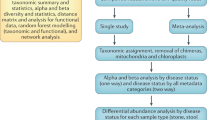
To address these questions of defining dysbiosis, we performed an independent analysis of microbiome data from multiple sources that spanned a cluster of chronic diseases that included USD, cardiovascular disease, obesity, diabetes, and kidney disease. Specifically, we quantified the use of antibiotics in disease populations compared to healthy controls, which can lead to a loss in microbial diversity relevant to disease processes33,34,35,36,37,38,39,40, quantified metrics of loss or gain of microbiota diversity associated with each disease state relative to stochastically defined populations, determined the similarities and differences in microbial taxa enriched and depleted in disease populations, and determined which of these taxa were differentially abundant more than expected given their genus-level diversity in samples. These diseases often exist as comorbidities in patients and have all been linked to the microbiome to some degree. We hypothesize that if the diseases in question are driven in part by dysbiosis, that there will be common metrics of dysbiosis that include antibiotic use, loss or gain of microbial diversity, and microbial taxa associated with disease.
Results
Datasets
For cardiovascular disease, obesity, diabetes, kidney disease, and healthy counterparts, 16S rRNA sequences from the stool and associated sample metadata were drawn from the April 26th, 2017 version of American Gut Project data (AGP), which has an extensive list of metadata that includes antibiotic history and the presence or absence of several disease states41. Samples for sequencing were collected between December 2012 and April 2017 from individuals from a global population. Healthy individuals were defined by the “I do not have this condition” entry for diabetes, cardiovascular disease, and kidney disease, as well as “normal” for body mass index (“BMI_CAT” in the metadata). Those with “diagnosed” for cardiovascular disease, diabetes, or kidney disease, or with “obese” for BMI_CAT were assigned to the appropriate chronic disease state. Health status for each defined term were based on self-reported medical diagnoses which may bias results41. A random number generator was used to assign random numbers to each sample that fit one of the defined health states for the AGP data and the samples with the lowest 300 randomly assigned numbers for each health status were extracted for further analysis, except for kidney disease which only had 111 entries. For kidney disease, we extracted samples with the lowest 100 numbers for further analysis. For USD, which is not represented in the AGP metadata, 16S rRNA sequences and associated sample metadata were downloaded from all publicly available metagenomic data from clinical USD studies published at the time of analysis18,20,42,43. Samples for USD studies were collected between 2015–2019 and originated in the United States, Canada, India, or China. Samples were defined as healthy or USD based on the criteria used in the original study. The entire combined dataset spanned a total of 1468 samples (Table 1).
Association of antibiotics and chronic disease
For all chronic disease states, individuals were significantly more likely to have taken antibiotics in the last year compared to healthy individuals (Fig. 1A). Furthermore, antibiotic use in the last year was significantly associated with microbiota composition as assessed by a weighted UniFrac analysis followed by a post-hoc PERMANOVA of the regularized AGP datasets (Fig. 1B)20,44.
The effect of antibiotics on chronic disease and the microbiota. (A) Antibiotic use within the last year for individuals with or without chronic disease. For diabetes, cardiovascular disease, kidney disease, obesity, and their healthy counterparts, antibiotic history was derived from the subset of AGP samples randomly selected for this study (N = 300 for each group except kidney disease which had 100 samples; Table 1). Only one study on the microbiome of USD patients included metadata associated with antibiotic use (N = 43 healthy individuals and 24 individuals with USD; Table 1). Proportions of antibiotic use were compared between chronic disease states and healthy populations with a relative risk ratio followed by a post-hoc Fisher’s exact test, which was Holm’s corrected for multiple comparisons. *p < 0.05; **p < 0.01; ***p < 0.001 compared to the healthy population. (B) PCoA plot based on a weighted UniFrac analysis the microbiome composition from the AGP data. Community composition based on antibiotic use was compared by PERMANOVA with 999 permutations. p = 0.006.
Nature of dysbiosis in chronic disease
The differential abundance of OTUs between healthy and disease populations was used to determine if each of the disease states were associated with a depletion or enrichment of microbial diversity compared to healthy controls, with the number of OTUs enriched or depleted in disease quantified for each pairwise comparison (Fig. 2). The average fold difference in OTUs enriched in healthy cohorts vs. disease cohorts reveals that for each disease state, there was significantly more OTUs enriched in healthy cohorts than disease cohorts, indicative of a loss of microbial diversity in the gut (Fig. 3). No significant bias for group 1 or 2 was detected when health status was stochastically assigned to samples through random assignment (see methods).
An example of differential abundance analysis for each of the disease states. Each dot represents an OTU. Grey dots are OTUs that do not exhibit significant differential abundance, while red dots are differentially abundant OTUs. (A) Stochastic metadata; (B) USD; (C) Cardiovascular disease; (D) Diabetes; (E) Obesity; (F) Kidney disease.
Average fold difference in the number of OTUs enriched in either the healthy group/stochastic group 1 or in the disease group/stochastic group 2. Positive values reflect greater enrichment in healthy group/stochastic group 1, whereas negative values reflect greater enrichment in disease group/stochastic group 2. Significance was determined with a one-sample t-test against an expected value of 1. *p < 0.05; **p < 0.01; ***p < 0.001.
Strength of dysbiotic taxa and commonalities across chronic disease states
The proportion of independent healthy x disease comparisons in which a particular genus had at least one OTU that was differentially abundant was plotted on a heatmap (Fig. 4). The heatmap reveals that the most common dysbiotic genera were the Coprococcus, Prevotella, and Bacteroides for OTUs enriched in the healthy cohorts. For OTUs enriched in disease cohorts, the Bacteroides, Ruminococcus, and Blautia genera were most common. Hierarchal clustering reveals statistically significant similarities between diabetes and kidney disease when considering potential health protective bacteria lost. When considering potential pathogenic bacteria acquired, obesity and USD exhibit a statistically significant cluster, which also clusters with cardiovascular disease. Diabetes again clustered with kidney disease with statistical significance. Additionally, from the heatmaps, it is apparent that each of the diseases is associated with a loss of diverse genera more so than the gain of microbial genera, largely corroborating analyses based on antibiotic use and the number of dysbiotic OTUs (Fig. 4).
Heatmaps showing the most common dysbiotic genera for each disease. Genera were counted for each independent population comparison had at least one dysbiotic OTU associated with it. The proportion of populations each genera showed up in is plotted. (A) Genera depleted in the disease populations (potential probiotic bacteria lost); (B) Genera enriched in disease populations (potential pathogenic bacteria). Hierarchal cluster analysis shows clustering of disease states with the approximately unbiased alpha levels (AU) and bootstrap probability (BP) provided for each cluster (AU/BP). AU values > 95 are considered significant and are bolded.
Impact of genus diversity on dysbiotic OTUs
When the total number of OTUs identified in each genus is plotted against the number of dysbiotic OTUs identified in each genus, there is a significant correlation between the two factors, reflecting the fact that dysbiosis is driven in part by the number of constituent OTUs in each genus (Fig. 5). However, the number of genera determined to be dysbiotic more than expected given genus level diversity ranged from one (USD) to 14 (kidney disease). Of these, the Bacteroides genus was more likely to be dysbiotic for all diseases and Corynebacterium for all but USD. The Anaerococcus genus was more likely to be dysbiotic than expected for three of the five disease states (Table 2).
Discussion
In the last ten years, there has been an explosion of metagenome-wide association studies (MWAS) that seek to determine if the microbiome contributes to disease processes. Importantly, these studies have revealed that there is both considerable diversity in the microbiome with hundreds to thousands of unique bacterial species in the gut alone28,29 and a high amount of inter-individual variability30,31. Given these unique characteristics of the microbiome, studies on any one population of individuals can be driven simply by the individual variability and are thus unreliable. Despite these limitations, one leading hypothesis has emerged that postulates that the loss of diversity from our microbiome is increasing the risk of developing chronic disease45,46,47. Several questions remain about defining the nature of dysbiosis for any one particular disease. First, are MWAS results driven by inter-individual variability and study design, or are there consistent responses by disease? Second, are different diseases consistently associated with the loss of beneficial bacteria or the gain of harmful bacteria? Finally, is dysbiosis driven by a general loss or gain of microbial taxa or are there specific taxa that are more important than others? To address these questions, we conducted a meta-analysis of MWAS data from multiple sources for a cluster of disease states that included USD, diabetes, cardiovascular disease, obesity, and kidney disease.
Some of the most striking results of the current study is that every disease in the cluster of chronic diseases exhibited a statistically significant association with past year antibiotic use and that antibiotics had a statistically significant association with gut microbiota composition up to a year after use, both of which supports the loss of function dysbiosis hypothesis (Fig. 1). Further evidence that a loss of microbial diversity increases risk of developing the chronic diseases studied here comes from the differential abundance analyses, in which individuals from each of the disease states exhibited a statistically significant propensity for the reduced presence of bacteria in their gut compared to their healthy counterparts (Fig. 3).
Past studies have shown both direct and indirect links between antibiotic use and the risk of developing USD20,35,48,49,50,51 and obesity37,38,39,52,53. For diabetes, the literature is mixed with some studies concluding that antibiotics increase the risk of diabetes54,55,56,57, some that conclude repeated antibiotic use but not single antibiotic use increases diabetes risk58, and some that conclude antibiotics protect against diabetes54,59,60. For cardiovascular disease, animal studies show that antibiotics lead to a reduction in microbial activity that promotes cardiovascular disease61. However, outcomes using antibiotic therapy to treat cardiovascular disease in a clinical setting to date have been mixed62,63. In addition to the specific diseases above, another meta-analysis of MWAS studies that spanned multiple unrelated diseases has been published. In that meta-analysis, it was found that some diseases such as enteric diarrheal disease, Clostridium difficile infection, and IBD were associated with loss of function dysbiosis, whereas diseases such as colorectal cancer, autism spectrum disorder, liver diseases, and Parkinson’s disease were associated with gain of function dysbiosis64.
Among the taxa that were most consistently found to be depleted in disease individuals were the Bacteroides, Coprococcus, Prevotella, Ruminococcus, and Sutterella (Fig. 5). However, subsequent analysis suggests that the association of some of these taxa with dysbiosis is closely tied to their number of constituent OTUs in the gut, indicative of a non-specific response. Some taxa, primarily Bacteroides, Corynebacterium, and Anaerococcus were identified as dysbiotic more than expected given their diversity, indicative of a more specific physiological interaction between these taxa and disease (Table 2). Past studies have concluded that Bacteroides either has a health-protective65,66,67 or health-antagonistic response68,69,70. Results from the current analysis show that this genus is strongly associated with both health and disease in terms of the number of independent populations it was found to be associated with (Fig. 4) and thus it is likely that some OTUs within the genus provide more of a protective effect and others more of a detrimental health impact. The Corynebacterium and Anaerococcus genera were only associated with the healthy individuals in our meta-analysis. Corynebacterium is most often associated with diphtheria, an illness that primarily affects the respiratory system71. The number of independent healthy populations associated with Corynebacterium here suggests that this genus of bacteria may play a more beneficial role in the context of the gut microbiota. Anaerococcus sp. are commonly associated with the normal flora of the skin, mouth, and gut, but are also often recovered from clinical specimens such as vaginal discharge and chronic infectious wounds72.
Numerous MWAS studies have demonstrated statistically significant interactions between the microbiome and disease processes. However, questions remain about the consistency of results obtained from single studies and whether there are common dysbiotic origins for related diseases. Importantly, our results show that for the cluster of chronic diseases that span USD, diabetes, kidney disease, obesity, and cardiovascular disease, there is a statistically significant association with antibiotic use and a loss of microbial diversity. Furthermore, several of the dysbiotic taxa are shared between different disease states suggesting that there are some common dysbiotic associations for these diseases. However, as the current data represents a retrospective analysis of data from a single time-point, prospective, longitudinal clinical studies are required to understand the underlying mechanisms between antibiotics, loss of function dysbiosis, and the onset of chronic disease.
Materials and Methods
Data processing
Raw sequencing data for all studies were demultiplexed and quality-controlled with default parameters in QIIME73. An open reference strategy was used to assign OTUs with 97% homology compared to a reference database composed of the Greengenes V13.8 dataset74,75,76. Datasets were combined prior to OTU assignment to allow for direct comparisons. OTUs that did not match any of sequences in the reference database were then classified de novo, which first clusters unclassified sequences based on homology, picks a representative sequence, and assigns taxonomy based on a 94% similarity threshold to the reference database77. Chloroplasts, mitochondria, and chimeric sequences were removed from datasets as well as those that had <10 representations across each dataset as previously described74,75. The final OTU count across all datasets was 35,582. Datasets were regularized with a negative binomial Wald test, executed by the DESeq2 algorithm, which fits the data to a negative binomial distribution, to account for differences in sequencing depth in each sample, but does not account for the compositional nature of the data78,79.
Association of antibiotics and chronic disease
To determine if antibiotics were associated with chronic disease, we quantified the proportion of individuals who had taken antibiotics in the last year for cardiovascular disease, obesity, diabetes, and kidney disease compared to their healthy counterparts in the AGP data. For USD, only one of the four studies included in our meta-analysis had metadata associated with antibiotic use20. Proportions were compared with a relative risk ratio followed by a post-hoc, Fisher’s exact test, using a Holm’s correction for multiple hypotheses.
In order for antibiotic use to have a meaningful impact on chronic disease, we expect that antibiotics would exhibit a statistically significant association with microbiota composition. Therefore, we also conducted a weighted UniFrac beta-diversity analysis on all regularized data from the AGP dataset, which were all collected and processed by the same group, followed by a post-hoc PERMANOVA against antibiotic use in the past year.
Defining dysbiosis for each disease state
To determine the nature of dysbiosis for each disease state, while considering the multi-dimensional and variable nature of microbiome data, we divided the samples into the following independent populations. For USD data, each study was considered an independent population and the healthy vs. USD groups were compared against each other within each study. Four populations were available for USD and their healthy counterparts. For AGP data, each category of health states was sub-divided into three independent sets of 100 samples by assigning samples to one of three groups through the lowest, middle, and highest 100 random numbers previously assigned. For kidney disease, only one subset was available. Subdivisions provided three independent populations for AGP healthy individuals, cardiovascular disease, obesity, and diabetes. One population was available for kidney disease. Additionally, random numbers were reassigned to AGP data independent of metadata. The random numbers generated were used to assign three independent populations each for stochastic groups 1 & 2. Groups with stochastic metadata were used to determine if our definition of dysbiosis for each disease state were driven by disease or some other stochastic process.
Following subdivisions, differential abundance analysis, calculated as the log fold-change divided by standard error of negative binomial regularized data (lfcse), was conducted on all relevant pairwise comparisons of independent populations using the DESeq2 algorithm78. All p-values for differentially abundant OTUs were false-discovery rate corrected, following the Benjamini-Hochberg method80. Differential abundance analysis was chosen to define dysbiosis because it gives the specific OTUs enriched in both healthy and disease populations rather than net differences in diversity or community composition as with alpha- and beta-diversity respectively. Thus, this metric is suitable for identifying situations in which a combination of loss and gain of function dysbiosis may contribute to disease processes. For USD data, since samples and data were collected independently by four different research groups, differential abundance analysis between healthy and USD populations within each study was conducted. For AGP data, since all samples were collected, processed, and sequenced by the same group, each independent healthy population was compared to each disease population. This resulted in nine pairwise comparisons for cardiovascular disease, diabetes, obesity, and stochastic groups 1 & 2, along with three pairwise comparisons for kidney disease.
From the results of independent differential abundance analyses, the fold difference in OTUs between the healthy and disease populations was calculated as the number of OTUs enriched in the healthy group divided by the number of OTUs enriched in the disease group. Absolute values were compared with a one-sample t-test against an expected value of 1, indicative of no difference, and p-values were Holm’s corrected for multiple comparisons. For purposes of plotting fold difference, raw values below 1 were inverted and made negative.
Strength of dysbiotic taxa and commonalities across chronic disease states
To determine the consistency of dysbiotic taxa identified and if there were common dysbiotic genera across the different chronic diseases, differentially abundant OTUs for each comparison were first reduced to genera, which allows for greater comparison between each disease state but comes with the limitation that physiologically disparate OTUs can exist within the same genus. The OTUs that were not classified at least to the genus level were removed from further analysis. We counted the number of times a microbial genus had at least one OTU that was enriched in either the healthy or disease populations across all pairwise comparisons for each disease. Genera enriched in the healthy populations were considered “potentially beneficial” whereas those enriched in the disease populations were considered “potentially pathogenic”. The proportion of independent analyses that each genus appeared in was plotted in a heatmap in R statistical software using the heatmap.2 function with dendograms generated by the hclust function to show clustering by disease state. The bootstrap probability and approximately unbiased alpha levels were calculated by the pvclust package with 1000 bootstraps81.
Influence of genus diversity on dysbiotic genera
Next, we sought to determine if dysbiosis was driven by a general loss or gain of bacteria in the gut or if some microbial genera had a greater association with disease than expected by the number of constituent OTUs within the genus. To do this, for each disease, we plotted the total number of OTUs detected per genus against the number of dysbiotic OTUs detected per genus and determined if there was a Spearman correlation significantly different than 0 with the cor.test function in R statistical software. Next, genera in which the number of dysbiotic OTUs was greater than three standard deviations above what was expected based on genus diversity were considered to be highly associated with dysbiosis, following the three-sigma rule82.
Data Availability
Sequence reads for USD are available at the Sequence Read Archive under Accession #’s SRP140641, SRP140933, PRJNA382644, PRJNA304689. American gut project data is available at the European Bioinformatics Institute under accession # PRJEB11419.
References
Scales, C. D., Smith, A. C., Hanley, J. M. & Saigal, C. S. & Project, U. D. i. A. Prevalence of kidney stones in the United States. European urology 62, 160–165 (2012).
Ng, M. et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. The lancet 384, 766–781 (2014).
Schnabel, R. B. et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: a cohort study. The Lancet 386, 154–162 (2015).
Menke, A., Casagrande, S., Geiss, L. & Cowie, C. C. Prevalence of and trends in diabetes among adults in the United States, 1988–2012. Jama 314, 1021–1029 (2015).
Loftus, P. A. & Wise, S. K. Epidemiology of asthma. Current opinion in otolaryngology & head and neck surgery 24, 245–249 (2016).
Ng, S. C. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. The Lancet 390, 2769–2778 (2017).
Caughey, G. E. et al. Comorbidity in the elderly with diabetes: Identification of areas of potential treatment conflicts. Diabetes research and clinical practice 87, 385–393 (2010).
Kartha, G. K., Li, I., Comhair, S., Erzurum, S. C. & Monga, M. Co-occurrence of asthma and nephrolithiasis in children. PloS one 12, e0168813 (2017).
Gudbjartsson, D. F. et al. Association of variants at UMOD with chronic kidney disease and kidney stones—role of age and comorbid diseases. PLoS genetics 6, e1001039 (2010).
Hand, T. W., Vujkovic-Cvijin, I., Ridaura, V. K. & Belkaid, Y. Linking the microbiota, chronic disease, and the immune system. Trends in Endocrinology & Metabolism 27, 831–843 (2016).
Blaser, M. J. & Falkow, S. What are the consequences of the disappearing human microbiota? Nature Reviews Microbiology 7, 887 (2009).
Lynch, S. V. & Pedersen, O. The human intestinal microbiome in health and disease. New England Journal of Medicine 375, 2369–2379 (2016).
Hawrelak, J. A. & Myers, S. P. The causes of intestinal dysbiosis: a review. Alternative medicine review 9, 180–198 (2004).
Liu, X. N. et al. Intestinal Epithelial Cell–Derived LKB1 Suppresses Colitogenic Microbiota. The Journal of Immunology, https://doi.org/10.4049/jimmunol.1700547 (2018).
Karin, M., Lawrence, T. & Nizet, V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell 124, 823–835 (2006).
Medzhitov, R. Origin and physiological roles of inflammation. Nature 454, 428 (2008).
Sokol, H. et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflammatory bowel diseases 15, 1183–1189 (2009).
Suryavanshi, M. V. et al. Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Scientific reports 6, 34712 (2016).
Ticinesi, A. et al. Understanding the gut–kidney axis in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut, gutjnl-2017–315734 (2018).
Zampini, A., Nguyen, A., Rose, E., Monga, M. & Miller, A. Reduced functional microbial network in patients with urolithiasis. (In revisions).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. nature 457, 480 (2009).
Schippa, S. et al. A distinctive’microbial signature’in celiac pediatric patients. BMC microbiology 10, 175 (2010).
Carroll, I. M., Ringel‐Kulka, T., Siddle, J. P. & Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea‐predominant irritable bowel syndrome. Neurogastroenterology & Motility 24, 521–e248 (2012).
Kang, D.-W. et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PloS one 8, e68322 (2013).
Abrahamsson, T. et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clinical & Experimental Allergy 44, 842–850 (2014).
Scher, J. U. et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis & rheumatology 67, 128–139 (2015).
Britton, R. A. & Young, V. B. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 146, 1547–1553 (2014).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Claesson, M. J. et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PloS one 4, e6669 (2009).
Booijink, C. C. et al. High temporal and inter‐individual variation detected in the human ileal microbiota. Environmental microbiology 12, 3213–3227 (2010).
Huttenhower, C. et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207 (2012).
Korthauer, K. et al. A practical guide to methods controlling false discoveries in computational biology. Genome biology 20, 118 (2019).
Vangay, P., Ward, T., Gerber, J. S. & Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell host & microbe 17, 553–564 (2015).
Ungaro, R. et al. Antibiotics associated with increased risk of new-onset Crohn’s disease but not ulcerative colitis: a meta-analysis. The American journal of gastroenterology 109, 1728–1738 (2014).
Tasian, G. E. J. et al. Oral antibiotic exposure and kidney stone disease. Journal of the American Society of Nephrology 29 (2018).
Sjlund, M., Wreiber, K., Andersson, D. I., Blaser, M. J. & Engstrand, L. Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Annals of internal medicine 139, 483–487 (2003).
Scott, F. I. et al. Administration of antibiotics to children before age 2 years increases risk for childhood obesity. Gastroenterology 151, 120–129. e125 (2016).
Riley, L. W., Raphael, E. & Faerstein, E. Obesity in the United States–dysbiosis from exposure to low-dose antibiotics? Frontiers in public health 1, 69 (2013).
Mueller, N. T. et al. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. International journal of obesity 39, 665 (2015).
Morgun, A. et al. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut, gutjnl-2014–308820 (2015).
McDonald, D. et al. American Gut: an Open Platform for Citizen Science Microbiome Research. mSystems 3, https://doi.org/10.1128/mSystems.00031-18 (2018).
Miller, A. W. C., Choy, D., Penniston, K. L. & Lange, D. Identification of oxalate microbiome in non-stone forming individuals. Kidney international (In review).
Tang, R. et al. 16S rRNA gene sequencing reveals altered composition of gut microbiota in individuals with kidney stones. Urolithiasis, 1–12 (2018).
Lozupone, C., Hamady, M. & Knight, R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics 7, 1 (2006).
Neuman, H., Forsythe, P., Uzan, A., Avni, O. & Koren, O. Antibiotics in early life: dysbiosis and the damage done. FEMS microbiology reviews 42, 489–499 (2018).
Cao, Y. et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut 67, 672–678 (2018).
Blaser, M. J. Missing microbes: how the overuse of antibiotics is fueling our modern plagues. (Macmillan, 2014).
Sidhu, H. et al. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. The Lancet 352, 1026–1029 (1998).
Mittal, R., Kumar, R., Bid, H. & Mittal, B. Effect of antibiotics on Oxalobacter formigenes colonization of human gastrointestinal tract. Journal of endourology 19, 102–106 (2005).
Siener, R. et al. The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney international 83, 1144–1149 (2013).
Sidhu, H. et al. Evaluating children in the Ukraine for colonization with the intestinal bacterium Oxalobacter formigenes, using a polymerase chain reaction-based detection system. Molecular Diagnosis 2, 89–97 (1997).
Cox, L. M. & Blaser, M. J. Antibiotics in early life and obesity. Nature Reviews Endocrinology 11, 182 (2015).
Janson, B. & Thursky, K. Dosing of antibiotics in obesity. Current opinion in infectious diseases 25, 634–649 (2012).
Hu, Y., Wong, F. S. & Wen, L. Antibiotics, gut microbiota, environment in early life and type 1 diabetes. Pharmacological research 119, 219–226 (2017).
Boursi, B., Mamtani, R., Haynes, K. & Yang, Y.-X. The effect of past antibiotic exposure on diabetes risk. European journal of endocrinology, EJE-14–1163 (2015).
Candon, S. et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PloS one 10, e0125448 (2015).
Mikkelsen, K. H., Knop, F. K., Frost, M., Hallas, J. & Pottegård, A. Use of antibiotics and risk of type 2 diabetes: a population-based case-control study. The Journal of Clinical Endocrinology & Metabolism 100, 3633–3640 (2015).
Mikkelsen, K. H. et al. Use of antibiotics in childhood and risk of Type 1 diabetes: a population‐based case–control study. Diabetic Medicine 34, 272–277 (2017).
Carvalho, B. et al. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55, 2823–2834 (2012).
Brugman, S. et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 49, 2105–2108 (2006).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57 (2011).
Grayston, J. T. Antibiotic Treatment of Atherosclerotic Cardiovascular Disease. Circulation 107, 1228–1230 (2003).
Frostegård, J. Immunity, atherosclerosis and cardiovascular disease. BMC medicine 11, 117 (2013).
Duvallet, C., Gibbons, S. M., Gurry, T., Irizarry, R. A. & Alm, E. J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nature communications 8, 1784 (2017).
Ochoa-Repáraz, J. et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. The Journal of Immunology, 1001443 (2010).
Ochoa-Reparaz, J. et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal immunology 3, 487 (2010).
Zhou, Y. & Zhi, F. Lower level of bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-analysis. BioMed research international 2016 (2016).
Bloom, S. M. et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell host & microbe 9, 390–403 (2011).
Sánchez, E., Laparra, J. & Sanz, Y. Discerning the role of Bacteroides fragilis in celiac disease pathogenesis. Applied and environmental microbiology, AEM. 00563–00512 (2012).
Wick, E. C. & Sears, C. L. Bacteroides spp. and diarrhea. Current opinion in infectious diseases 23, 470 (2010).
Pichichero, M. E. et al. Combined tetanus, diphtheria, and 5-component pertussis vaccine for use in adolescents and adults. Jama 293, 3003–3011 (2005).
Murphy, E. C. & Frick, I.-M. Gram-positive anaerobic cocci–commensals and opportunistic pathogens. FEMS microbiology reviews 37, 520–553 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336 (2010).
Miller, A. W., Dale, C. & Dearing, M. D. Microbiota Diversification and Crash Induced by Dietary Oxalate in the Mammalian Herbivore <em>Neotoma albigula</em>. mSphere 2, https://doi.org/10.1128/mSphere.00428-17 (2017).
Miller, A. W., Dale, C. & Dearing, M. D. The Induction of Oxalate Metabolism In Vivo Is More Effective with Functional Microbial Communities than with Functional Microbial Species. MSystems 2, e00088–00017 (2017).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal 6, 610 (2012).
Navas-Molina, J. A. et al. In Methods in enzymology Vol. 531 371-444 (Elsevier, 2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 1 (2014).
McMurdie, P. J. & Holmes, S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10, e1003531 (2014).
Benjamini, Y. & Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society, Series B (Methodological) 57 (1995).
Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Systematic biology 51, 492–508 (2002).
Pukelsheim, F. The three sigma rule. The American Statistician 48, 88–91 (1994).
Acknowledgements
This study was funded with seed funds from Lerner Research Institute and grants from the Research Program Committee at The Cleveland Clinic Foundation and the Endourology Society Summer Student Scholarship Program. The funding bodies had no role in the design of the study.
Author information
Authors and Affiliations
Contributions
A.W.M., M.M. and L.J.W. contributed to the design of the study. L.J.M. and A.W.M. conducted data analysis. All authors contributed to writing the manuscript and have approved the final version.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wilkins, L.J., Monga, M. & Miller, A.W. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci Rep 9, 12918 (2019). https://doi.org/10.1038/s41598-019-49452-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-49452-y
This article is cited by
-
Gold nanoparticles exhibit anti-osteoarthritic effects via modulating interaction of the “microbiota-gut-joint” axis
Journal of Nanobiotechnology (2024)
-
Exploring the potential effects of forest urbanization on the interplay between small mammal communities and their gut microbiota
Animal Microbiome (2024)
-
Deficient butyrate metabolism in the intestinal microbiome is a potential risk factor for recurrent kidney stone disease
Urolithiasis (2024)
-
Microbial Mechanisms of Rheumatoid Arthritis Pathogenesis
Current Rheumatology Reports (2024)
-
Fine-scale spatial variation shape fecal microbiome diversity and composition in black-tailed prairie dogs (Cynomys ludovicianus)
BMC Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.