Abstract
Sideritis scardica Giseb. is a subalpine/alpine plant species endemic to the central part of the Balkan Peninsula. In this study, we combined Amplified Fragment Length Polymorphism (AFLP) and environmental data to examine the adaptive genetic variations in S. scardica natural populations sampled in contrasting environments. A total of 226 AFLP loci were genotyped in 166 individuals from nine populations. The results demonstrated low gene diversity, ranging from 0.095 to 0.133 and significant genetic differentiation ranging from 0.115 to 0.408. Seven genetic clusters were revealed by Bayesian clustering methods as well as by Discriminant Analysis of Principal Components and each population formed its respective cluster. The exception were populations P02 Mt. Shara and P07 Mt. Vermio, that were admixed between two clusters. Both landscape genetic methods Mcheza and BayeScan identified a total of seven (3.10%) markers exhibiting higher levels of genetic differentiation among populations. The spatial analysis method Samβada detected 50 individual markers (22.12%) associated with bioclimatic variables, among them seven were identified by both Mcheza and BayeScan as being under directional selection. Four bioclimatic variables associated with five out of seven outliers were related to precipitation, suggesting that this variable is the key factor affecting the adaptive variation of S. scardica.
Similar content being viewed by others
Introduction
Adaptive genetic variation evolves as a result of changing environmental conditions imposing selective pressure on individuals living in heterogeneous habitats1. Elucidating the genetic basis of species adaptations to their environments, the nature of selection imposed by environmental changes and the capacity of a species to respond by evolutionary adaptation2,3 is one of the challenging tasks of evolutionary biology. Such information may direct conservation strategies for the long-term persistence of species, especially for those facing rapid climate changes and are threatened with extinction and population decline.
The potential of species adaptation to rapidly changing environments might be assessed by detecting genetic signatures of adaptation to prevailing or historic environmental conditions4. Genotyping of numerous random loci across entire genomes enables distinguishing the effects of the evolutionary forces that act on the entire genome (e.g., demographic events, inbreeding or bottleneck) from those influencing only individual loci (e.g., natural selection)5,6,7. Loci that are characterized by higher than expected levels of genetic differentiation are referred to as outlier loci or candidate loci and are considered to be under strong natural selection7. Such loci that are exposed to divergent selection pressures across studied populations can be recognized by a higher pairwise index of genetic differentiation values (FST) if compared to the genome-wide pairwise-FST8. In non-model organisms with a large number of individuals included in a study, Amplified Fragment Length Polymorphism (AFLP) technique has been frequently used in search of such genome regions under selection, when no prior knowledge of their position or importance exists9,10. For AFLP genome scans, two analyses methods are most commonly applied: the frequentist method of Beaumont and Nichols11 as implemented in LOSITAN12 and Mcheza13 programs and the hierarchical Bayesian method as implemented in BayeScan program14. Between the two methods, it has been suggested that BayeScan is more efficient in recognition of outlier loci with low false positive rate15. In addition, by using both geo-referenced environmental data and individual molecular genetic data, a locus-based approach as implemented in Samβada16 is often used17,18,19,20. This approach correlates individual loci occurrence with environmental variables, thus aiming to determine whether each investigated molecular marker is selected by one or a set of specific environmental variables16. The described methods provide the opportunity to identify adaptive genetic variation in non-model organisms for which the genomic data are scarce and species are of conservation concern21.
Various investigations have been conducted in order to determine the genetic variation of different plant species in relation to the landscape, e. g. Arabis alpina L.22,23, Gentiana nivalis L.24, alpine plant species25, Eruca sativa Mill.26, Keteleeria davidiana var. formosana (Hayata) Hayata27, Cotinus coggygria Scop.28, Picea abies (L.) H. Karst.29, Liriodendron chinense (Hemsl.) Sarg.30, Geropogon hybridus (L.) Sch. Bip.21, Diplotaxis hara (Forssk.) Boiss.31, etc. Nosil et al.32 in the literature review state that the available studies usually report 5–10% of loci to be outliers, while Strasburg et al.33 reported that the proportion of loci identified as outliers ranges from 0.4 to 35.5% with an average value of 8.9%. However, both studies pointed out that the results from different investigations are difficult to compare, as they differ in methodologies used and significant thresholds chosen.
Manel et al.25 suggested that genetic variation that appears to be caused by natural selection might be the results of isolation by distance, which limits gene flow among populations or the result of secondary contact of populations that survived isolated in glacial refugia. Past historic climatic oscillations have had a considerable impact on present-day biota34,35,36,37. Numerous phylogeographical investigations carried out over the recent years, have confirmed the latter, elucidating the impact of both ancient and recent history on current European flora and fauna distribution patterns and of genetic variations38,39,40,41,42.
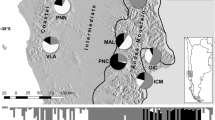
Sideritis scardica Griseb is an outcrossing diploid (2n = 32)43 and perennial subalpine/alpine herbaceous plant, endemic to the central parts of the Balkan Peninsula. The species is distributed in southwest parts of Albania, North Macedonia, Bulgaria and Greece44. It is a plant of the alpine zone, occurring in dry stony meadows, mainly at altitudes 1600–2300 m a.s.l., and only occasionally down to 500 m a.s.l. on limestone45,46. In Greece, most abundant populations of S. scardica can be found in mountains of the North and Central regions such as Vermion, Voras, Tzena, Paikon, Pangeon, Menikio, Falakro, Mt. Olympus, Mt. Ossa, etc.46. In North Macedonia, it occurs in mountains of Central and Western parts of the country47, namely in Mt. Shara, Mt. Suva Gora, Mt. Bistra, Mt. Ilina, Mt. Kozuf and Mt. Kajmakchalan.
Sideritis scardica has successfully established populations in contrasting environmental conditions and therefore represents an ideal system to examine the effect of specific environmental variables on genetic variation found between the genomes of individuals.
In this study, we selected nine S. scardica from southern parts of Balkan Peninsula that were exposed to contrasting environmental conditions. Altitudinal and climatic data were used to characterize the environmental conditions at each collecting site. To test our hypothesis that signals of natural selection (i.e. FST outlier loci) can be found across chosen S. scardica populations, we performed AFLP fingerprinting followed by comprehensive analyses of adaptive genetic variation and underlying drivers of adaptive divergence. In addition, possible influences of past climatic oscillations on population genetic structure and diversity of studied populations were addressed.
Results
Within population diversity
The proportion of polymorphic markers varied from 25.37% in population P05 Mariovo to 50.00% in P07 Mt. Vermio. Private alleles were detected in all the analyzed populations and P08 Mt. Olympus had the highest number, 15 respectively. Shannon’s information index (I) indicated that the total diversity (Ht) was 0.417 with average intrapopulation diversity (Hp) being 0.238. Similar levels of diversity within populations (Hp/Ht) (57.20%) and among populations (42.80%) were determined. Shannon’s index per population ranged from 0.175 (P05 Mariovo) to 0.305 (P07 Mt. Vermio) (Fig. 1a). Estimates of frequency down-weighted marker values (DW) were above average for five populations (P04, P06-P09) (Fig. 1b). Expected heterozygosity (HE) ranged from 0.095 (P06 Mt Kozuf) to 0.133 (P07 Mt. Vermio) (Table 1).
Genetic diversity and structure of 12 Sideritis scardica populations from North Macedonia and Greece: Shannon’s information index (a), Frequency down-weighted marker values (b), Genetic structure derived from Bayesian analysis using STRUCTURE at K = 2 (c) and K = 7 (d). In (a) and (b), the size of the dots is directly proportional to the depicted values.
Genetic relationships and population structure
The coefficient of genetic differentiation (FST) ranged from 0.115 between P02 Mt. Shara and P03 Mt. Suva Gora to 0.408 between P01 Mt. Skopska Crna Gora and P06 Mt. Kozuf, with an average value of 0.263. The Analysis of Molecular Variance (AMOVA) further showed that most of the genetic diversity was attributable to differences among individuals within populations (63.25%). However, φST value for the among-populations component was significant (φST = 0.367, P < 0.0001) (Table 2), suggesting the existence of population differentiation. Pairwise φST values ranged from 0.202 between P02 Mt. Shara and P03 Mt. Suva Gora to 0.534 between P01 Mt. Skopska Crna Gora and P06 Mt. Kozuf.
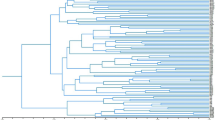
The Neighbor-joining tree based on Dice’s distance matrix showed that all the individuals belonging to the same population were consistently grouped together (Supplementary Fig. S1). Bootstrap support was strong (>90%) in the case of population clusters P05 Mariovo, P06 Mt. Kozuf, P08 Mt. Olympus and P09 Mt. Paggaio, all sampled in the south-eastern part of the studied area. Bootstrap values were moderate for P04 Mt. Ilinska (81%) and weak for P03 Mt. Suva Gora (57%), while all the other population clusters were not supported, having bootstrap values lower than 50%. The cluster comprised of populations P01 Mt. Skopska Crna Gora, P02 Mt. Shara, P03 Mt. Suva Gora, P04 Mt. Ilinska, and P07 Mt. Vermio showed weak bootstrap support (69%).
Model-based clustering analysis using STRUCTURE revealed the same pattern of grouping as the distance-based method. Average estimates of the likelihood of the data, conditional on a given number of clusters, ln [Pr(X|K)], kept increasing with higher K as did the standard deviations among different runs for each K. The highest ΔK was observed for K = 2 (826.03), followed by that of K = 7 (9.89) (Supplementary Fig. S2). At K = 2, all the individuals belonging to populations sampled in the north-western part of the studied area, i. e. P01 Mt. Skopska Crna Gora, P02 Mt. Shara and P03 Mt. Suva Gora were assigned to cluster A; while cluster B included all the individuals from P05 Mariovo, P06 Mt. Kozuf, P08 Mt. Olympus and P09 Mt Paggaio, sampled in the south-eastern part of the studied area (Fig. 1c). North-western montane population P04 Mt. Ilinska and south-eastern colline population P07 Mt. Vermio were admixed between the two clusters.
At K = 7, each population formed its respective cluster, except for the populations P02 Mt. Shara and P07 Mt. Vermio, that were admixed between two clusters (Fig. 1d). Population P02 Mt. Shara was admixed between clusters A consisting of individuals from population P01 Mt. Skopska Crna Gora and B including individuals from population P03 Mt. Suva Gora, while the individuals from population P07 were assigned either to cluster A or to cluster C together with all the individuals from P04 Mt. Ilinska.
BAPS (Bayesian Analysis of Population Structure) mixture analyses with or without spatially informative priors yielded nearly identical results to those obtained by STRUCTURE at K = 7 (Supplementary Fig. S3). The best partitions received log marginal likelihoods of -17761,59 at P = 1 (without using geographic coordinates as informative priors) and -17937,18 at P = 1 (spatial clustering). In both cases, each population formed its respective cluster, except for the populations P02 Mt. Shara and P07 Mt. Vermio, that were admixed between two clusters.
In both Discriminant Analyses of Principal Components (DAPC) performed with and without prior information of individual population membership we retained 50 principal components representing 77.97% of total genetic variation. The scatterplot of the first analysis (with prior; Fig. 2a) showed that along the first principal component the populations P05 Mariovo, P06 Mt. Kozuf, P08 Mt. Olympus and P09 Mt. Paggaio were separated from the remaining populations, that were positioned close to each other. In the second analysis (without prior) we covered a range of possible clusters from 1 to 10. The lowest BIC value (556) corresponded to K = 7 (Supplementary Fig. S4). The clusters A, B and C consisted of individuals from more than one population: cluster A grouped individuals belonging to P01 Mt. Skopska Crna Gora, P02 Mt. Shara and P07 Mt. Vermio, cluster B from P02 Mt. Shara and P03 Mt. Suva Gora, while cluster C grouped individuals from P04 Mt. Ilinska and P07 Mt. Vermio (Fig. 2b). On the other hand, clusters D, E, F and G corresponded completely with the population memberships of the individuals: D (P05 Mariovo), E (P06 Mt. Kozuf), F (P08 Mt. Olympus) and G (P09 Mt. Paggaio). In congruence with the results of model-based clustering analysis using both STRUCTURE (at K = 7) and BAPS, the DAPC analysis without prior information of individual population membership revealed that populations P02 Mt. Shara and P07 Mt. Vermio consisted of individuals grouped in more than one cluster, while the rest of the populations belonged to a single genetic cluster.
Scatterplot of Sideritis scardica samples fromNorth Macedonia and Greece on the two principal components of Discriminant Analysis of Principal Components (DAPC) obtained with prior information of individual population membership (a: nine populations) and without that information (b: seven clusters). Eigenvalues of the analysis are shown as an inset for each graph with dark gray bars representing those used in the scatterplot.
Isolation by distance
The Mantel test was significant, and we found a positive correlation between pairwise genetic distances and spatial distances among all nine populations (r = 0.397, PMantel = 0.017), and 15.7% of the genetic variation could be explained by geographical distances (Supplementary Fig. S5).
Principal Component Analysis
The 19 bioclimatic variables used in the study were highly inter-correlated. Out of 171 pairwise examinations, a strong positive correlation (r > 0.70) was found in 61 cases, while in 58 cases a strong negative correlation (r < −0.70) was identified (Supplementary Table S1). A Principal Component Analysis based on the correlation matrix revealed that the first three principal components had an eigenvalue ˃1, jointly explaining 94.39% of the total variation. Ten temperature-related variables (BIO01, BIO02, BIO04-BIO11) exhibited a strong positive correlation (r > 0.70), while seven precipitation-related variables (BIO12-BIO14, BIO016-BIO019) exhibited a strong negative correlation (r < −0.70) with the first principal component (PC1) that explained as much as 76.52% of the total variation (Table 3). With the second principal component (PC2), explaining 11.68% of the total variation, only a single variable was strongly negatively correlated [Precipitation Seasonality (BIO15)]. The biplot constructed by the two principal components showing populations and bioclimatic variables (as vectors) is presented in Fig. 3. Along the PC1 the north-western montane populations (P02 Mt. Shara, P03 Mt. Suva Gora and P04 Mt. Ilinska) were clearly separated from colline populations (P01 Mt. Skopska Crna Gora, P05 Mariovo, P07 Mt. Vermio), while south-eastern montane populations (P06 Mt. Kozuf, P08 Mt. Olympus, P09 Mt. Paggaio) were positioned near the center of the plot. The sampling sites of the north-western montane populations were characterized by substantially higher precipitation and lower temperature, in comparison to the sampling sites of colline populations while the sampling sites of south-eastern montane populations displayed the intermediate values. The PC2 separated populations P08 Mt. Olympus and P09 Mt. Paggaio from the rest as those two sampling sites exhibited above-average values of Precipitation Seasonality (BIO15).
Biplot of Principal Component Analysis (PCA) based on 19 bioclimatic variables of nine Sideritis scardica sampling sites. The temperature-related variables (BIO01-BIO11) are shown as red vectors while the precipitation-related variables (BIO12-BIO19) are shown as blue vectors. The colline populations (P01, P05, P07) are shown in red, the north-western montane populations (P02, P03, P04) in blue and the south-eastern montane populations (P06, P08, P09) in green.
Adaptive genetic variation
A total of 226 AFLP markers were used to identify the outlier loci (after the removal of markers with a frequency below 3% or above 97%). With a confidence level set to 99%, Mcheza detected a total of 38 outlier loci (16.81%), possibly under selection, among which 12 (5.31%) are/were under directional and 26 (11.50%) under balancing selection (Fig. 4a). BayeScan identified seven loci (3.10%) exceeding the threshold for very strong evidence of selection [False Discovery Rate (FDR < 0.01; posterior odds (PO) = 37; log10 (PO) = 1.697], none of them under stabilizing selection. All seven loci were also detected by Mcheza as outliers (Fig. 4b). After calculating logistic regressions between all possible marker/bioclimatic variable pairs (a total of 4,294 models), Samβada detected 190 (4.42%) significant models involving 50 markers (22.12%) correlated with one up to 17 bioclimatic variables. Bioclimatic variables associated with 15 or more markers were Isothermality (BIO03), Precipitation Seasonality (BIO15), Precipitation of Driest Quarter (BIO17) and Precipitation of Warmest Quarter (BIO18). Out of 50 markers detected by Samβada, seven were identified by both Mcheza and BayeScan (Table 4). Thus, out of a total of 226 markers, seven (3.10%) were identified across three methods as presented in the Venn diagram illustrating the overlap in outlier detection (Fig. 4c). All 19 bioclimatic variables were associated with at least one outlier marker. In general, precipitation-related variables (BIO12-BIO19) were associated with more outlier markers (from one to five) than temperature-related variables (BIO01-BIO11; from one to three). All four bioclimatic variables associated with five out of seven outlier markers were related to precipitation: Annual Precipitation (BIO12), Precipitation of Driest Month (BIO14), Precipitation of Driest Quarter (BIO17) and Precipitation of Warmest Quarter (BIO18).
Identification of FST outlier loci using (a) Mcheza and (b) BayeScan, and (c) the Venn diagram summarizing the number of loci identified as FST outlier loci by Mcheza and BayeScan and significantly associated with environmental variables by Samβada. In (A) FST values were plotted against its heterozygosity (HE). The dashed lines represent the 99% confidence intervals. Loci under positive selection are indicated as black dots, those under balancing selection as white dots and neutral as grey dots. Loci under positive selection detected also by BayeScan are underlined while those identified by Samβada are shown in italics. In (B) FST values were plotted against the log10 of the posterior odds (PO). The vertical line shows the critical PO used for identifying outlier markers [FDR < 0.01; PO = 49.76; log10(PO) = 1.697]. Loci under positive selection detected also by Mcheza are underlined while those identified by Samβada are shown in italics.
Discussion
Genetic diversity and structure
The obtained results demonstrated relatively low within population genetic diversity and significant population differentiation. Within population diversity in terms of polymorphic markers (average 36.21%) and gene diversity (average 0.11) was somewhat higher for the south-eastern S. scardica populations. However, overall gene diversity levels were lower than the values reported by Nybom48 for the endemic plants using dominant markers (HE = 0.20). Private bands were detected in all analyzed populations, ranging from 3 to 15. Generally, south-eastern populations tend to have more private bands and higher DW values. In concordance with detected private bands, genetic differentiation among analyzed populations was significant (FST = 0.263), indicating moderate population differentiation. Sampled S. scardica populations are distributed among fragmented mountain ranges, separated by deep valleys and consequently, pollen and seed dispersal among them is limited. A positive and significant correlation between genetic and geographical distances revealed a pattern of isolation by distance across the distribution range of S. scardica and further confirmed limited gene flow between analyzed populations, which facilitates the establishment of local adaptations. However, more continuous distribution in the recent past and historical gene flow among analyzed populations is a possibility. Evidence for historical gene flow between neighboring populations of S. scardica is provided from both distance and model-based methods, i. e. populations with closer proximity to each other are generally also genetically closer. During the Quaternary, suitable habitats for mountain species became limited and less available40 and it can be assumed that the species survived by altitudinal shifts to adjacent lowlands, as it has been proposed for other mountain biota34,49, followed by postglacial remigration to higher altitudes. Such elevational shifts have been documented for other cold-adapted taxa, i. e. arctic and alpine species which escaped the postglacial warming to cooler climates by altitudinal and/or latitudinal range shifts50. Furthermore, it is possible that during these colder periods when the species presumably occupied significantly lower altitudes than today, inter-population hybridization involving different gene pools easily occurred, at least in some areas. As a consequence, in areas where genetically divergent groups of populations came into contact, admixed population-genetic patterns comprising of elements from different gene pools (07 Mt. Vermio) may be observed. During the post-glacial period, remigration to higher altitudes resulted in inter-population isolation. However, this recolonization happened relatively recently and there was not enough time for the development of more pronounced differentiation among populations. Some of the obtained results support this hypothesis, e. g. population P09 Mt. Paggaio, which is geographically isolated from populations P04 Mt. Ilinska, P05 Mariovo and P06 Mt. Kozuf in present days originates from a common ancestral population as revealed by STRUCTURE analysis at K = 2. For population P09 Mt. Paggaio, we presume that it survived isolated for a longer period. Our estimates of DW values (201.60) and detected number of private alleles (13) support this idea as their higher values are expected in long-term isolated populations where rare markers accumulate due to mutations41.
When discussing the detected levels of differentiation, it can also be assumed that because of already discussed climatic oscillations and resulting migrations along the altitudinal gradient, at least some of these populations have experienced severe fluctuations in size and consequently a strong genetic drift. Since the genetic drift, as well as its most extreme form (i.e. the bottleneck), is a stochastic fluctuation in allele frequencies, it could not only cause the loss of the genetic variation, but also increase the amount of genetic differentiation among populations51,52. When a population is founded by a limited number of individuals (i.e. through either bottleneck or founder effect), it is expected that its genetic composition may substantially differ from source population and other populations founded through the same processes, and from the identical source population, simply because of stochastic nature of genetic drift53,54. Consequently, it is possible to assume that at least some of the detected genetic differentiation originates from genetic drift, followed by the absence of inter-population gene flow, rather than solely natural selection.
Overall, the results implicate that connectivity among populations might have been maximal during the glaciation periods and interrupted in the post-glacial period with remigration to higher altitudes, resulting in significant and moderate levels of genetic differentiation among populations.
Adaptive divergence
A total of 226 AFLP markers were analyzed to find the signals of divergent selection among nine natural S. scardica populations sampled in environments reflecting variation in altitude, precipitation and temperature, by performing widely used FST-outlier methods Mcheza and BayeScan, and a genotype-environment association analysis with Samβada. Mcheza detects loci with unusually high or low FST values using the frequentist method, while BayeScan uses the Bayesian approach and directly estimates the posterior probability that a given locus is under selection by defining two alternative models (with and without the effect of selection) (See Materials and Methods Section). In our study, Mcheza revealed 16.81% loci possibly under selection, among which 5.31% of loci exhibited higher FST values and 11.50% of loci exhibited lower FST values than the majority of loci, while BayeScan detected the lower proportion of loci under divergent selection (3.10%) than Mcheza. In the AFLP study on Ceracris kiangsu Tsai & P., the authors found the proportion of outliers detected by Dfdist to be 7.6% and by BayeScan 6.7%, indicating the more conservative nature of Bayesian method20. The comparison between the performance of the two software by different authors resulted in the identical conclusion that BayeScan performs more efficiently, as it detects a high percentage of outlier loci with a low proportion of false positives15,55,56 (discussed further in the chapter).
The three implemented methods (i. e. Mcheza, BayeScan and Samβada) identified seven outlier loci (3.1%) of high reliability in studied S. scardica populations, as they were identified by complementary and exhaustive methods. This proportion is in concordance with findings of Hoffmann and Willi3 which reported less than 5% of outlier loci, but bellow the values reported by Nosil et al.32, 5–10% respectively. Similar research on an invasive weed Mikania micrantha Kunth detected 14 outlier loci by both Dfdist and BayeScan corresponding to 2.9% of 483 investigated loci57. In Eruca sativa Mill. nine candidate loci were identified, however, only three of them (1.6%) were detected by more than one method26. A significantly higher proportion of outliers was detected in Geropogon hybridus (L.) Sch. Bip. Eleven outlier loci (8.9%) were detected by Mcheza and BayeScan as well as spatial analysis method (SAM) to be associated with precipitation21.
It is expected that outlier loci occur only sporadically throughout the genome, thus involving only its minor part in a process of divergent selection58,59. Consequently, the results obtained in this study, which indicate that only 3.1% of the studied genome was influenced by intense natural selection, are not surprising. Since no prior knowledge of the genome structure of the studied species exists, we do not yet know the location or function of the detected outlier loci.
In addition, although it is expected that AFLP loci mostly originate from non-coding regions of genome60,61, it can be assumed that because of the ‘hitchhiking effect’ at least some of the detected loci are linked to the actual target60 and can thus be treated as the carriers of selection signature. Nevertheless, based on obtained results from PCA and Samβada analysis, we concluded that divergent selection detected among studied populations likely emerged because of their exposure to contrasting precipitation conditions. Although AFLP technique targets genome-wide anonymous loci that represent only a small fraction of the whole genome, it can be assumed that detected outlier loci are representatives of those genomic regions that evolved more rapidly than the remaining parts of the genome, thus enabling organisms’ adaptation to contrasting precipitation conditions and divergence of ecotypes. Temperature and precipitation are the parameters that strongly affect the survival of alpine species62 and probably act as the main triggers of their selective responses63. Numerous studies have revealed an important role of environmental factors in driving species adaptation. For example, temperature, precipitation and radiation were the three main factors affecting the local adaptation of Liriodendron chinense (Hemsl.) Sarg. whereas nine AFLP loci showed evidence of being outliers for population differentiation in both Dfdist and BayeScan detection methods30. Temperature and precipitation were the main drivers of adaptive genetic variation in 13 alpine plant species collected across the entire European range25. In Cotinus coggygria Scop. five ecologically relevant microsatellite alleles related to the precipitation were identified28. Investigations on Arabis alpina L. sampled on 208 locations across French and Swiss Alps identified 78 loci of ecological relevance that were mainly related to mean annual minimum temperature23. In Picea abies L. Karst sampled in the South-Eastern Alps 13 potentially adaptive loci mainly associated with precipitation were identified29.
The number of marker loci required to infer the genetic structure and to detect local adaptation is especially important in case of dominant markers having a lower information content as compared to co-dominant markers. Bonin et al.64 reported that using 150 AFLP loci instead of 300 had a negligible effect on the estimates of genetic differentiation (FST). Nelson and Anderson65 used simulated dominant marker sets to determine the number of loci needed to obtain satisfactory results from the analysis of molecular variance (AMOVA) and Bayesian model-based cluster analysis (STRUCTURE). They concluded that the minimum number of loci needed depends on the level of genetic differentiation among populations as measured by AMOVA’s φST and that if φST is 0.3 or greater, adequate results can be achieved with only 45 to 90 loci. Leipold et al.66 investigated the minimum number of individuals and the required number of AFLP loci to describe a population’s genetic diversity using resampling procedure based on real data of 15 plant species and reported that approximately 120 loci were sufficient for a stable estimation of genetic diversity, while 14 individuals per population were needed to cover 90% of the total genetic diversity. Although simulation-based studies used to assess the power to detect markers under selection14,67 did not discuss the minimum number of markers, it is clear that FST-outlier analysis is univariate in nature but there must be a sufficient number of markers to obtain a reliable estimate of allele frequencies in populations and subsequently an unbiased estimate of FST (and other population genetic parameters). Thus, if the number of loci is sufficient to infer the population genetic parameters, the same would hold also for outlier detection. The problem that remains is that with the decreasing number of markers, the chance of not finding any being an outlier increases. However, the modest number of markers should not increase the probability of false-positives as compared to genome-wide association studies with millions of SNPs. Hoban et al.68 highlighted the importance of a sufficient number of sampled locations and individuals to maximize the power of local adaptation studies and stated that the methods based on allele frequencies require more than 10 samples from each sampling site. The authors also pointed out that the most empirical studies involve from 100 to 1,000 individuals from 5 to 40 locations.
The occurrence of false positives is a typical problem encountered with the FST outlier detection methods26,33,69 and can be the result of weak genome coverage of AFLP markers21, statistical departures from the model assumptions, population structure70, and demographic processes. Demographic processes, such as bottlenecks, secondary contacts, allele surfing during population range expansion, and isolation by distance can create similar genomic patterns that imitate selection1,71, and complicate the process of distinguishing selection from demography72. The effective strategy to reduce the rate of false positives and increase confidence in identified outliers is simultaneous use of methods based on different assumptions and parameters57, as it has been done in this study. The comparison of the results obtained with these different approaches represents the cross-validation or a double-check and increases the reliability of the identified loci potentially under selection22. To further minimize the false discovery rate, we set the stringent threshold levels permitted by the outlier detection methods (see Materials and Methods section). Seven outlier loci (3.1%) that were concordantly detected by all three implemented methods (Mcheza, BayeScan and Samβada) were regarded as of adaptive relevance. However, further exploration of functional mechanism operating at putatively adaptive loci is required.
According to De Mita73 spatial analyses methods used to measure associations between allele frequencies and environmental variables are prone to false positives, as they do account for population structure. As stated by Muller et al.21 the existence of isolation by distance pattern can result in false positive associations between environment and gene frequencies. To overcome this problem and increase the power to detect selected loci and minimize the occurrence of false positive associations, Lotterhos and Whitlock69 suggest the sampling design based on comparing geographically closer populations, that have the similar genetic constitution for neutral genes, but thrive in different environmental conditions and are differentiated by selection. In our study, the sampling scheme was as far as possible followed by that proposed by Lotterhos and Whitlock69, i.e. comparison between geographically nearby populations (P01/P02, P05/P06 and P07/P08) sampled in different environmental conditions and altitudes was performed.
Conclusion and further implications
The present work was focused on the assessment of genetic and adaptive diversity, and population differentiation in Sideritis scardica, as well as the identification of environmental factors that are responsible for the revealed genetic structuring. We believe that the obtained results could be applied in future conservation management, whose development is of utmost importance for S. scardica, for two general reasons. First, as an alpine species it is especially vulnerable to the ongoing climate changes, as highlighted for a number of cold-adapted species74,75,76, and second, as an important medicinal plant, it is endangered, mostly because of human-mediated overexploitation77.
Two differentiation-based methods (Mcheza and BayeScan) and genotype-environment association analysis implemented in Samβada enabled us to identify loci that are potentially linked to the genes under selection, and precipitation as the key environmental driver of selection in S. scardica. AFLP markers are widely employed in the genomic investigations of non-model species9,10. However, due to their dominant nature and the fact that Hardy–Weinberg equilibrium cannot be tested, but it must be assumed to assess allele frequencies, sampling errors are likely to occur78. Additionally, inaccurate allele frequency estimates could also be generated due to a modest sampling size68. Therefore, the interpretation of the results should be treated with caution and the identified candidate loci could provide a priori hypotheses for further comprehensive analysis of the adaptive divergence.
Further work needs to be done to characterize the identified outliers, i. e. to identify chromosomal locations and functional mechanisms operating at loci of adaptive relevance. As climate changes are manifested through precipitation and temperature changes, validating genetic and phenotypic variation of different abiotic stresses tolerance in S. scardica populations is of utmost importance. This could be done through genotype-phenotype association studies, where genotypic variation could be linked to phenotypic variation in drought and temperature tolerance related traits, and other mechanisms that affect the persistence of the species. The proposed approach could help conservationist to assess adaptive phenotypes and thereby improve the effectiveness of conservation practices. However, considering the fact that this research involved only a part of the natural geographical distribution of S. scardica the obtained results should be validated further for the entire distribution range of the species.
For S. scardica in situ strategies should also be developed in order to protect its natural habitats, thereby conserving the overall genetic diversity of the species. Populations that are the most endangered and at imminent risk of extinction should be prioritized for conservation. Ex situ strategies should be carried out by storing germplasm in ex situ field collections and long-term germplasm storage facilities. It is crucial that principles and procedures for the sustainable collection of the species from the wild are adopted by collectors and all the stakeholders involved. Moreover, the genetic analysis should support further breeding research, contributing thus to the successful introduction of the species into cultivation and its sustainable exploitation as it is a promising candidate due to valuable medicinal properties and long tradition of use. Also, identified adaptive genotypes should be introduced to the breeding programs to conserve the adaptive potential of S. scardica.
Materials and Methods
Sampling
Leaves from nine Sideritis scardica natural populations were collected throughout species distribution range in North Macedonia and Greece (Codes MKD and GRC, respectively). The sampling sites were chosen to represent different habitats. Three populations (P02 Mt. Shara, P03 Mt. Suva Gora, P04 Mt. Ilinska) were sampled in the north-western montane region of the studied area, at an altitude above 1,400 m a.s.l. Three populations (P06 Mt. Kozuf, P08 Mt. Olympus, P09 Mt. Paggaio) were sampled in the south-eastern montane region, at an altitude above 1,400 m a.s.l. Furthermore, three colline populations were sampled from lower altitudes (<1,000 m a.s.l.); thus, one population (P01 Mt. Skopska Crna Gora) was sampled in the north-western part and two populations (P05 Mariovo, P07 Mt. Vermio) in the south-eastern part of the studied area (Table 1).
Altitude was measured by a GPS device and bioclimatic data was extracted from the WorldClim database (http://www.worldclim.org/) for each collecting site. The sampling sites were described with 19 temperature and precipitation bioclimatic variables (BIO01-BIO19), representing the annual trends, seasonal variations, and extremes in temperature and precipitation (Table 3).
DNA extraction and AFLP analysis
Total genomic DNA was isolated from individual young top fresh leaves collected in situ using the DNeasy Plant Mini Kit (Sigma Aldrich®, St. Louis, Missouri, USA). The DNA concentrations were measured with a Qubit Fluorometer (InvitrogenTM, Carlsbad, California).
The AFLP analysis was carried out following the original protocol described by Vos et al.79, with minor modifications Carović-Stanko et al.80. The restriction, ligation and all amplification reactions were performed in GeneAmp PCR System 9600 (Applied Biosystems, Foster City, CA, USA). For selective amplification, four primers combinations were used: FAM-EcoRI-ACA + MseI-CAC, NED-EcoRI-AGA + MseI-CAC; VIC-EcoRI-ACG + MseI-CGA, PET-EcoRI-ACC + MseI-CGA.
The amplified fragments were separated by capillary electrophoresis in an ABI3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Data analysis
To construct a binary matrix, the obtained AFLP fragments were scored as being present (1) or absent (0). Within-population molecular diversity was assessed in terms of the proportion of polymorphic markers, the number of private markers (Npr) and Shannon’s information index (I). Shannon’s information index was computed by using the following formula I = −Σ (pi log2 pi), where pi is the phenotypic frequency81,82. Shannon’s information index was used to determine the total diversity (Ht), average intra-population diversity (Hp) and the proportions of diversity within (Hp/Ht) and among populations [(Ht–Hp)/Ht]. AFLPdat83 was used for assessing the frequency down-weighted marker values (DW)41.
To calculate the allelic frequencies at AFLP marker loci, a Bayesian approach suggested by Zhivotovsky84 as implemented in AFLP-Surv v. 1.085 was used, assuming Hardy-Weinberg equilibrium due to the outcrossing nature of S. scardica. The calculated allelic frequencies were further used in the analysis of genetic diversity within and between populations86. The total gene diversity (HT), the average gene diversity within populations (HW), the average gene diversity among populations in excess of that observed within populations (HB), and Wright’s FST statistics were assessed to describe the population genetic structure.
Dice’s distance between individual plants was obtained as 1-Dice’s similarity index87 and a Neighbour Joining88 tree was constructed. Statistical support was tested with bootstrap analysis using 1,000 replicates89. The calculations were made using PAST version 3.2290. Scores between 50 and 74 bootstrap percentages (BS) were defined as weak support, scores between 75 and 89% BS as moderate, and scores greater than 90% BS as strong support.
Total genetic variation among and within populations was analyzed by AMOVA91 with the software Arlequin ver. 3.592. The variance components were tested using 10,000 permutations. Pairwise population comparisons resulted in values of φST that are equivalent to the proportion of the total variance that is partitioned between two populations and could be interpreted as the inter-population distance average between any two populations93.
Bayesian model-based cluster analysis was performed on multilocus AFLP data by using the software STRUCTURE ver. 2.3.394. Ten runs per each K were performed by setting the number of clusters (K) from 1 to 11. Each run consisted of a burn-in period of 200,000 steps followed by 106 MCMC (Monte Carlo Markov Chain) replicates assuming admixture model and correlated allele frequencies. The calculations were carried out on the Isabella computer cluster at the University of Zagreb, University Computing Centre (SRCE). The optimal number of clusters (K) was assessed by calculating an ad hoc statistic ΔK95 as implemented in STRUCTURE HARVESTER v0.6.9496.
The software BAPS 6.097,98,99 was applied for the population mixture analysis without the geographic origin of the samples used as an informative prior (‘clustering of individuals’) and with this prior (‘spatial clustering of individuals’)100. BAPS software was run setting the maximum number of clusters (K) to 10 and each run was replicated 10 times. Results of the mixture analysis were used as input for population admixture analysis98, with the default settings.
As an alternative to Bayesian clustering, population structure was also assessed using Discriminant Analysis of Principal Components (DAPC)101 as implemented in the package adegenet v. 2.0.1102 for the software R v. 3.3.1103. The analyses were carried out both with and without prior information of individual population membership. The analysis without prior information was performed using the function ‘find clusters’ and the optimal number of clusters was selected based on Bayesian Information Criterion (BIC).
To test the significance of the isolation by distance (IBD)104 among S. scardica populations we carried out a Mantel test with 10,000 permutations as implemented in NTSYS-pc ver. 2.10s105.
A Principal component analysis (PCA) was performed on 19 bioclimatic variables obtained from the Worldclim database for Sideritis scardica L. sampling sites. We constructed the biplot with two principal components (PC) displaying sampled populations and bioclimatic variables (as vectors).
The detection of candidate loci under selection was carried out using two basic approaches: by identification of FST outlier loci and by correlating genetic variation with environmental variables. The identification of FST outlier loci was performed using the frequentist method11 implemented in Mcheza13 and the Bayesian method implemented in BayeScan ver. 2.0114. The associations between genetic variation and environmental variables were assessed using the spatial analysis method106 as implemented in Samβada107. AFLP markers with a low minor allele frequency (below 5%) were removed from the dataset since they systematically bias the FST estimates108. The frequentist method for the identification of FST outlier loci implemented in Mcheza detects loci with unusually high or low FST values. Loci under directional selection are expected to display significantly higher FST values than the majority of neutral loci in a sample, while loci with significantly lower FST values are considered to be under stabilizing selection. The neutral distribution of FST values was simulated using 106 iterations with ‘Neutral mean FST’ and ‘Force mean FST’ options. Results were corrected for multiple comparisons by setting the confidence interval (CI) to 99% and the false discovery rate (FDR) to 0.1.
Bayesian approach for the identification of FST outlier loci implemented in BayeScan directly estimates the posterior probability that a given locus is under selection by defining two alternative models, one with and other without the effect of selection. Twenty pilot runs of 5,000 iterations were used to adjust the proposal distribution to acceptance rates between 0.25 and 0.45 for the runs. A burn-in of 50,000 iterations was used, followed by 500,000 iterations using a thinning interval of 50. We used prior odds of 10, corresponding to a prior belief that the model with selection is ten times less likely that the model without selection. The logarithm of Posterior Odds [log10(PO)] higher than 1.5 was taken as a ‘very strong’ evidence for selection14,109. The multitest correction on false discovery rates (FDR) was set at 0.01 to avoid overestimating the percentage of outliers.
A spatial analysis method implemented in Samβada uses the logistic regression approach to estimate the probability that an individual carries a specific genetic marker given the environmental conditions of its sampling site. The associations between genetic markers and environmental variables were assessed with both log-likelihood G ratio and Wald test using Bonferroni correction for multiple hypothesis testing (P < 0.01).
Data Availability
The datasets generated during and/or analyzed during this investigation are available from the corresponding author upon request.
References
Schoville, S. D. et al. Adaptive Genetic Variation on the Landscape: Methods and Cases. Annu Rev Ecol Evol S 43, 23–43 (2012).
Nielsen, R. Molecular signatures of natural selection. Annu Rev Genet 39, 197–218 (2005).
Hoffmann, A. A. & Willi, Y. Detecting genetic responses to environmental change. Nat Rev Genet 9, 421–432 (2008).
Dillon, S. et al. Characterisation of Adaptive Genetic Diversity in Environmentally Contrasted Populations of Eucalyptus camaldulensis Dehnh. (River Red Gum). Plos One 9, https://doi.org/10.1371/journal.pone.0103515 (2014).
Lewontin, R. C. & Krakauer, J. Distribution of Gene Frequency as a Test of Theory of Selective Neutrality of Polymorphisms. Genetics 74, 175–195 (1973).
Luikart, G., England, P. R., Tallmon, D., Jordan, S. & Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat Rev Genet 4, 981–994 (2003).
Bonin, A., Taberlet, P., Miaud, C. & Pompanon, F. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frog (Rana temporaria). Mol Biol Evol 23, 773–783 (2006).
Storz, J. F. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol 14, 671–688 (2005).
Stinchcombe, J. R. & Hoekstra, H. E. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100, 158–170 (2008).
Ekblom, R. & Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107, 1–15 (2011).
Beaumont, M. A. & Nichols, R. A. Evaluating loci for use in the genetic analysis of population structure. P Roy Soc B-Biol Sci 263, 1619–1626 (1996).
Antao, T., Lopes, A., Lopes, R. J., Beja-Pereira, A. & Luikart, G. LOSITAN: A workbench to detect molecular adaptation based on a F(st)-outlier method. Bmc Bioinformatics 9, https://doi.org/10.1186/1471-2105-9-323 (2008).
Antao, T. & Beaumont, M. A. Mcheza: a workbench to detect selection using dominant markers. Bioinformatics 27, 1717–1718 (2011).
Foll, M. & Gaggiotti, O. A Genome-Scan Method to Identify Selected Loci Appropriate for Both Dominant and Codominant Markers: A Bayesian Perspective. Genetics 180, 977–993 (2008).
Pérez-Figueroa, A., Garcia-Pereira, M. J., Saura, M., Rolan-Alvarez, E. & Caballero, A. Comparing three different methods to detect selective loci using dominant markers. J Evolution Biol 23, 2267–2276 (2010).
Stucki, S. et al. High-performance computation of landscape genomic models including local indicators of spatial association. Mol Ecol Resour 17, 1072–1089 (2017).
Pariset, L., Joost, S., Marsan, P. A., Valentini, A. & Ec. Landscape genomics and biased FST approaches reveal single nucleotide polymorphisms under selection in goat breeds of North-East Mediterranean. Bmc Genet 10, https://doi.org/10.1186/1471-2156-10-7 (2009).
Nunes, V. L., Beaumont, M. A., Butlin, R. K. & Paulo, O. S. Multiple approaches to detect outliers in a genome scan for selection in ocellated lizards (Lacerta lepida) along an environmental gradient. Mol Ecol 20, 193–205 (2011).
Cerwenka, A. F., Brandner, J., Geist, J. & Schliewen, U. K. Strong versus weak population genetic differentiation after a recent invasion of gobiid fishes (Neogobius melanostomus and Ponticola kessleri) in the upper Danube. Aquat Invasions 9, 71–86 (2014).
Feng, X. J., Jiang, G. F. & Fan, Z. Identification of outliers in a genomic scan for selection along environmental gradients in the bamboo locust, Ceracris kiangsu. Sci Rep-Uk 5, https://doi.org/10.1038/srep13758 (2015).
Muller, C. M. et al. Geropogon hybridus (L.) Sch.Bip. (Asteraceae) exhibits micro-geographic genetic divergence at ecological range limits along a steep precipitation gradient. Plant Syst Evol 303, 91–104 (2017).
Manel, S., Poncet, B. N., Legendre, P., Gugerli, F. & Holderegger, R. Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Mol Ecol 19, 3824–3835 (2010).
Poncet, B. N. et al. Tracking genes of ecological relevance using a genome scan in two independent regional population samples of Arabis alpina. Mol Ecol 19, 2896–2907 (2010).
Bothwell, H. et al. Identifying genetic signatures of selection in a non-model species, alpine gentian (Gentiana nivalis L.), using a landscape genetic approach. Conserv Genet 14, 467–481 (2013).
Manel, S. et al. Broad-scale adaptive genetic variation in alpine plants is driven by temperature and precipitation. Mol Ecol 21, 3729–3738 (2012).
Westberg, E., Ohali, S., Shevelevich, A., Fine, P. & Barazani, O. Environmental effects on molecular and phenotypic variation in populations of Eruca sativa across a steep climatic gradient. Ecology and evolution 3, 2471–2484 (2013).
Fang, J. Y. et al. Divergent Selection and Local Adaptation in Disjunct Populations of an Endangered Conifer, Keteleeria davidiana var. formosana (Pinaceae). Plos One 8, https://doi.org/10.1371/journal.pone.0070162 (2013).
Lei, Y. K., Wang, W., Liu, Y. P., He, D. & Li, Y. Adaptive genetic variation in the smoke tree (Cotinus coggygria Scop.) is driven by precipitation. Biochem Syst Ecol 59, 63–69 (2015).
Di Pierro, E. A. et al. Climate-related adaptive genetic variation and population structure in natural stands of Norway spruce in the South-Eastern Alps. Tree Genet Genomes 12, https://doi.org/10.1007/s11295-016-0972-4 (2016).
Yang, A. H., Wei, N., Fritsch, P. W. & Yao, X. H. AFLP Genome Scanning Reveals Divergent Selection in Natural Populations of Liriodendron chinense (Magnoliaceae) along a Latitudinal Transect. Front Plant Sci 7, https://doi.org/10.3389/Fpls.2016.00698 (2016).
Oberprieler, C., Zimmer, C. & Bog, M. Are there morphological and life-history traits under climate-dependent differential selection in S Tunesian Diplotaxis harra (Forssk.) Boiss. (Brassicaceae) populations? Ecology and evolution 8, 1047–1062 (2018).
Nosil, P., Funk, D. J. & Ortiz-Barrientos, D. Divergent selection and heterogeneous genomic divergence. Mol Ecol 18, 375–402 (2009).
Strasburg, J. L. et al. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos T R Soc B 367, 364–373 (2012).
Comes, H. P. & Kadereit, J. W. The effect of quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3, 432–438 (1998).
Taberlet, P., Fumagalli, L., Wust-Saucy, A. G. & Cosson, J. F. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7, 453–464 (1998).
Hofreiter, M. & Stewart, J. Ecological Change, Range Fluctuations and Population Dynamics during the Pleistocene. Curr Biol 19, R584–R594, https://doi.org/10.1016/j.cub.2009.06.030.
Feliner, G. N. Southern European glacial refugia: A tale of tales. Taxon 60, 365–372 (2011).
Podnar, M., Mayer, W. & Tvrtkovic, N. Mitochondrial phylogeography of the Dalmatian wall lizard, Podarcis melisellensis (Lacertidae). Org Divers Evol 4, 307–317 (2004).
Paun, O., Schonswetter, P., Winkler, M., Tribsch, A. & Consortium, I. Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Mol Ecol 17, 4263–4275 (2008).
Schönswetter, P., Stehlik, I., Holderegger, R. & Tribsch, A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol Ecol 14, 3547–3555 (2005).
Schönswetter, P. & Tribsch, A. Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae). Taxon 54, 725–732 (2005).
Surina, B., Schonswetter, P. & Schneeweiss, G. M. Quaternary range dynamics of ecologically divergent species (Edraianthus serpyllifolius and E. tenuifolius, Campanulaceae) within the Balkan refugium. J Biogeogr 38, 1381–1393 (2011).
Esra, M., Duman, H. & Ünal, F. Karyological studies onsection Empedoclia of Sideritis (Lamiaceae) from Turkey. Caryologia 62, 180–197 (2009).
Petrova, A. & Vladimirov, V. Red List of Bulgarian Vascular. Plants. Phytol Balcan 15, 63–94 (2009).
Strid, A., Tan, K. Mountain Flora of Greece, Volume 2. (eds Strid, A. & Tan, K.) 89–90 (Edinburgh University Press, 1991).
Papanikolaou, K., Kokkini, S. A taxonomic revision of Sideritis L. Section Empedoclia (Rafin) Bentham (Labiatae) in Greece in Aromatic Plants: Basic and Applied Aspects (ed. Margaris, N.) 101–128 (Martinus Nijhoff, 1982).
Petreska, J. et al. Potential bioactive phenolics of Macedonian Sideritis species used for medicinal “Mountain Tea”. Food Chem 125, 13–20 (2011).
Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13, 1143–1155 (2004).
Schmitt, T. Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool 4, https://doi.org/10.1186/1742-9994-4-11 (2007).
Varga, Z. S. & Schmitt, T. Types of oreal and oreotundral disjunctions in the western Palearctic. Biol J Linn Soc 93, 415–430 (2008).
Ellstrand, N. C. & Elam, D. R. Population Genetic Consequences of Small Population-Size - Implications for Plant Conservation. Annu Rev Ecol Syst 24, 217–242 (1993).
Young, A., Boyle, T. & Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11, 413–418 (1996).
Kolbe, J. J., Leal, M., Schoener, T. W., Spiller, D. A. & Losos, J. B. Founder Effects Persist Despite Adaptive Differentiation: A Field Experiment with Lizards. Science 335, 1086–1089 (2012).
Funk, W. C. et al. Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis). Mol Ecol 25, 2176–2194 (2016).
Narum, S. R. & Hess, J. E. Comparison of F-ST outlier tests for SNP loci under selection. Mol Ecol Resour 11, 184–194 (2011).
Vilas, A., Perez-Figueroa, A. & Caballero, A. A simulation study on the performance of differentiation-based methods to detect selected loci using linked neutral markers. J Evolution Biol 25, 1364–1376 (2012).
Wang, T., Chen, G. P., Zan, Q. J., Wang, C. B. & Su, Y. J. AFLP Genome Scan to Detect Genetic Structure and Candidate Loci under Selection for Local Adaptation of the Invasive Weed Mikania micrantha. Plos One 7, https://doi.org/10.1371/journal.pone.0041310 (2012)
Galindo, J., Moran, P. & Rolan-Alvarez, E. Comparing geographical genetic differentiation between candidate and noncandidate loci for adaptation strengthens support for parallel ecological divergence in the marine snail Littorina saxatilis. Mol Ecol 18, 919–930 (2009).
Kuchma, O. & Finkeldey, R. Evidence for selection in response to radiation exposure: Pinus sylvestris in the Chernobyl exclusion zone. Environ Pollut 159, 1606–1612 (2011).
Schlötterer, C. Hitchhiking mapping - functional genomics from the population genetics perspective. Trends Genet 19, 32–38 (2003).
Tollenaere, C., Duplantier, J. M., Rahalison, L., Ranjalahy, M. & Brouat, C. AFLP genome scan in the black rat (Rattus rattus) from Madagascar: detecting genetic markers undergoing plague-mediated selection. Mol Ecol 20, 1026–1038 (2011).
Körner, C. Alpine Plant Life. Functional Plant Ecology of High Mountain Ecosystems 2nd edn, (Springer Science & Business Media, 2003).
Chaves, M. M., Maroco, J. P. & Pereira, J. S. Understanding plant responses to drought - from genes to the whole plant. Funct Plant Biol 30, 239–264 (2003).
Bonin, A., Ehrich, D. & Manel, S. Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol 16, 3737–3758 (2007).
Nelson, M. F. & Anderson, N. O. How many marker loci are necessary? Analysis of dominant marker data sets using two popular population genetic algorithms. Ecology and evolution 3, 3455–3470 (2013).
Leipold, M., Tausch, S., Hirtreiter, M., Poschlod, P. & Reisch, C. Sampling for conservation genetics: how many loci and individuals are needed to determine the genetic diversity of plant populations using AFLP? Conservation Genetics Resources, https://doi.org/10.1007/s12686-018-1069-1 (2018).
Fischer, M. C., Foll, M., Excoffier, L. & Heckel, G. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis). Mol Ecol 20, 1450–1462 (2011).
Hoban, S. et al. Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions. Am Nat 188, 379–397 (2016).
Lotterhos, K. E. & Whitlock, M. C. The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Mol Ecol 24, 1031–1046 (2015).
Excoffier, L., Hofer, T. & Foll, M. Detecting loci under selection in a hierarchically structured population. Heredity 103, 285–298 (2009).
Hahn, M. W. Toward a selection theory of molecular evolution. Evolution 62, 255–265 (2008).
Li, H. P. A New Test for Detecting Recent Positive Selection that is Free from the Confounding Impacts of Demography. Mol Biol Evol 28, 365–375 (2011).
De Mita, S. et al. Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol Ecol 22, 1383–1399 (2013).
Thuiller, W. Biodiversity - Climate change and the ecologist. Nature 448, 550–552 (2007).
Engler, R. et al. 21st century climate change threatens mountain flora unequally across Europe. Global Change Biol 17, 2330–2341 (2011).
Kutnjak, D. et al. Escaping to the summits: Phylogeography and predicted range dynamics of Cerastium dinaricum, an endangered high mountain plant endemic to the western Balkan Peninsula. Mol Phylogenet Evol 78, 365–374 (2014).
Koutsos, T., Chatzopoulou, P. Sideritis species in Greece: the current situation in Report of a Working Group on Medicinal and Aromatic Plants (ed. Lipman, E.) 112–114 (Biodiversity International, 2009).
Murray, M. C. & Hare, M. P. A genomic scan for divergent selection in a secondary contact zone between Atlantic and Gulf of Mexico oysters, Crassostrea virginica. Mol Ecol 15, 4229–4242 (2006).
Vos, P. et al. Aflp - a New Technique for DNA-Fingerprinting. Nucleic Acids Res 23, 4407–4414 (1995).
Carović-Stanko, K. et al. Molecular and chemical characterization of the most widespread Ocimum species. Plant Syst Evol 294, 253–262 (2011).
Shannon, C. E., Weaver, W. The Mathematical Theory of Communication, (University of Illinois Press, 1949).
Lewontin, R. C. The apportionment of human diversity. Evolution Biology 6, 381–398 (1972).
Ehrich, D. AFLPDAT: a collection of R functions for convenient handling of AFLP data. Mol Ecol Notes 6, 603–604 (2006).
Zhivotovsky, L. A. Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8, 907–913 (1999).
Vekemans, X., Beauwens, T., Lemaire, M. & Roldan-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11, 139–151 (2002).
Lynch, M. & Milligan, B. G. Analysis of Population Genetic-Structure with Rapd Markers. Mol Ecol 3, 91–99 (1994).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Saitou, N. & Nei, M. The Neighbor-Joining Method - a New Method for Reconstructing Phylogenetic Trees. Mol Biol Evol 4, 406–425 (1987).
Felsenstein, J. Confidence-Limits on Phylogenies - an Approach Using the Bootstrap. Evolution 39, 783–791 (1985).
Hammer, Ø., Harper, D. A. T. & Ryan, P. D. PAST: paleontological statistics software package for education and data analysis. Palaeontolo Electron 4, 9 (2001).
Excoffier, L., Smouse, P. E. & Quattro, J. M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 (1992).
Excoffier, L., Laval, G. & Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform 1, 47–50 (2005).
Huff, D. R. RAPD characterization of heterogeneous perennial ryegrass cultivars. Crop Sci 37, 557–564 (1997).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14, 2611–2620 (2005).
Earl, D. A. & Vonholdt, B. M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4, 359–361 (2012).
Corander, J., Waldmann, P. & Sillanpaa, M. J. Bayesian analysis of genetic differentiation between populations. Genetics 163, 367–374 (2003).
Corander, J. & Marttinen, P. Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15, 2833–2843 (2006).
Corander, J., Marttinen, P., Siren, J. & Tang, J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. Bmc Bioinformatics 9, https://doi.org/10.1186/1471-2105-9-539 (2008a).
Corander, J., Siren, J. & Arjas, E. Bayesian spatial modeling of genetic population structure. Computation Stat 23, 111–129 (2008b).
Jombart, T., Devillard, S. & Balloux, F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. Bmc Genet 11, https://doi.org/10.1186/1471-2156-11-94 (2010).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
R Core Team. R: A Language and Environment for Statistical. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/ (2017).
Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228 (1997).
Rohlf, F. J. NTSYS-pc. Numerical Taxonomy and Multivariate Analysis System, version 2.1. User Guide (Exeter Publications, 2000).
Joost, S. et al. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol 16, 3955–3969 (2007).
Stucki, S. & Joost, S. Samβada: User manual, Version v0.5.1. URL, http://lasig.epfl.ch/sambada (2015).
Roesti, M., Salzburger, W. & Berner, D. Uninformative polymorphisms bias genome scans for signatures of selection. Bmc Evol Biol 12, 94, https://doi.org/10.1186/1471-2148-12-94 (2012).
Jeffreys, H. Theory of Probability, 3rd edn, (Clarendon Press, Oxford, 1961).
Acknowledgements
This study was supported by the SEE - ERA.NET PLUS - ERA 135/01 MOUNTEA-CONSE project.
Author information
Authors and Affiliations
Contributions
Z.Š., M.G. and I.R. designed the study; Z.Š., P.C., G.S., P.R. collected the samples; Z.L., I.R. conducted the laboratory work; Z.Š. analyzed the data; M.G., Z.Š., I.R. and K.C.S. wrote the manuscript. All the authors read and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grdiša, M., Radosavljević, I., Liber, Z. et al. Divergent selection and genetic structure of Sideritis scardica populations from southern Balkan Peninsula as revealed by AFLP fingerprinting. Sci Rep 9, 12767 (2019). https://doi.org/10.1038/s41598-019-49097-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-49097-x
This article is cited by
-
Altitudinal differences in cytogenetic traits of common dandelion during its invasion in high altitude areas
Biologia (2024)
-
Analyses of genetic diversity and population structure of endemic and endangered species Sideritis gulendamii (Lamiaceae) and implications for its conservation
Genetic Resources and Crop Evolution (2024)
-
Phenotypic, chemical component and molecular assessment of genetic diversity and population structure of Morinda officinalis germplasm
BMC Genomics (2022)
-
Plant phylogeography of the Balkan Peninsula: spatiotemporal patterns and processes
Plant Systematics and Evolution (2022)
-
Population structure and adaptive variation of Helichrysum italicum (Roth) G. Don along eastern Adriatic temperature and precipitation gradient
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.