Abstract
A chemical study of the EtOAc extract of Nemania bipapillata (AT-05), an endophytic fungus isolated from the marine red alga Asparagopsis taxiformis - Falkenbergia stage, led to the isolation of five new botryane sesquiterpenes, including the diastereomeric pair (+)-(2R,4S,5R,8S)-(1) and (+)-(2R,4R,5R,8S)-4-deacetyl-5-hydroxy-botryenalol (2), (+)-(2R,4S,5R,8R)-4-deacetyl-botryenalol (3), one pair of diastereomeric botryane norsesquiterpenes bearing an unprecedented degraded carbon skeleton, (+)-(2R,4R,8R)-(4) and (+)-(2R,4S,8S)-(5), which were named nemenonediol A and nemenonediol B, respectively, in addition to the known 4β-acetoxy-9β,10β,15α-trihydroxyprobotrydial (6). Their structures were elucidated using 1D and 2D NMR, HRESIMS and comparison with literature data of similar known compounds. The absolute configurations of 2, 3 and 4 were deduced by comparison of experimental and calculated electronic circular dichroism (ECD) spectra, while those of 1 and 5 were assigned from vibrational circular dichroism (VCD) data. Compound 4 weakly inhibited acetylcholinesterase, whereas compound 1 inhibited both acetylcholinesterase and butyrylcholinesterase. Compounds 1, 3, 5 and 6 were tested against two carcinoma cell lines (MCF-7 and HCT-116), but showed no significant citotoxicity at tested concentrations (IC50 > 50 µM).
Similar content being viewed by others
Introduction
Recent studies on marine-derived endophytic microorganisms have shown their potential as a source of new bioactive natural products1. As part of our ongoing efforts to identify new secondary metabolites produced by endophytic fungi isolated from the red alga Asparagopsis taxiformis (Falkenbergia stage)2, we have investigated extracts of laboratory cultures of the fungus Nemania bipapillata. This chemical study led to the isolation of one known and five new botryane terpenoids, including two pairs of diastereomeric compounds.
The genus Nemania belongs to the family Xylariaceae, one of the largest of Ascomycota phylum3, and has shown interesting applications associated with its bioactive small molecules as well as in biocatalysis. Nemania serpens was isolated as a fungal endophyte from Anemopsis californica, a plant used as a traditional medicine to treat infections due to its antibacterial activity against Staphylococcus aureus and Pseudomonas aeruginosa4. Nemania aenea SF10099-1, isolated from a forest soil sample was used as a bioacatalyst in the regio- and stereoselective synthesis of (−)-β-caryophyllene oxide using β-caryophyllene as substrate in a liquid–liquid interface bioreactor5.
Acetylcholinesterase (EC 3.1.1.7) (AChE) and butyrylcholinesterase (EC 3.1.1.8) (BChE) have gained attention due to their roles in Alzheimer disease (AD) and other Central Nervous System (CNS) conditions, such as delirium and traumatic brain injuries, associated with deficient levels of acetylcholine (ACh)6. Galanthamine, a natural alkaloid first isolated from the plant Galanthus spp. (Amaryllidaceae), is one of the few drugs currently available for the treatment of AD, which inhibits the AChE activity, increasing the levels of ACh in the brain and prolonging cholinergic functions7. Diterpenoids have been shown to be cholinesterase inhibitors8,9,10,11, which stimulated our interest in investigating the potential anticholinesterase activity of the new terpenoids isolated from Nemania bipapillata.
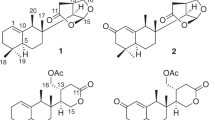
The search for new cancer chemotherapeutic agents has increased in the last years due to the drug resistance in cancer treatment. Natural sesquiterpenes have demonstrated cytotoxic activity12, including botrydial, a botryane sesquiterpene, and its derivatives13. In this paper, we describe the isolation and structure elucidation of three new botryane sesquiterpenes (1–3) and two norsesquiterpenes with a new degraded carbon skeleton (4 and 5) (Fig. 1), the determination of their absolute configurations, and their activities as cholinesterase inhibitors and cytotoxic agents.
For the determination of the absolute configuration of compounds 1–5, electronic and vibrational circular dichroism spectroscopies (ECD and VCD, respectively) were used with the aid of quantum chemical calculations. ECD and VCD methods are based on the differential absorption of left- and right-circularly polarized UV or IR radiation, respectively, by a chiral molecule during electronic or vibrational transitions. Both chiroptical methods have been successfully used over the years for absolute configuration assignments of a large array of chiral molecules including natural products14,15.
Results and Discussion
The EtOAc extract of Nemania bipapillata (AT-05) cultures was fractionated using a reversed-phase flash chromatography column to give 4 fractions (AT-05-F1 - AT-05-F4). Further purification of fraction AT-05-F2 using reversed-phase HPLC yielded five new compounds 1–5, in addition to the known compound 6 (Fig. 1). Their structures were elucidated via detailed analysis of NMR and mass spectrometry data, in addition to ECD and VCD analyses to determine their absolute configurations.
Compound 1 gave a [M + Na]+ ion at m/z 291.1576 in the ESITOFHRMS appropriate for a molecular formula of C15H24O4 (calcd for C15H24O4Na 291.1572) requiring 4 sites of unsaturation. The 1H NMR (Table 1) and HSQC spectra revealed the presence of four methyl groups, associated with three singlets (δ 1.20/H3-12, 1.00/H3-13 and 1.40/H3-14) and one doublet (δ 1.03, J11-2 6.6 Hz, H3-11), one hydroxymethine proton at δ 3.73 (dd, J4-3a 4.2, J4-3b 12.0 Hz, H-4) and a set of nonequivalent oxymethylene protons at δ 3.71 (d, J15a-15b 10.2 Hz, Ha-15)/3.57 (d, J15b-15a 10.2 Hz, Hb-15), in addition to one aldehyde proton (δ 10.28, s, H-10). The 1H‒1H COSY spectrum (Supplementary Fig. S4) showed correlations of a methine proton at δ 2.73 (m, H-2) to methylene hydrogens at δ 1.81 (m, Ha-3, 1H) and δ 1.67 (m, Hb-3, 1H) and to H3-11 (Fig. 2). The 13C NMR data (Table 2) exhibited one carbonyl carbon at δ 197.1 (C-10), which correlated with the proton at δ 10.28, as shown in the HSQC spectrum (Supplementary Fig. S5), and two quaternary sp2 carbons (δ 141.8/C-1, 164.7/C-9), suggesting the presence of an α,β-unsaturated carbonyl group, in addition to three quaternary sp3 (δ 44.6/C-6, 47.9/C-8, 83.1/C-5) hybridized carbons. The HMBC spectrum (Supplementary Fig. S6) evidenced correlations of H-10 to C-1 and C-2 (δ 31.5), of Ha-3 to C-1, C-4 (δ 69.9) and C-5, of Hb-3 to C-2, C-4 (δ 69.9) and C-11 (δ 20.9), of H3-12 to C-5, C-6, C-7 (δ 53.4) and C-13 (δ 26.6), of H3-14 to C-7, C-8 and C-9, as well as correlations of Ha-15 and Hb-15 to C-7 and C-9, respectively (Fig. 2). The 1H NMR spectrum of compound 1 obtained in DMSO-d6 displayed signals for the hydroxyl protons at δ 5.40 (t, J15-0H-15 4.8 Hz, 15-OH, 1H), 4.33 (s, 5-OH, 1H), and 4.23 (d, J4-OH-4 7.2 Hz, 4-OH, 1H) (Supplementary Table S1 and Fig. S7). ROESY (DMSO-d6) correlations revealed the relative configuration of 1 (Supplementary Fig. S9). The 4-OH and 5-OH signals showed ROESY correlations to H3-12 (δ 1.09 s, 3H), whereas the H-4 signal (δ 3.53, ddd, J4-3a 4.5, J4-4-OH 7.5, J4-3b 12.3 Hz, 1H) showed ROESY correlations to H-2 (δ 2.60, m, 1H) and H3-13 (δ 0.90, s, 3H). Finally, a ROESY correlation observed between H3-13 and H3-14 (δ 1.33, s, 3H) (Fig. 3) established the relative configuration at C-8, completing the assignment of the constitution and relative configuration of 1.
Compound 2 was assigned the same molecular formula as 1 (C15H24O4) on the basis of HRMS (ESI-Q-TOF, m/z: [M + Na]+ calcd for C15H24O4Na 291.1572; found 291.1563) data. Its 1H and 13C NMR data (Tables 1 and 2) were similar to those of 1, except for the chemical shifts of C-2 (δH/C 2.91/27.4) and C-4 (δH/C 4.10/71.9). Furthermore, the H-4 signal exhibited different coupling constants (J4-3a 3.6, J4-3b 6.9 Hz, 1H) from those observed for compound 1, suggesting that 2 was a stereoisomer of 1 with a configurational difference at C-4, which was confirmed from 2D NMR data analyses. The HMBC spectrum of 2 (Supplementary Fig. S16) showed correlations of H-10 (δ 10.26, s, 1H) to C-1 (141.9) and C-2, of H-3 (δ 1.80, m, 2 H) to C-1, C-4 and C-5 (δ 83.9) (Fig. 2), as observed in compound 1. The relative configuration of 2 was established via 1D NOE experiments (Supplementary Fig. S17). Irradiation of H-4 gave NOEs in H3-11 (δ 1.05, J11-2 7.2 Hz, 3H) and H3-12 (δ 1.15, s, 3H) consistent with a cis configuration between H-4 and H3-11, which was different from the cis configuration between H-4 and H-2 in 1. Irradiation of H3-14 gave an NOE in H3-13 confirming that C-8 had the same relative configuration in both 1 and 2 (Fig. 3).
The molecular formula of compound 3 was deduced as C15H24O3 from HRMS data (ESI-Q-TOF, m/z: [M + Na]+ calcd for C15H24O3Na 275.1623; found 275.1613). Comparison of the NMR data obtained for 3 (Tables 1 and 2) with those of 1 and 2 revealed that compound 3 bears a similar structure. However, its 1H NMR spectrum had a double doublet at δ 2.33 (J5-2 3.3, J5-4 8.7 Hz, 1H), which was assigned to H-5 after observation of a 1H‒1H COSY correlation to H-4 (δ 3.62, ddd, J4-3a 3.9, J4-5 8.7, J4-3b 11.1 Hz, 1H) (Fig. 2) and a HSQC correlation to the signal at δ 62.7 (C-5) (Supplementary Fig. S22). HOMODEC spectrum evidenced its additional coupling to H-2 (δ 2.79, m, 1H) and confirmed the coupling constant (J5-2 3.3 Hz), upon irradiation of H-2 (Supplementary Fig. S23). The HMBC spectrum (Supplementary Figs S24 and S25) exhibited correlations of Hb-7 (δ 1.40, d, J7b-7a 13.2 Hz, 1H), H3-12 (δ 0.98, s, 3H) and H3-13 (δ 1.25, s, 3H) to C-5 (Fig. 2), coherent with the structure proposed for compound 3. NOE experiments disclosed the relative configuration of 3 (Supplementary Fig. S26), evidencing the spatial correlation of H-4, H-5, H-2 and H3-13 upon irradiation of H-5. The spatial correlation between H3-12 and H3-14 (δ 1.44, s, 3H) was also observed upon irradiation of H3-14, which indicated the cis stereochemistry between such methyl groups in compound 3 (Fig. 3).
The molecular formula of compound 4 was determined as C14H22O3 by HRMS (ESI-TOF, m/z: [M + Na]+ calcd for C14H22O3Na 261.1467; found 261.1466). Its 1H NMR spectrum (Table 1) revealed the absence of an aldehyde proton, which differs from compounds 1, 2 and 3. On the other hand, its 13C NMR spectrum (Table 2) presented signals at δ 203.9 (C-1), 170.3 (C-5) and 140.5 (C-9), which indicated the presence of an α,β-unsaturated carbonyl group. Further similarities with compounds 1–3 included the presence of four methyl groups in the 1H NMR (Table 1) and HSQC (Supplementary Fig. S31) spectra, associated to one doublet (δ 1.10, J10-2 7.2 Hz, H3-10) and three singlets (δ 1.19/H3-13, 1.20/H3-12 and 1.32/H3-11), in addition to the signals for one oxymethine hydrogen at δ 4.43 (t, J4-3 3.0 Hz, H-4), and two doublets for one hydroxymethylene group at δ 3.63 (d, J14a-14b 10.8 Hz, 1H, Ha-14) and 3.42 (d, J14b-14a 10.8 Hz, 1H, Hb-14). The position of the α,β-unsaturated carbonyl group was deduced from HMBC correlations of H3-10 to C-1, of H3-11 and H3-12 to C-5 and of Hb-14 to C-9 (Fig. 2), which were consistent with the structure of an unprecedented botryane norsesquiterpene carbon skeleton for compound 4. Its relative configuration was established from NOE experiments (Supplementary Fig. S33). Irradiation of H3-10 induced an NOE in H-4, whereas irradiation of H-4 induced an NOE in H3-12, indicating a cis relative configuration among H3-10, H-4 and H3-12 (Fig. 3). Conversely, the spatial correlation between Hb-14 and H3-11 was observed upon irradiation of Hb-14.
Compound 5 was established as a stereoisomer of 4, based on the same molecular formula, C14H22O3, determined by HRMS (ESI-Q-TOF, m/z: [M + H]+ calcd for C14H23O3 239.1642; found 239.1648) in addition to key similarities on their 1H and 13C NMR (Tables 1 and 2) spectra, except for the resonances assigned to C-2 (δH/C 2.43/43.5) and C-4 (δH/C 4.69/68.1), and coupling constants for the H-4 signal (J4-3a 4.8, J4-3b 10.2). The HMBC spectrum (Supplementary Fig. S39) showed correlations of H3-10 (δ 1.10, d, J10-2 6.6, 3H) to C-1 (δ 203.4), of H3-11 (δ 1.38, s, 3H) and H3-12 (δ 1.31, s, 3H) to C-5 (δ 174.6), and of Hb-14 (δ 3.54, d, J14b-14a 10.8, 1H) to C-9 (δ 140.2) (Fig. 2), which are associated with an α,β-unsaturated carbonyl moiety as in compound 4. Similar results from NOE experiments corroborated the proposed relative configuration for 5 (Supplementary Fig. S40), including the observation of NOEs among H-4, H3-12 and H3-13 (δ 1.13, s, 3H) upon irradiation of H3-12, as well as between H-4 and H-2 (δ 2.43, m), upon irradiation of H-4, which is consistent with a cis relative configuration among H-4, H-2, H3-12 and H3-13. Such data differentiate compound 5 from 4, which presented a cis configuration between H-4 and H3-10 (Fig. 3).
It is worth noting the high chemical shift of C-9 for compounds 1–3 and for C-5 in case of compounds 4 and 5, which are consistent with the β-carbon shift of α,β-unsaturated carbonyl moieties16,17.
Compound 6 was identified as 4β-Acetoxy-9β,10β,15α-trihydroxyprobotrydial, based on NMR and HRMS data, and comparison with those reported in the literature18.
Due to the amount of sample available, VCD was used for the absolute configuration assignments of compounds 1 and 5 only, while ECD was carried out for compounds 2–4. Regarding compounds 1 and 5, the similarity between observed and density functional theory (DFT) predicted IR/VCD spectra, at the B3LYP/PCM(MeOH)/6-31G(d) level, led to the unambiguous assignment of their absolute configuration as (+)-(2R,4S,5R,8S)-1 and (+)-(2R,4S,8S)-5 (Fig. 4). In line with previous results from our group19, the agreement between theoretical and observed data in methanol-d4 solution was greatly improved by considering deuterated hydroxyl groups for the calculations. As for compounds 2–4, the correlation of experimental and time-dependent DFT (TD-DFT) simulated ECD spectra, at the CAM-B3LYP/PCM(MeOH)/TZVP level, allowed the determination of their absolute configuration as (+)-(2R,4R,5R,8S)-2, (+)-(2R,4S,5R,8R)-3, and (+)-(2R,4R,8R)-4 (Fig. 5).
(Left) Experimental and calculated [2R,4R,5R,8S; CAM-B3LYP/PCM(MeOH)/TZVP//B3LYP/PCM(MeOH)/6-31G(d)] UV and ECD spectra of (+)-2 (Center) Experimental and calculated [2R,4S,5R,8R; CAM-B3LYP/PCM(MeOH)/TZVP//B3LYP/PCM(MeOH)/6-31G(d)] UV and ECD spectra of (+)-3 (Right) Experimental and calculated [2R,4R,8R; CAM-B3LYP/PCM(MeOH)/TZVP//B3LYP/PCM(MeOH)/6-31G(d)] UV and ECD spectra of (+)-4. For structures of the lowest-energy conformers, please see Supplementary Information.
In the specific case of compound 2, DFT and TD-DFT calculations were carried out for both configurations possible at C-5, as its relative configuration were not readily available from NMR data. As a result, the best agreement between experimental and calculated UV/ECD data was observed for 2R,4R,5R,8S-2 (Fig. 5 and Supplementary Figs S46 and S47).
Compounds 1–3 are structurally related to botryenalol, previously isolated from Botrytis cinerea, a phytopathogenic ascomycete found in grapes in a Domecq vineyard, Jerez de La Frontera, Cádiz16. However, these compounds have different substituents, especially compounds 1 and 2, which bear a hydroxy group on carbon 5, a structural feature not described yet for botryane terpenoids.
Biogenetically, botryane derivatives have farnesyldiphosphate (FPP) as a precursor, and the key intermediate A is generated after cyclization, rearrangement, and hydroxylation. Subsequent hydroxylation and acetylation reactions could transform A to 4β-acetoxy-9β,10β,15α-trihydroxyprobotrydial (6), an intermediate in the biosynthesis of botrydial and its derivatives20,21. Further steps in the biogenetic proposal include cleavage of the vicinal diol in A to give the dialdehyde B followed by reduction, dehydration, and hydroxylation to generate 1 and 2. As suggested by Collado et al., a retro Aldol reaction might epimerize C-8 to give intermediate C21. Reduction, dehydration, and hydroxylation can convert intermediate C into 3 and intermediate D. Allylic rearrangement and oxidative decarboxylation would convert 1 into 5 and D into 4, the two new N. bipapillata metabolites with unprecedented rearranged and degraded terpenoid carbon skeletons (Fig. 6).
Some bothryane derivatives showed phytotoxic effects18, as well as antibacterial and cytotoxic activities13,21,22. Botryane metabolites were also previously described from marine microorganisms, including a fungal strain of Geniculosporium sp. isolated from the red alga Polysiphonia sp., which afforded eleven new botryane terpenoids with algicidal, antibacterial and fungicidal activity against Chlorella fusca, Bacillus megaterium and Microbotryum violaceum, respectively17. Furthermore, the first botrydiol-coumarin hybrid, hypocrolide A23, and novel heterodimeric botryane ethers24 were isolated from Hypocrea sp., an insect-associated fungus, isolated from a Septobasidium-infected insect, Serrataspis sp.
The isolated metabolites 1–6 were tested for their ability to inhibit cholinesterase (ChEIs) (Table 3). The inhibition assays were carried out using an immobilized capillary enzyme reactor (ICER) based on acetylcholinesterase human recombinant and/or butyrylcholinesterase from human serum, respectively huAChE-ICER and huBChE-ICER (in accordance with published procedure)25,26,27. Compounds 2–6 were more active towards huAChE than huBChE, indicating a selective cholinesterase inhibition, and 4 was the most active compound, with 27.7% (100 μM) inhibition against huAChE. Compound 1 was considered a non-selective inhibitor, as it showed similar inhibitory potentials against both huAChE and huBChE (19.9 and 14.1%, respectively), while its stereoisomer (compound 2) inhibited only huAChE (18.3%). The results represent only modest inhibition of huAChE or huBChE, but they suggest that botryane terpenoids could act as lead compounds for synthetic development of more potent selective cholinesterase ligands.
Compounds 1, 3, 5 and 6 were tested against colorectal carcinoma HCT-116 and breast adenocarcinoma MCF-7 cell lines using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide tetrazolium reduction (MTT) assay, and presented no significant toxicixity at tested concentrations (IC50 > 50 µM) against both cell lines. Previous studies have shown that the presence of the 1,5-dialdehyde moiety, as in botrydial, is an important structural feature for cytotoxicity, including the S configuration at C-113,21. Our results corroborate previous findings as compounds 1, 3, 5 and 6 do not bear the 1,5-dialdehyde unit, which possibly resulted in the lack of cytotoxic activity. In addition, C-1 in compounds 1, 3 and 5 is an sp2 carbon as part of α,β-unsaturated carbonyl system and therefore the S configuration at C-1 requirement is also not met.
In summary, this study expands the known chemodiversity of marine microorganisms, since it is the first to describe the isolation of metabolites from a fungal strain of Nemania bipapillata, as well as the occurrence of botryane terpenoids in this genus. Compounds 1–5 are new members of the botryane family, highlighting compounds 4 and 5, which bear a new norsesquiterpene skeleton and were named in accordance with structural functions and the genus Nemania. ECD and VCD quantum chemical calculations were essential techniques to determine the absolute configuration of this class of compounds since there were no studies of similar compounds in the literature to be used as models. The cholinesterase inhibitory activities of compounds 1 and 2, although relatively modest in potency, suggest that they may be lead structures for developing more potent analogs.
Methods
General experimental procedure
Optical rotations were determined with a Polartronic H (Schmidt + Haensch) spectrometer. UV spectra were recorded with a Perkin Elmer Lambda 1050 spectrophotometer. ECD and UV spectra were recorded in methanol using a JASCO J-815 spectrometer. Parameters were set as follows: band width 1 nm; response 1 sec; scanning speed 100 nm min−1; 3 accumulations; room temperature; 0.1 cm path lentgh cell; concentration 0.3–0.5 mg mL−1. IR and VCD spectra of compounds 1 and 5 were measured simultaneously with a ChiralIR-2X FT-VCD spectrometer (BioTools) equipped with a single photoelastic modulator (PEM) at a resolution of 4 cm−1 for 8 h. The optimum retardation of the ZnSe PEM was set at 1400 cm−1. Baselines were corrected by subtracting the VCD spectrum of compounds 1 and 5 from that obtained for the solvent. Vibrational chiroptical spectra were measured in a BaF2 cell with 100 μm path length in methanol-d4. Samples concentration: compound 1, 4 mg/120 μL of CD3OD, compound 5, 3 mg/120 μL of CD3OD. 1D and 2D NMR experiments were recorded on Bruker Avance III HD 600 and Bruker Avance 600 (14,1 T) spectrometers with a 5 mm TCI cryoprobe. Chemical shifts were reported in δ (ppm) and were referenced to the residual DMSO-d6 and CD3OD (δ 2.49 and 3.31, respectively, for 1H chemical shifts; δ 39.5 and 49.0 for 13C chemical shifts). High resolution ESI-TOF and ESI-Q-TOF-MS were recorded with Waters/Micromass LCT and Bruker Maxix Impact spectrometers, respectively. Semi-preparative HPLC was performed on a Shimadzu equipment containing two pumps LC-6AD, a SPD-M20A photodiode array detector and a system controller CBM-20A, and using a C-18 (2) Phenomenex Luna (250 × 10 mm; 5 μm; 100 Å) column and HPLC grade solvents. Chromatography analyses were carried out using reversed phase silica column (C-18, 40–75 μm, 60 Å; Sorbent Technologies). Silica gel plates on aluminum (silica gel 60 F254 -Merck) were used for analytical thin layer chromatography (TLC).
Collection of alga
Asparagopsis taxiformis (Falkenbergia stage) specimens were collected at the rocky shore of Fortaleza Beach (Ubatuba, SP, Brazil, GPS: 23°21′38″S, 44°49′63″W), in June 2013. A voucher specimen (SP 428533) was deposited in the herbarium of the Institute of Botany (SMA/SP). After washing carefully with seawater, the algal specimens were kept in previously prepared flasks containing sterile seawater and antibiotic chloramphenicol (200 mg L−1).
Fungal material
Algal specimens were immersed in a 1% NaOCl solution (v/v) during 6 seconds and 70% ethanol solution (v/v) during 2 seconds, followed by seawater rinsing as a surface sterilization procedure. Then, each alga was fragmented and small pieces were spread on PDA (potato, dextrose and agar) plates prepared with sterilized seawater containing the antibiotic chloramphenicol (200 mg L−1). A fungal strain codified as AT-05 was isolated after successive replication in PDA plates. Total DNA was extracted using fresh mycelium with an extraction kit (ZR Fungal/Bacterial DNA MicroPrep) following the manufacturer’s recommendations. Molecular identification was carried out by sequencing of ITS (primers ITS1 and ITS4)28. The amplicons were enzymatically purified (Agencourt AMPure XP, Beckman Coulter Inc.) and sequenced by the Sanger method (BigDye Terminator v.3.1 Cycle Sequencing). The obtained sequences were uploaded to Genbank database29 (accession number MH025760) and the comparison with sequences from the GenBank database was carried out using the nBLAST tool. The fungal strain was identified as Nemania bipapillata30. Voucher specimen was deposited at the Endophytic Fungi Collection in our laboratory.
Fermentation, extraction and isolation
Small agar slices bearing mycelia of the AT-05 strain were transferred to 15 Erlenmeyer flasks (500 mL) containing 300 mL of PDB (potato, dextrose broth) medium prepared with deionized water and incubated for 28 days at 25 °C. Thereafter, the medium was separated from the mycelium by filtration and extracted with EtOAc (3 × 50% of each medium volume). Evaporation of EtOAc yielded 1.74 g of AT-05 crude extract.
An aliquot of AT-05 crude extract (1.51 g) was fractionated on a C-18 derivatized silica column (150 g, Φ = 3.5 cm) using MeOH:H2O gradient (1:1, 60:40, 75:25, 100:0) as eluent and yielded 4 fractions (360 mL each; AT-05-F1 - AT-05-F4). Fraction AT-05-F2 (200 mg) was chromatographed by semi-preparative HPLC using a linear gradient (30 to 80% MeOH:H2O) for 40 minutes and a flow rate of 4.5 mL min−1, which afforded compounds 1 (tR 23.5 min; 5.8 mg), 2 (tR 21.1 min; 1.0 mg), 3 (tR 25.0 min; 3.0 mg), 4 (tR 21.9 min; 1.4 mg), 5 (tR 27.3 min; 3.6 mg) and 6 (tR 26.4 min; 13.8 mg).
(+)-(2R,4S,5R,8S)-4-deacetyl-5-hydroxy-botryenalol (1): colorless oil; [α]D23 + 55.9 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 248 nm (3.80); CD (MeOH, 1.86 mmol L−1) λext (Δε) 337 (+2.11), 246 (−2.87), 206 nm (+4.59); 1H NMR data see Table 1; 13C NMR data see Table 2; HRMS (ESI-TOF) m/z: [M + Na]+ calcd for C15H24O4Na 291.1572; found 291.1576.
(+)-(2R,4R,5R,8S)-4-deacetyl-5-hydroxy-botryenalol (2): colorless oil; [α]D23 + 33.7 (c 0.13, MeOH); UV (MeOH) λmax (log ε) 249 nm (3.01); CD (MeOH, 1.86 mmol L−1) λext (Δε) 341 (+0.97), 264 (+0.82), 220 nm (+1.43); 1H NMR data: see Table 1; 13C NMR data: see Table 2; HRMS (ESI-Q-TOF) m/z: [M + Na]+ calcd for C15H24O4Na 291.1572; found 291.1563.
(+)-(2R,4S,5R,8R)-4-deacetyl-botryenalol (3): colorless oil; [α]D23 + 22.2 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 250 nm (3.49); CD (MeOH, 1.98 mmol L−1) λext (Δε) 330 (+0.91), 270 (+0.29), 237 nm (–0.93); 1H NMR data see Table 1; 13C NMR data see Table 2; HRMS (ESI-Q-TOF) m/z: [M + Na]+ calcd for C15H24O3Na 275.1623; found 275.1613.
Nemenonediol A [(+)-(2R,4R,8R)-4]: colorless oil; [α]D23 + 6.86 (c 0.14, MeOH); UV (MeOH) λmax (log ε) 246 nm (3.77); CD (MeOH, 1.26 mmol L−1) λext (Δε) 309 (– 0.40), 253 (+1.67), 224 (– 1.93); 1H NMR data see Table 1; 13C NMR data see Table 2; HRMS (ESI-TOF) m/z: [M + Na]+ calcd for C14H22O3Na 261.1467; found 261.1466. IUPAC nomenclature: (3R,5R,7R)-7-hydroxy-3-(hydroxymethyl)-1,1,3,5-tetramethyl-2,3,6,7-tetrahydro-1H-inden-4(5H)-one.
Nemenonediol B [(+)-(2R,4S,8S)-5]: colorless oil; [α]D23 + 8.6 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 247 nm (3.65); CD (MeOH, 2.10 mmol L−1) λext (Δε) 251 (– 4.06), 215 nm (+6.09); 1H NMR data see Table 1; 13C NMR data see Table 2; HRMS (ESI-Q-TOF) m/z: [M + H]+ calcd for C14H23O3 239.1642; found 239.1648. IUPAC nomenclature: (3S,5R,7S)-7-hydroxy-3-(hydroxymethyl)-1,1,3,5-tetramethyl-2,3,6,7-tetrahydro-1H-inden-4(5H)-one.
4β-Acetoxy-9β,10β,15α-trihydroxyprobotrydial (6): colorless oil; 1H NMR and 13C NMR data were consistent with those previously reported18; HRMS (ESI-Q-TOF) m/z: [M + Na]+ calcd for C17H28O5Na 335.1834; found 335.1828.
Calculations
This section was adapted from previously published methods19,31. All DFT simulations were carried using Gaussian 09 software32. Solvation in methanol was treated implicitly using the polarizable continuum model (PCM) in its integral equation formalism version (IEFPCM). For the calculations, the following configurations were arbitrarily chosen: (2R,4S,5R,8S)-1; (2R,4R,5S,8S)- and (2R,4R,5R,8S)-2; (2R,4S,5R,8R)-3; (2R,4R,8R)-4; and (2R,4S,8S)-5. Conformational searches were performed using molecular mechanics with the Monte Carlo algorithm and MM + force field available in HyperChem 8.0.10 software package. For (2R,4S,5R,8S)-1, 10 conformers with relative energy (rel E.) within 6 kcal mol−1 were selected and had their geometry optimized at the B3LYP/PCM(MeOH)/6-31G* level. The five conformers with rel E. <2.0 kcal mol−1 (>98% of the total Boltzmann distribution) were selected for IR and VCD spectral calculations. Regarding (2R,4R,5S,8S)- and (2R,4R,5R,8S)-2, 2 conformers each, with rel E. within 6 kcal mol−1 were selected and had their geometry optimized at the B3LYP/PCM(MeOH)/6-31 G* level. One and two conformers, respectively (rel E. <2.0 kcal mol−1) were used for UV and ECD spectral calculations. For (2R,4S,5R,8R)-3, 8 conformers (rel E. within 6 kcal mol−1) were selected and had their geometry optimized at the B3LYP/PCM(MeOH)/6-31G* level. Two conformers with rel E. <2.0 kcal mol−1 were used during UV and ECD spectral calculations. In the case of (2R,4R,8R)-4, 2 conformers (rel E. within 6 kcal mol−1) were selected and had their geometry optimized at the B3LYP/PCM(MeOH)/6-31G* level. These two conformations (rel E. <2.0 kcal mol−1) were selected for UV and ECD spectral calculations. For (2R,4S,8S)-5, 2 conformations (rel E. within 6 kcal mol−1) were selected and had their geometry optimized at the B3LYP/PCM(MeOH)/6-31G* level. These same conformers were used for IR and VCD calculations. Replacement of some hydrogen atoms with deuterium atoms was performed using GaussView 5.0.9 software for the same conformer population identified for each configuration. Dipole and rotational strengths were used to create IR and VCD spectra, in M−1 cm−1 units, These quantities were calculated at the same level of theory used during geometry optimization. Spectra were plotted as a sum of Lorentzian bands with HWHM of 6 cm−1. The calculated wavenumbers were multiplied with a scaling factor of 0.975. The final spectra were generated as Boltzmann averages of the lowest-energy conformers identified and plotted using Origin8 software. No imaginary frequencies were obtained after vibrational analysis at the B3LYP/PCM(MeOH)/6-31G* level thus confirming the considered conformers as real minima. Subsequently, TD-DFT was used to calculate excitation energies (in nm) and rotatory strengths (R) in dipole velocity form (Rvel in cgs units: 10−40 esu2 cm2), at the CAM-B3LYP/PCM(MeOH)/TZVP level. The calculated rotatory strengths from the first 30 singlet → singlet electronic transitions were simulated into an ECD curve using Gaussian bands with bandwidth σ 0.25 eV. Calculated wavelength transitions were multiplied with a scaling factor of 1.04, which was determined based on the agreement between experimental and calculated UV data. The final spectra were generated as simple averages of the lowest-energy conformers and plotted using Origin8 software.
Cholinesterase inhibition screening assays
Test samples were submitted to a punctual cholinesterase inhibition screening assay. huAChE and huBChE were immobilized independently onto fused silica capillary (0.1 mm I.D × 0.375 mm × 30 cm) using glutaraldehyde as spacer (huAChE-ICER and huBChE-ICER)26,27.
huAChE-ICER and huBChE-ICER were interfaced to the LC–IT-MS/MS as alow affinity and high selectivity biochromatography column. The LC system (Nexera, Shimadzu) consisted of two LC-20AD pumps, a SIL20A autosampler with a 50 μL loop, a DGU-20A5 degasser and a CBM-20A interface. The LC system was coupled to an Amazon Speed Ion Trap (IT) mass spectrometer (Bruker Daltonics) equipped with an ESI source, operating in a positive mode (scan 50–250 m/z). Data acquisition was carried out using the Bruker Data Analysis Software (version 4.3). All analyses were performed at room temperature (21 °C). The enzymatic reaction was monitored by direct quantification of acetylcholine hydrolysis product, Ch (m/z: [M + H]+ 104.17)26,27.
2 U mL−1 of huAChE and 0.5 U mL−1 of huBChE were immobilized and the apparent kinetic constant (KMapp = 70 μM) was obtained independently using acetylcholine (ACh) as substrate by varying the substrate concentration while measuring the Ch [M + H]+ 104.17 m/z formation. GraphPad Prism 5 software was used to obtain Michaelis–Menten plots by nonlinear regression analysis27.
Ammonium acetate solution (15 mM, pH 8.0) was used as running buffer at a flow rate of 50 μL min−1and a second pump delivered methanol after the huBChE-ICER or huAChE-ICER and before the IT-MS throughout a “T” shaped connection, at the same flow rate as the running buffer. Total analysis time was 3 min. IT-MS parameters were: 4.000 V capillary voltage, 500 V end plate voltage, 8.0 Lmin−1 drying gas, 275 °C drying temperature and 35 psi nebulizer. Isolation width was set at ±m/z 0.5. Fragmentation amplitude was set to 120%. Nitrogen was used as sheath gas and helium as collision gas27.
Galanthamine was used as standard cholinesterase inhibitor. Methanol stock solutions (1 mM) were prepared for each tested compound. Reaction mixtures (100 μL) were prepared by mixing 100 μM of tested compound with ACh 70 μM. Final volume was completed with ammonium acetate solution (15 mM, pH 8.0). Each reaction mixture (10 μL) was injected into the LC-MS system and the percentage of inhibition was calculated in accordance with Eq. 1.
where, P is the attained peak area of Ch produced: (i) in the presence of the tested compound; (0) in the absence of the tested compound25,26,27.
Any possible interference at the 104 m/z range from the tested sample was evaluated injecting a reaction mixture without the substrate.
Cytotoxic activity
The cytotoxicity of compounds 1, 3, 5 and 6 was measured through the MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) assay33, that is based in the conversion of the tetrazolium salt to a formazan product by viable cells. Two cell lines obtained from American Type Culture Collection (Manassas, Virginia, EUA) were used: HCT-116 (colorectal carcinoma, ATCC® CCL-247™) and MCF-7 (breast adenocarcinoma, ATCC® HTB-22). The assay was conducted essentially as described by Monteiro et al.34. The HCT-116 and MCF-7 cells (1 × 104 cells well−1) were seeded in 96-well plates and, after 24 h, different concentrations (5 µM and 50 µM) of the compounds were added and incubated for 72 h. Doxorubicin (0.003 to 10 µM) and DMSO (0.5%) were used as a positive and negative controls, respectively. At the end of incubation, the supernatant was replaced with fresh medium containing MTT (0.5 mg/mL) for 3 h. The supernatant was, then, removed, and the MTT formazan product was dissolved in 150 μL DMSO. The absorbance was measured using a multiplate reader (Multiskan™ FC, Thermo Scientific, Finland) at 570 nm. All experiments were performed in duplicate. IC50 values, along with 95% confidence intervals, were calculated by non-linear regression using GraphPad Prism 5.0 (Intuitive Software for Science, La Jolla, CA, USA).
Data Availability
The authors declare availability to all data, materials or information contained in this manuscript.
References
Flewelling, A. J., Currie, J., Gray, C. A. & Johnson, J. A. Endophytes from marine macroalgae: promising sources of novel natural products. Curr. Sci. 109, 88–111 (2015).
Medina, R. P. et al. Aromatic compounds produced by endophytic fungi isolated from red alga Asparagopsis taxiformis - Falkenbergia stage. Nat. Prod. Res. 33, 443–446 (2019).
Kuhnert, E. et al. Lenormandins A-G, new azaphilones from Hypoxylon lenormandii and Hypoxylon jaklitschii sp. nov., recognised by chemotaxonomic data. Fungal Divers. 71, 165–184 (2015).
Bussey, R. O. et al. Comparison of the chemistry and diversity of endophytes isolated from wild-harvested and greenhouse-cultivated yerba mansa (Anemopsis californica). Phytochem. Lett. 11, 202–208 (2015).
Oda, S., Fujinuma, K., Inoue, A. & Ohashi, S. Synthesis of (−)-β-caryophyllene oxide via regio- and stereoselective endocyclic epoxidation of β-caryophyllene with Nemania aenea SF 10099-1 in a liquid-liquid interface bioreactor (L-L IBR). J. Biosci. Bioeng. 112, 561–565 (2011).
Giacobini, E. Cholinesterase inhibitors: new roles and therapeutic alternatives. Pharmacol. Res. 50, 433–440 (2004).
Heinrich, M. & Teoh, H. L. Galanthamine from snowdrop - the development of a modern drug against Alzheimer’s disease from local Caucasian knowledge. J. Ethnopharmacol. 92, 147–162 (2004).
Houghton, P. J., Ren, Y. & Howes, M.-J. Acetylcholinesterase inhibitors from plants and fungi. Nat. Prod. Rep. 23, 181 (2006).
Hostettmann, K., Borloz, A., Urbain, A. & Marston, A. Natural product inhibitors of acetylcholinesterase. Curr. Org. Chem. 10, 825–847 (2006).
Sangnoi, Y. et al. Acetylcholinesterase-inhibiting activity of pyrrole derivatives from a novel marine gliding bacterium, Rapidithrix thailandica. Mar. Drugs 6, 578–586 (2008).
Murray, A. P., Faraoni, M. B., Castro, M. J., Alza, N. P. & Cavallaro, V. Natural AChE inhibitors from plants and their contribution to Alzheimer’s disease therapy. Curr. Neuropharmacol. 11, 388–413 (2013).
Wibowo, M. et al. Cytotoxic sesquiterpenes from the endophytic fungus Pseudolagarobasidium acaciicola. Phytochemistry 122, 126–138 (2016).
Reino, J. L. et al. Chemical transformations on botryane skeleton. Effect on the cytotoxic activity. J. Nat. Prod. 66, 344–349 (2003).
Pescitelli, G., Di Bari, L. & Berova, N. Conformational aspects in the studies of organic compounds by electronic circular dichroism. Chem. Soc. Rev. 40, 4603 (2011).
Batista, J. M. Jr., Blanch, E. W., Bolzani, V. & da, S. Recent advances in the use of vibrational chiroptical spectroscopic methods for stereochemical characterization of natural products. Nat. Prod. Rep. 32, 1280–1302 (2015).
Collado, I. G., Hernandez-Galan, R., Prieto, V., Hanson, J. R. & Rebordinos, L. G. Biologically active sesquiterpenoid metabolites from the fungus Botrytis cinerea. Phytochemistry 41, 513–517 (1996).
Krohn, K. et al. Botryane metabolites from the fungus Geniculosporium sp. isolated from the marine red alga Polysiphonia. J. Nat. Prod. 68, 400–405 (2005).
Collado, I. G., Hernández-Galán, R., Durán-Patrón, R. & Cantoral, J. M. Metabolites from a shake culture of Botrytis cinerea. Phytochemistry 38, 647–650 (1995).
Sprenger, R. et al. Solution-state conformations of natural products from chiroptical spectroscopy: the case of isocorilagin. Org. Biomol. Chem. 14, 3369–3375 (2016).
Durán-Patrón, R., Colmenares, A. J., Hernández-Galán, R. & Collado, I. G. Some key metabolic intermediates in the biosynthesis of botrydial and related compounds. Tetrahedron 57, 1929–1933 (2001).
Collado, I. G., Sánchez, A. J. M. & Hanson, J. R. Fungal terpene metabolites: biosynthetic relationships and the control of the phytopathogenic fungus Botrytis cinerea. Nat. Prod. Rep. 24, 674–686 (2007).
Kuhnert, E., Surup, F., Wiebach, V., Bernecker, S. & Stadler, M. Botryane, noreudesmane and abietane terpenoids from the ascomycete Hypoxylon rickii. Phytochemistry 117, 116–122 (2015).
Yuan, Y. et al. A botryane metabolite with a new hexacyclic skeleton from an entomogenous fungus Hypocrea sp. Org. Lett. 15, 6050–6053 (2013).
Ren, F. et al. Hypocriols A-F, heterodimeric botryane ethers from Hypocrea sp., an insect-associated fungus. J. Nat. Prod. 79, 1848–1856 (2016).
Vanzolini, K. L., Vieira, L. C. C., Corrêa, A. G., Cardoso, C. L. & Cass, Q. B. Acetylcholinesterase immobilized capillary reactors-tandem mass spectrometry: an on-flow tool for ligand screening. J. Med. Chem. 56, 2038–2044 (2013).
Vilela, A. F. L., Seidl, C., Lima, J. M. & Cardoso, C. L. An improved immobilized enzyme reactor-mass spectrometry-based label free assay for butyrylcholinesterase ligand screening. Anal. Biochem. 549, 53–57 (2018).
Seidl, C., Vilela, A. F. L., Lima, J. M., Leme, G. M. & Cardoso, C. L. A novel on-flow mass spectrometry-based dual enzyme assay. Anal. Chim. Acta 1072, 81–86 (2019).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR Protocols: A Guide to Methods and Applications (eds Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T. J.) 315–322 (Academic Press, 1990).
NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46, D8–D13 (2018).
Sánchez-Ballesteros, J. et al. Phylogenetic study of Hypoxylon and related genera based on ribosomal ITS sequences. Mycologia 92, 964–977 (2000).
Frisch, M. J. et al. Gaussian, Inc., Wallingford, CT, USA (2009).
Sarria, A. L. F. et al. Dimeric chalcones derivatives from Myracrodruon urundeuva act as cathepsin V inhibitors. Phytochemistry 154, 31–38 (2018).
Mossman, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Monteiro, A. F. et al. Oxidative functionalization of a halimane diterpenoid achieved by fungal transformation. Bioorg. Chem. 86, 550–556 (2019).
Acknowledgements
We acknowledge CAPES (Coordination for the Improvement of Higher Education Personnel) for a scholarship to R. P. M. (#3334/15-5), CNPq (#474270/2012-2) and FAPESP (Grants #2013/07600-3; 2014/50926-0; 2014/25222-9; 2014-50299-5 - Bruker Daltonics Amazon Speed mass spectrometer; 2013/01710-1; 2015/17177-6) for financial support and fellowships. This research was also supported by resources supplied by the Centre for Scientific Computing (NCC/GridUNESP) of the São Paulo State University (UNESP). We also acknowledge Professor Marcos A. Soares for submitting the sequence of AT-05 fungal strain to GenBank.
Author information
Authors and Affiliations
Contributions
R.P.M. isolated the fungal strain AT-05, prepared its cultivation and extraction, isolated and identified all compounds and wrote the manuscript. A.R.A. supervised the fungal strain isolation, fermentation and extraction. J.M.B. Junior carried out computational calculations, VCD experiments and determined the absolute configurations. C.S. and A.F.L.V. performed the ChEIs screening assays and C.L.C. designed and supervised those experiments. H.V.D. performed the cytotoxic assay and L.V.C.L. supervised the experiment. R.J.A. identified the compounds and wrote/revised the manuscript. D.H.S.S. supervised all the experiments and wrote/revised the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Medina, R.P., Araujo, A.R., Batista, J.M. et al. Botryane terpenoids produced by Nemania bipapillata, an endophytic fungus isolated from red alga Asparagopsis taxiformis - Falkenbergia stage. Sci Rep 9, 12318 (2019). https://doi.org/10.1038/s41598-019-48655-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48655-7
This article is cited by
-
An update on chemistry and bioactivities of secondary metabolites from the marine algal-derived endophytic fungi
Phytochemistry Reviews (2023)
-
Fungal diversity notes 1611–1716: taxonomic and phylogenetic contributions on fungal genera and species emphasis in south China
Fungal Diversity (2023)
-
Recent progress in biodiversity research on the Xylariales and their secondary metabolism
The Journal of Antibiotics (2021)
-
Vibrational Circular Dichroism for the Assignment of Absolute Configuration of Natural Products in Brazil
Revista Brasileira de Farmacognosia (2021)
-
Arthrinins E–G, Three Botryane Sesquiterpenoids from the Plant Endophytic Fungus Arthrinium sp. HS66
Natural Products and Bioprospecting (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.