Abstract
d-Amino acid oxidase (DAAO) is a valuable flavoenzyme capable of being used in various practical applications, such as in determining d-amino acids and producing a material for semisynthetic cephalosporins, requiring higher thermal stability, higher catalytic activity, and broad substrate specificity. In this study, we isolated the thermophilic fungus Rasamsonia emersonii strain YA, which can grow on several d-amino acids as the sole nitrogen source, from a compost and characterized DAAO (ReDAAO) of the fungus. ReDAAO expressed in Escherichia coli exhibited significant oxidase activity against various neutral and basic d-amino acids, in particular hydrophobic d-amino acids. In addition, the enzyme also significantly acted on cephalosporin C, a starting material for semisynthetic antibiotics, and d-Glu, a general substrate for d-aspartate oxidase but not for DAAO, showing its unique and practically useful substrate specificity. The apparent kcat and Km values of the enzyme toward good substrates were comparable to those of higher catalytic fungal DAAOs, and the thermal stability (T50 value of ~60 °C) was comparable to that of a thermophilic bacterial DAAO and significantly higher than that of other eukaryotic DAAOs. These results highlight the great potential of ReDAAO for use in practical applications.
Similar content being viewed by others
Introduction
d-Amino acid oxidase (DAAO, EC 1.4.3.3) catalyzes the oxidative deamination of neutral and basic d-amino acids with flavin adenine dinucleotide (FAD) as a cofactor. The same reaction of acidic d-amino acids is catalyzed by another flavin enzyme, d-aspartate oxidase (DDO, EC 1.4.3.1). DAAO was first found in the pig kidney and, subsequently, in various eukaryotic organisms as a peroxisomal protein and a part of prokaryotic organisms1. In eukaryotic organisms, DAAO has been known to play a role as a detoxifying reagent in the metabolic degradation of exogenous and endogenous d-amino acids2. It has also been reported to be responsible for helping to utilize d-amino acids as nutritional and energy sources for cell growth in fungi3. In the human brain, DAAO is suggested to be involved in the central nervous system and its related disorders, such as schizophrenia and amyotrophic lateral sclerosis (ALS), through the metabolism of d-Ser, which is a co-agonist of the N-methyl d-aspartate receptor4,5,6,7. Decreased and increased levels of d-Ser have been observed in schizophrenia and ALS patients, respectively8,9, suggesting the possibility of an early diagnosis of these diseases by measuring d-Ser levels in vivo.
DAAO is an enzyme that is valuable for use in various applications10,11,12, such as the optical resolution of a racemic mixture of amino acids13; detection and quantification of d-amino acids14; and production of α-keto acids, which can be used in pharmaceutical materials15. In particular, the use of this enzyme is most important in the production of 7-aminocephalosporanic acid (7-ACA), an initial material of approximately 50 types of β-lactam antibiotics, from cephalosporin C (CPC)12. DAAOs of the mesophilic fungi Rhodotorula gracilis (RgDAAO) and Trigonopsis variabilis (TvDAAO) have been known to be useful in several practical applications because of their high catalytic activity and broad substrate specificity16; however, their poor stability inhibits application over a wide spectrum. To obtain a highly thermostable DAAO, we have previously identified a DAAO homologous gene in the thermophilic bacterium Rubrobacter xylanophilus (RxDAAO), and the recombinant RxDAAO has been shown to have the highest thermal stability among the known DAAOs17; however, the substrate specificity of this thermophilic bacterial enzyme is narrow and limited to branched-chain d-amino acids, such as d-Val and d-Ile. Moreover, the catalytic activity is much lower than those of the fungal RgDAAO and TvDAAO.
Thermophilic fungi have been an attractive source of industrially useful enzymes, such as lipase, protease, xylanase, and cellulase, because enzymes from thermophilic fungi are usually highly stable18. We have also recently reported that DDO from the thermophilic fungus Thermomyces dupontii NBRC 30541 has high thermal stability than that of other DDOs and high catalytic activity19. However, DAAO has not been identified and isolated from thermophilic fungi. One of thermophilic fungi, Rasamsonia emersonii (phylum, Ascomycota), has been found in composts20 and grows well at 45–50 °C21. This fungus also produces several biotechnologically valuable enzymes with high thermal stability, such as β-glucosidase, α-glucuronidase, and FAD-dependent glucose dehydrogenase20,22,23, suggesting it to be a promising source of various thermostable enzymes including DAAO.
In this study, we isolated the thermophilic fungus R. emersonii strain YA that can utilize d-amino acids for cell growth from a compost and isolated a DAAO homologous gene of the fungus. The gene product (ReDAAO) expressed in Escherichia coli exhibited oxidase activity against various neutral and basic d-amino acids. The enzyme also significantly acted on CPC and, interestingly, an acidic d-amino acid, d-Glu, which is a general substrate for DDO but not for DAAO. Additionally, the enzyme showed higher catalytic activity than most bacterial and animal DAAOs and higher thermal stability than that of eukaryotic DAAOs. Altogether, ReDAAO, which was the first characterized thermophilic fungal DAAO, showed unique broad substrate specificity and more advantageous properties for practical applications than other DAAOs.
Results
Isolation of thermophilic fungi that can utilize d-amino acids for cell growth
To isolate thermophilic fungi, we inoculated a suspension of various composts onto PDA medium and cultivated them at 50 °C and 60 °C for 2 weeks. Three fungi were obtained at 50 °C from bark and cattle manure composts—strain YA and strains PB and PE, respectively; whereas, no fungi growth was observed at 60 °C. These thermophilic fungi were able to grow on a minimal medium containing d-Ala, d-Asn, d-Gln, or d-His as the sole nitrogen source, which are better substrates of broad substrate specificity DAAOs such as RgDAAO and TvDAAO16 (Fig. 1). In particular, strain YA grew better on all the d-amino acids, especially on d-Gln, than the other strains. The nucleotide sequence of the ITS region of strain YA showed a high sequence identity with that of the thermophilic fungi R. emersonii strain CBS 393.64 (99.7%) and strain CBS 396.64 (100%); which suggests that it belongs to R. emersonii and to which we assigned it (Supplementary Fig. 1).
Identification of DAAO homologous gene of strain YA
To identify the DAAO homologous gene of strain YA, we first searched for the gene encoding putative DAAO protein in the genome sequence of a type strain of R. emersonii strain CBS 393.64 using the amino acid sequence of fungal DAAOs. The search gave a gene (T310_5354) that is annotated to code for putative DAAO. The genomic gene was predicted to comprise six exons spanning 1,443 bp and to encode a protein of 368 amino acids (Fig. 2). Based on the nucleotide sequence of strain CBS 393.64, we obtained a DAAO homologous gene of strain YA using PCR. The nucleotide sequence and the exon-intron structure of the strain YA homologous gene was identical to those of strain CBS 393.64 except for one base (G instead of A in the gene of strain CBS 393.64) near the 3′-end in the ORF, which causes a difference in the amino acid sequence as follows: Arg363 in strain YA DAAO homolog instead of Lys363 in strain CBS 393.64 DAAO (Fig. 2).
Multiple sequence alignment of ReDAAO with those of other known DAAOs and DDOs. DAAOs of Rhodotorula gracilis (Swiss-Prot, P80324), Trigonopsis variabilis (GenBank, CAA90322), pig kidney (GenBank, NP_999231), human (GenBank, NP_001908), and Rubrobacter xylanophilus (GenBank, BAP18969). DDOs of Cryptococcus humicola (GenBank, BAD13387) and Thermomyces dupontii (Fungal Genomes, Talth1p4_002474). The closed circles show the nucleotide-binding motif. The open circles indicate the position of the gate-anchoring residues, Tyr55 and Tyr314, in mammalian DAAOs. The wavy line shows the dimerization loop of RgDAAO. The open triangle shows the amino acid residues conserved in fungal DAAOs (Asn residue) and DDOs (His residue). The closed triangles show the amino acid residues interacting with the α-carboxy and α-amino groups of the substrates. The open square indicates the position of Gln residue that fixes the α-amino groups of substrates through a water molecule. The active site lid of pkDAAO is underlined. Met213 of RgDAAO and Cys108 of TvDAAO are in squares.
In the amino acid sequence of the putative DAAO of strain YA (ReDAAO), a nucleotide-binding motif (GXGXXG, where “X” is any amino acid) was observed in the N-terminal region (Fig. 2). The Tyr and Arg residues that interact with the α-carboxy group of the substrates in DAAOs were conserved at positions 244 and 308 of ReDAAO, respectively, and the Gly residue that interacts with the α-amino group of the substrates was found at position 335. An Asn and a Gln residues that are conserved in the active site of fungal DAAOs and fixe the α-amino group of substrates via hydrogen bonding through a water molecule in RgDAAO were found at positions 53 and 339, respectively19,24. In the corresponding position of Met213 of RgDAAO, which plays an important role in the substrate specificity of RgDAAO25, an Ile residue was present at position 232. A consensus type-1 peroxisome-targeting consensus sequence [STAGCN]-[RKH]-[LIVMAFY] (PROSITE, PS00342) was found in the C-terminus. These findings in the amino acid sequence suggested that ReDAAO gene could encode functional DAAO.
Expression in E. coli and purification of ReDAAO
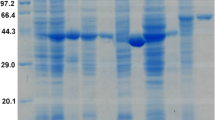
To confirm that ReDAAO gene encodes functional DAAO, ReDAAO was expressed in E. coli as a histidine-tagged fusion protein. The recombinant protein was efficiently expressed as a soluble protein in E. coli and was estimated to account for ~2% of the soluble proteins in the crude extract using SDS-PAGE (Fig. 3). The crude extract exhibited oxidase activity against neutral and basic d-amino acids, in particular d-Ile and d-Val (Supplementary Table 1). Interestingly, the crude extract also exhibited a significant activity against an acidic d-amino acid, d-Glu, a general substrate of DDO but not of DAAO. ReDAAO was purified approximately 25-fold with a yield of 25.7% from the crude extract, which gave 1.43 mg purified enzyme from 600 ml culture broth (Supplementary Table 2). The purified protein migrated as a single band with a molecular mass of 42 kDa using SDS-PAGE (Fig. 3) and 185 kDa, both at 0.01 and 0.1 mg/ml protein concentrations, using a gel filtration column, which suggested that the enzyme is a tetramer. The purified ReDAAO showed a specific activity of 70.0 and 112 U/mg at 37 °C and 55 °C, respectively, toward d-Ile as a substrate. The results suggested that the recombinant protein is DAAO with broad substrate specificity and high catalytic activity.
SDS-PAGE of purified ReDAAO. Lanes: M, protein marker; 1, crude extract; 2, purified ReDAAO. The original uncropped gel image is shown in Supplementary Fig. 3.
Cofactor analyses
To determine whether ReDAAO contains flavin, we analyzed its absorption spectrum (Fig. 4a). The protein solution was yellow and exhibited a typical absorption spectrum of flavoproteins, with maximum absorption intensities of 273, 370, and 455 nm. The maximum visible absorption disappeared by adding d-Ile to the protein solution. After removing the thermally denatured protein, the supernatant was also yellow and within the flavin absorption spectrum (data not shown), which indicated a noncovalent association between flavin and the protein. The TLC analysis of the supernatant showed a single fluorescent spot that was identical to that of the FAD standard, suggesting that the prosthetic group could be FAD (Fig. 4b). These results suggested that ReDAAO contains noncovalently bound FAD that functions as a cofactor.
Cofactor analysis of ReDAAO. (a) Absorption spectra of ReDAAO. 20 μM ReDAAO was analyzed before (solid line) and after (dashed line) incubating with 20 mM d-Ile at 40 °C for 10 min. (b) TLC analysis of ReDAAO flavin. Lanes: 1. 0.1 mM FMN; 2, 0.1 mM FAD; 3, flavin from 0.185 mM ReDAAO. The original uncropped TLC image is shown in Supplementary Fig. 4.
Substrate specificity and kinetic parameters
We analyzed the substrate specificity of purified ReDAAO using various d-amino acids, some l-amino acids, and CPC as substrates (Table 1). The enzyme showed high activities against various d-amino acids, especially d-Val and d-Ile, but not against the corresponding l-isomers. As in the case of the crude extract, the purified enzyme also exhibited significant oxidase activities toward d-Glu as well as CPC, which were approximately 56% and 14%, respectively, of that against d-Val, the best substrate. These results revealed that ReDAAO might have novel broad substrate specificity and might be potentially valuable for practical applications.
Next, we determined the kinetic parameters of ReDAAO for some neutral d-amino acids as well as for d-Glu and d-Arg (Table 2). The enzyme exhibited typical Michaelis–Menten kinetics for each substrate (Supplementary Fig. 2). The highest apparent catalytic rate constant (kcat) was observed for d-Val, followed by d-Ala, d-Met, d-Glu, and d-Arg, in decreasing order, and the lowest apparent Michaelis constant (Km) was obtained for d-Met, followed by d-Val, d-Ala, d-Glu, and d-Arg, in increasing order, thus, providing the highest apparent catalytic efficiency (kcat/Km) for d-Met followed by d-Val, d-Ala, d-Glu, and d-Arg, in decreasing order.
Effect of pH, temperature, protein concentration, and FAD
The enzyme exhibited higher activity at basic pHs with the highest at pH 8.0, but markedly lower activity at neutral and acidic pHs (e.g., <60% of that at pH 8.0) (Fig. 5a). The enzyme was stable between pH 6.0 and pH 10 for 1 h, where >80% of the activity at pH 8.0 was remained. The enzyme exhibited a higher activity at 50 –60 °C, with the highest at 55 °C (Fig. 5b). It was stable up to 50 °C for 1 h but gradually inactivated at >55 °C, and its activity was completely lost at 65 °C. T50 (the temperature at which it loses 50% activity) was ~60 °C. The thermal stability was only slightly decreased (approximately 12%) with decreasing the protein concentration from 0.1 mg/ml to 0.01 mg/ml, and the addition of exogenous FAD increased the stability at the lower protein concentration (Fig. 5c). In the case of the effect of FAD on pH stability, the addition of exogenous FAD significantly stabilized the enzyme at an alkaline pH but, strangely, conversely destabilized at an acidic pH (Fig. 5d).
Effect of pH, temperature, protein concentration, and FAD. (a) Effect of pH on activity (open circles) and stability (closed circles). (b) Effect of temperature on activity (open circles) and stability (closed circles). (c) Effect of protein concentration and exogenous FAD on thermal stability. (d) Effect of exogenous FAD on pH stability. For the determination of stability, the residual activity was determined after incubation at 45 °C for 1 h in the presence and absence of exogenous 0.1 mM FAD. Each data point represents the mean ± the standard deviation of three measurements.
Effect of DAAO and DDO competitive inhibitors
The effect of DAAO and DDO competitive inhibitors on ReDAAO activity was analyzed (Supplementary Table 3). With 10 mM competitive inhibitors, the enzyme was most inhibited by the DDO inhibitor, d-malate, but only by 13.5%. The other competitive inhibitors decreased the activity by <9%. With 50 mM competitive inhibitors, the activity was significantly inhibited by DAAO inhibitors and one DDO inhibitor as follows: benzoate, crotonate, and d-malate decreased the activity by 13.2%, 22.6%, and 17.2%, respectively. These results showed that ReDAAO is sensitive to both DAAO and DDO inhibitors.
Discussion
DAAO is a valuable enzyme for various practical applications, such as determining and quantifying d-amino acids and the production of materials for semisynthetic cephalosporins; therefore, the enzyme having high stability, high catalytic activity, and broad substrate specificity is desired. In this study, we isolated a strain of the thermophilic fungus R. emersonii that can efficiently grow on several d-amino acids and further isolated the fungal DAAO gene. The recombinant DAAO exhibited unique broad substrate specificity (i.e., the enzyme efficiently catalyzed the reaction of an unusual substrate, d-Glu and CPC) and high thermal stability as well as high catalytic activity. These results showed that ReDAAO has valuable and unique properties that are useful for various practical applications.
To isolate a practically useful DAAO which has high thermal stability, high catalytic activity, and broad substrate specificity, we first aimed to isolate the thermophilic fungi that can grow well on d-Asn, d-Gln, and d-His as the sole source of nitrogen source, because the practically useful fungal DAAOs, RgDAAO and TvDAAO, which have high catalytic activity and broad substrate specificity and can be used in the production of 7-ACA from CPC, can efficiently act on these d-amino acids16. Additionally, to our knowledge, no other enzymes, except DAAO, have been reported to accept d-Asn, d-Gln, and d-His as substrates in fungi26; therefore, the growth of the thermophilic fungi on the d-amino acids was predicted to be supported mainly by DAAO, as shown in other fungal DAAOs27. However, the growth level of R. emersonii strain YA on each d-amino acid appeared to be not fully comparable to the respective activities of ReDAAO (Fig. 1, Table 1). Other factors, such as the different uptake efficiencies among the d-amino acids, might also affect the growth on these d-amino acids. Nevertheless, as we successfully obtained a targeted DAAO, other novel fungal DAAOs that have unique and useful substrate specificities could be obtained by the screening based on the growth of fungi on d-amino acids.
The optimal pH of ReDAAO was in alkaline region similar to those of RgDAAO and pkDAAO but not to that of TvDAAO (Fig. 5a), which has the optimal pH within the neutral region17,28. Additionally, the pH stability of ReDAAO was somewhat different from that of the fungal DAAOs, RgDAAO and TvDAAO: The fungal DAAOs are stable at pH 6.0, while ReDAAO was slightly unstable at the pH. ReDAAO had high catalytic activity at 55 °C, which was comparable to that of other fungal DAAOs and higher than mammalian and bacterial DAAOs, although the assay temperature in this study was different from that in the assay of other DAAOs. Mammalian DAAOs typically have a long loop covering the active site, called active-site lid, that opens and closes to accept substrates and to release products29. This conformational change is slow and thereby lowers the overall turnover rate30. The fungal RgDAAO lacks this lid, which has been considered to be the reason for its higher catalytic activity24. In pig kidney DAAO (pkDAAO), the active site lid is formed by 13 amino acid residues, Thr216–Tyr228 (Fig. 2). In the corresponding region of ReDAAO as well as RgDAAO, a relatively small number of amino acid residues (10 and 9 amino acid residues, respectively) were found in the sequence alignment, suggesting the possibility that ReDAAO might also lack the lid; however, the same number of amino acid residues as ReDAAO was also observed in the corresponding lid region of the low catalytic bacterial RxDAAO, while a comparable or a larger number of residues than that of mammalian DAAOs were found in the lid region of the higher catalytic TvDAAO (12 amino acid residues) and DDO of the yeast Cryptococcus humicola (15 amino acid residues) (Fig. 2). Thus, further studies, such as those on determining their crystal structures, might be necessary to clarify the relationship between the catalytic turnover rate and the loop length.
ReDAAO accommodated various d-amino acids and CPC as substrates, similar to the fungal RgDAAO and TvDAAO, but there were some differences in their substrate use (Table 1). RgDAAO showed relatively moderate activity against d-Trp and d-Pro, whereas ReDAAO showed much lower activity against these d-amino acids. Conversely, RgDAAO showed much lower activity against d-Arg, whereas, as with TvDAAO, ReDAAO showed moderate activity against the basic d-amino acid25,31. The most intriguing feature of ReDAAO was its high activity against d-Glu, which is a typical substrate of DDO but not of DAAO (Table 1). The apparent kcat/Km value for d-Glu of ReDAAO at an optimum temperature was approximately 600-fold higher than that of RgDAAO25. Such a DDO property of ReDAAO was also confirmed in its sensitivity against DDO competitive inhibitors (Supplementary Table 3). These results suggested that ReDAAO might be a novel DAAO having both DAAO and DDO activities. The amino acid residues located at the upper side of the active site, including those of the active site lid, are involved in the substrate specificity of DAAO25,32. In RgDAAO, Met213 in the active site plays a most important role in the substrate specificity, and the mutation of Met213 into Arg residue gave the oxidase activity against d-Asp25, a general substrate of DDO. In the spatially corresponding position of ReDAAO model, Ser233 is likely to be located instead of Ile232 that was predicted by amino acid sequence alignment described above (Figs 2 and 6b), suggesting that Ser233 might be involved in the substrate specificity. In addition, the substitution of a part of the lid of pkDAAO with the corresponding part of human DDO, which contains an Arg residue, also provided DDO activity32. In mouse DDO, Arg216 in the active site is vital for accepting acidic d-amino acids as substrates by possibly interacting with the β-carboxy side chain of the substrates (Fig. 6a)33. In the model structure of ReDAAO, an Arg residue at position 97 was found at the upper side of the active site (Fig. 6b); whereas, in RgDAAO, a Gln residue is present in the corresponding spatial position (Fig. 6c), which suggested the possibility that Arg97 might play an important role in the higher activity against d-Glu.
Active site structures of ReDAAO, RgDAAO, pkDAAO, and mouse DDO. Oxygen, nitrogen, and sulfur atoms are shown in red, blue, and yellow, respectively. Carbon atoms of the amino acid residues of proteins and substrates are shown in gray or green and pink, respectively. FAD is shown in orange. (a) Active site model of mouse DDO. (b) Active site model of ReDAAO. The hydrophobic amino acid residues covering the side chain of substrates are indicated by green. (c) Active site of RgDAAO crystal structure (PDB accession no. 1c0p). (d) Active site of pkDAAO crystal structure (PDB accession no. 1kif). Each image was rendered using PyMOL (https://pymol.org/2/).
ReDAAO exhibited higher activity, in particular, toward hydrophobic d-amino acids (Table 1). In the active site of ReDAAO model, the hydrophobic amino acid residues Tyr54, Leu55, Pro56, Val57, Val94, and Phe144 are located at the position covering the side chain of substrates (Fig. 6b). In mammalian DAAOs, the corresponding hydrophobic region, mainly formed by the amino acid residues around Tyr55 with Tyr314 in pkDAAO (Figs 2 and 6d), is called the hydrophobic secondary binding pocket and, besides the active site lid, play an important role in the substrate specificity, activity, product egress, and inhibitor binding34,35. The van der Waals and hydrophobic interactions in this region of ReDAAO might also contribute to the higher activity toward hydrophobic substrates and the substrate specificity. The importance of hydrophobic interactions could also be explained by considerably lower activities against d-Thr than that against d-Val and also against d-Asn than that against d-Leu (Table 1), which are similar in size and shape but different in hydrophobicity. In addition, the shape of the side chain of substrates might affect the activity, because the activities against d-Thr and d-allo-Thr as well as d-Ile and d-allo-Ile were largely different (Table 1). The substitutions of Phe54 of TvDAAO and Tyr55 of pkDAAO with other amino acid residues change their substrate specificities (Figs 2 and 6d)36. Val57 was likely to be placed at the corresponding position in ReDAAO (Figs 2 and 6b), suggesting that Val57 and also adjacent amino acid residues might cause the different activities toward the isomers possibly by their steric hindrance. Tyr55 of mammalian DAAOs, which are the corresponding amino acid residues of Val57 of ReDAAO, plays a role in controlling access the active site, together with Y224, Y228, and Y314 (https://doi.org/10.1371/journal.pone.0198990). The substitution of Tyr55 with a smaller amino acid residue Ala improve the activity toward bulky substrates such as d-Trp, which might be due to significantly improved access to the active site by the substitution. From these findings, we first supposed that the broad substrate specificity of ReDAAO was due to the smaller amino acid residue Val57 instead of a Tyr or Phe residue found at the corresponding position in other DAAOs (Fig. 2). However, as described above, ReDAAO was less active on d-Trp (Table 1), while RgDAAO, which has a Phe residue (Phe58 in Fig. 6c) at the corresponding position, is moderately active on the bulky substrate37. The further analysis, such as the determination of the crystal structure and mutational analysis, might be however needed to more clearly understand the structural factors that govern the substrate specificity.
The T50 of ReDAAO (60 °C) was much higher than those of mammalian and other fungal DAAOs, which are 44 °C, 39 °C, and 54 °C for pkDAAO, RgDAAO, and TvDAAO, respectively, at 0.1 mg/ml protein concentration and after incubating for 30 min28, and comparable to that of the thermophilic bacterial RxDAAO (64 °C) at 0.1 mg/ml protein concentration and after incubating for 1 h17. These comparisons showed that ReDAAO has much higher thermal stability than known DAAOs, except for thermophilic bacterial RxDAAO. FAD and subunit interactions have been shown to affect the thermal stability of DAAO17,38,39. In this study, ReDAAO was more thermally stable in the presence of an excess amount of exogenous FAD (Fig. 5c), suggesting that the interaction with FAD might be involved in the higher thermal stability. In the effect of exogenous FAD on pH stability, we obtained an intriguing result that exogenous FAD considerably destabilized ReDAAO at an acidic condition (Fig. 5d). As the enzyme was stabilized by exogenous FAD at an alkaline condition, the protonation of exogenous FAD could cause the decreased stability at the acidic condition. Because hydrogen bonds are important for binding to FAD40, the protonation of FAD might impair the hydrogen-bonding interactions, leading to the enhanced destabilization of the enzyme. Additionally, as ReDAAO was a tetramer, the subunit interaction might be also involved in the higher thermal stability. The thermal stability of DAAOs is usually depended on the protein concentration: the decrease in protein concentration markedly decreased the thermal stability due to subunit dissociation41. However, the thermal stability of ReDAAO was only slightly decreased by decreasing protein concentration (Fig. 5c). As the tetramer formation of ReDAAO was still observed at the lower protein concentration, tight subunit interactions might also contribute to the higher thermal stability. It has been also reported that the oxidation of Cys108 in TvDAAO, which is not located in the active site, significantly decreases the stability by affecting the protein environment of the bound FAD42. In the corresponding position of ReDAAO, a Cys residue was not observed (Fig. 2), suggesting the lack of destabilizing Cys residue might be also a reason for the higher stability.
Methods
Materials
d-Leu and d-His were purchased from Wako Pure Chemical Industries (Osaka, Japan), and d-Ile was from the Peptide Institute (Osaka, Japan). d-Asp was a gift from Tanabe Pharmaceutical (Osaka, Japan). All other d-amino acids were purchased from Nacalai Tesque (Kyoto, Japan). DNA modification enzymes were obtained from Toyobo (Osaka, Japan) or Takara Bio (Shiga, Japan). All other chemicals were of analytical reagent grade and were from Wako Pure Chemical Industries, Nacalai Tesque, or Sigma-Aldrich (St. Louis, MO, USA). Primers were synthesized by Eurofins (Tokyo, Japan). Composts were purchased from local markets.
Microbial strains and cultivation media
E. coli strain DH5α and strain BL21(DE3) pLysS were used as the host for DNA manipulation and protein expression, respectively. Thermophilic fungi were grown on potato dextrose agar (PDA) medium (Merck KGaA, Darmstadt, Germany). E. coli cells were cultivated in LB. For solid media, 2.0% or 1.5% (w/v) agar was added to the media for fungi and E. coli, respectively.
Isolation of thermophilic fungi
One gram of compost was suspended in 10 ml saline solution. An aliquot of the suspensions was spread onto PDA medium containing 0.5 g/l chloramphenicol and 0.05 g/l gentamicin and incubated at 50 °C and 60 °C for 2 weeks.
D-Amino acid-assimilating ability of thermophilic fungi
Thermophilic fungi were inoculated onto synthetic agar media composed of a yeast carbon base (Becton Dickinson Co., Franklin Lakes, NJ, USA) containing 50 mM d-Ala, d-Asn, d-Gln, or d-His as the sole nitrogen source and incubated at 50 °C for 1 week.
Identification of thermophilic fungi
The genomic DNA of thermophilic fungi was isolated as described by Yu et al.43. The internal transcribed spacer (ITS) region was amplified using Tks Gflex DNA polymerase with the primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′)44 and genomic DNA as a template (Takara Bio, Shiga, Japan). The reaction mixture was preheated at 98 °C for 1 min, followed by 30 cycles at 98 °C for 10 s, 55 °C for 10 s, and 68 °C for 15 s, with a final extension at 68 °C for 10 min. The product was ligated into the pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced. NCBI-BLAST was used to conduct the homology search of the ITS region of strain YA. A multiple sequence alignment of the ITS region was conducted using T-Coffee (https://tcoffee.vital-it.ch/apps/tcoffee/do:regular) and was edited using Gblocks v. 0.91b (http://molevol.cmima.csic.es/castresana/Gblocks_server.html). A phylogenetic tree was constructed using the maximum likelihood method with MEGA 7.0 (https://www.megasoftware.net/home).
Isolation DAAO homologous gene of R. emersonii strain YA
The DAAO homologous gene (T310_5354) of R. emersonii strain CBS 393.64 was identified by a BLAST search using amino acid sequences of RgDAAO and TvDAAO in the fungal genome database Ensembl Fungi (http://fungi.ensembl.org/). The corresponding homologous gene of strain YA (ReDAAO) was amplified using Tks Gflex DNA polymerase with the primers ReDAAO-F (5′-ATGGCAACCAATAACATCG-3′) and ReDAAO-R (5′-TTACAGCCTCGCCTCTCC-3′) and the genomic DNA of strain YA as a template. The reaction mixture was preheated at 98 °C for 1 min, followed by 30 cycles at 98 °C for 10 s, 50 °C for 15 s, and 68 °C for 84 s, with a final extension at 68 °C for 10 min. The resulting product was inserted into the pGEM-T Easy vector and sequenced. ReDAAO ORF was synthesized based on the codon usage of E. coli by GenScript (Piscataway, NJ, USA) and ligated into pET15b.
Expression and purification of ReDAAO in E. coli
E. coli BL21(DE3) pLysS harboring ReDAAO was precultivated in LB medium containing 100 µg/ml ampicillin and 34 µg/ml chloramphenicol at 37 °C for 14 h. One milliliter of preculture was transferred to 200 ml LB medium containing the same antibiotics and cultivated at 37 °C. When the optical density at 660 nm reached approximately 0.5, isopropyl β-d-thiogalactopyranoside (IPTG) was added to the medium at a final concentration of 0.1 mM, and the cells were further cultivated at 37 °C for 4 h. The cells were collected after centrifuging at 5,000 × g for 10 min at 4 °C and were resuspended in 50 mM potassium phosphate (KPi) buffer (pH 8.0). The cells were sonicated 25 times for 30 s with the UD-201 sonicator (Tomy Seiko, Tokyo, Japan) at an output of 10 and a duty cycle of 50%. The homogenate was centrifuged at 20,000 × g for 30 min at 4 °C, and the supernatant was filtrated through a 0.45-µm membrane filter (Omnipore; Millipore, Billerica, MA, USA). After adding imidazole at a final concentration of 10 mM, the supernatant was applied to a TALON metal-affinity column (Clontech, Mountain View, CA, USA). The column was washed once with 50 mM KPi buffer (pH 8.0) containing 300 mM NaCl and 30 mM imidazole, and the bound proteins were eluted with the same buffer containing 150 mM imidazole. The eluate was concentrated using an Amicon Ultra-15 centrifugal filter device with a 10 K molecular–weight cutoff membrane (Millipore) and dialyzed overnight against 50 mM KPi buffer (pH 8.0) containing 5% (v/v) glycerol.
SDS-PAGE and molecular determination
The homogeneity of purified ReDAAO was determined using 12.5% SDS-PAGE. The native molecular mass was determined by gel filtration on a Superdex 200 HR 10/30 column (GE Healthcare, Tokyo, Japan) equilibrated with 50 mM KPi buffer (pH 8.0) containing 150 mM NaCl and 5% (v/v) glycerol at a flow rate of 0.3 ml/min. The molecular mass standards used were catalase (232 kDa), aldolase (158 kDa), conalbumin (75.0 kDa), chymotrypsinogen A (25.0 kDa), and RNase A (13.7 kDa).
Enzyme assay and protein determination
Enzyme activity was determined using a horseradish peroxidase (HRP)–coupled reaction with phenol and 4-aminoantipyrine (4-AAP) or using 2,4-dinitrophenylhydrazine (DNPH). The HRP-coupled reaction mixture contained 20 mM amino acid (2 mM d-Tyr), 2 mM phenol, 1.5 mM 4-AA, 2.5 U highly stabilized HRP (Sigma-Aldrich), and 0.02 mM FAD in 50 mM KPi buffer (pH 8.0). The enzyme activity was determined at 505 nm using a molar extinction coefficient for quinonimine of 5.86 × 103 M−1 cm−1 45. A DNPH assay was conducted as described previously17. The protein concentration was determined using the BioRad protein assay kit (Bio-Rad, Hercules, CA, USA) with fetal bovine serum albumin as a standard.
Substrate specificity and kinetic parameters
The substrate specificity and the apparent kinetic parameters were determined using the HRP-coupled method at 37 °C and 55 °C, respectively. For determining the apparent kinetic parameters, enzyme assays were conducted with 0.5–100 mM d-amino acids. The apparent kinetic parameters were determined by fitting the Michaelis–Menten equation to the initial reaction rates using SigmaPlot 12.5 (Systat Software, San Jose, CA, USA).
Absorption spectra
The absorption spectra of the enzyme (0.16 mg/ml) in 50 mM KPi buffer (pH 8.0) were measured before and after incubation with 50 mM d-Val at 40 °C for 10 min using a Shimadzu UV-3100PC spectrophotometer (Kyoto, Japan).
Thin-layer chromatography
ReDAAO cofactor was analyzed by thin-layer chromatography (TLC) as described by Yamada et al.46. The purified enzyme (0.185 mM) in 50 mM KPi buffer (pH 8.0) containing 5% (v/v) glycerol was incubated at 100 °C for 10 min in the dark. After centrifugation at 20,000 × g at 4 °C for 30 min, the supernatant (1.0 µl) was subjected to TLC on silica gel 60 F-254 plates (2-mm thickness; Merck) and developed in 5% Na2HPO4 (w/v) as the solvent phase. FAD and flavin mononucleotide (FMN; 1 µl, 0.1 mM each) in 50 mM KPi buffer (pH 8.0) containing 5% (v/v) glycerol were used as the standards and monitored under UV light (312 nm).
Effect of temperature, pH, protein concentration, and FAD
The optimum pH was determined by measuring the activity at different pH values in 50 mM GTA buffer (16.7 mM 3,3-dimethyl glutaric acid, 16.7 mM tris, and 16.7 mM 2-amino-2-methyl-1,3-propanediol). The optimum temperature was determined by measuring the activity at different temperatures. The pH stability was determined by measuring the residual activity after incubating 0.1 mg/ml enzyme at 45 °C for 1 h in 50 mM GTA buffer at different pH values. The thermal stability was determined by measuring the residual activity after incubating 0.1 mg/ml enzyme in 50 mM KPi buffer (pH 8.0) for 1 h at different temperatures. The effect of protein concentration and the effect of exogenous 0.1 mM FAD on thermal stability or pH stability were analyzed after incubation at 45 °C for 1 h. The enzyme assays for pH and thermal stability were conducted using the HRP-coupled method at 37 °C with 20 mM d-Val as a substrate, and those for optimum pH and temperature were conducted using the DNPH method at 55 °C with 20 mM d-Val as a substrate.
Effect of competitive inhibitors
The effects of competitive inhibitors were analyzed by assaying the activity in the presence of 10 mM or 50 mM each inhibitor. These assays were conducted using the DNPH method at 55 °C with 20 mM d-Val as a substrate.
Construction of three-dimensional model structures
The three-dimensional model structures of ReDAAO and mouse DDO were generated using I-TASSER Server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) and evaluated using PROCHECK47.
Nucleotide sequence accession numbers
The DNA sequences of the ITS region of strain YA, the genomic DNA with introns, and the synthesized ORF DNA of ReDAAO gene were deposited in DDBJ/EMBL/GenBank under accession numbers LC436776, LC436777, and LC437087, respectively.
References
Takahashi, S., Abe, K. & Kera, Y. Bacterial d-amino acid oxidases: Recent findings and future perspectives. Bioengineered 6, 237–241, https://doi.org/10.1080/21655979.2015.1052917 (2015).
Pilone, M. S. d-Amino acid oxidase: new findings. Cell Mol Life Sci 57, 1732–1747, https://doi.org/10.1007/pl00000655 (2000).
Pollegioni, L., Porrini, D., Molla, G. & Pilone, M. S. Redox potentials and their pH dependence of d-amino-acid oxidase of Rhodotorula gracilis and Trigonopsis variabilis. Eur J Biochem 267, 6624–6632, https://doi.org/10.1046/j.1432-1327.2000.01757.x (2000).
Mothet, J. P. et al. d-Serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc Natl Acad Sci USA 97, 4926–4931, https://doi.org/10.1073/pnas.97.9.4926 (2000).
Schell, M. J., Molliver, M. E. & Snyder, S. H. d-Serine, an endogenous synaptic modulator: localization to astrocytes and glutamate-stimulated release. Proc Natl Acad Sci USA 92, 3948, https://doi.org/10.1073/pnas.92.9.3948 (1995).
Tsai, G., Yang, P., Chung, L.-C., Lange, N. & Coyle, J. T. D-Serine Added to Antipsychotics for the Treatment of Schizophrenia. Biol Psychiatry 44, 1081–1089, https://doi.org/10.1016/S0006-3223(98)00279-0 (1998).
Chumakov, I. et al. Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 99, 13675, https://doi.org/10.1073/pnas.182412499 (2002).
Hashimoto et al. Decreased Serum Levels of d-Serine in Patients with Schizophrenia: Evidence in Support of the N-Methyl-d-Aspartate Receptor Hypofunction Hypothesis of Schizophrenia. Arch Gen Psychiatry 60, 572–576, https://doi.org/10.1001/archpsyc.60.6.572 (2003).
Sasabe, J., Suzuki, M., Imanishi, N. & Aiso, S. Activity of D-amino acid oxidase is widespread in the human central nervous system. Front Synaptic Neurosci 6, 14, https://doi.org/10.3389/fnsyn.2014.00014 (2014).
Pollegioni, L. & Molla, G. New biotech applications from evolved D-amino acid oxidases. Trends Biotechnol 29, 276–283, https://doi.org/10.1016/j.tibtech.2011.01.010 (2011).
Pollegioni, L. et al. Properties and applications of microbial d-amino acid oxidases: current state and perspectives. Appl Microbiol Biotechnol 78, 1–16, https://doi.org/10.1007/s00253-007-1282-4 (2008).
Khoronenkova, S. V., Tishkov, V. I. & D-Amino Acid Oxidase: Physiological Role and Applications. Biochemistry (Mosc) 73, 1511–1518, https://doi.org/10.1134/S0006297908130105 (2008).
Patel, R. N. Enzymatic synthesis of chiral intermediates for Omapatrilat, an antihypertensive drug. Biomol Eng 17, 167–182, https://doi.org/10.1016/S1389-0344(01)00068-5 (2001).
Sacchi, S., Pollegioni, L., Pilone, M. S. & Rossetti, C. Determination of D-amino acids using a D-amino acid oxidase biosensor with spectrophotometric and potentiometric detection. Biotechnol Techniques 12, 149–153, https://doi.org/10.1023/A:1008896602544 (1998).
Garcia-Garcia, M., Martinez-Martinez, I., Sanchez-Ferrer, A. & Garcia-Carmona, F. Production of the Apoptotic Cellular Mediator 4-Methylthio-2-oxobutyric Acid by Using an Enzymatic Stirred Tank Reactor with in Situ Product Removal. Biotechnol Prog 24, 187–191, https://doi.org/10.1021/bp0702424 (2008).
Tishkov, V. I. & Khoronenkova, S. V. D-Amino Acid Oxidase: Structure, Catalytic Mechanism, and Practical Application. Biochemistry (Mosc) 70, 40–54, https://doi.org/10.1007/s10541-005-0050-2 (2005).
Takahashi, S. et al. A Highly Stable d-Amino Acid Oxidase of the Thermophilic Bacterium Rubrobacter xylanophilus. Appl Environ Microbiol 80, 7219–7229, https://doi.org/10.1128/AEM.02193-14 (2014).
Mouchacca, J. Thermophilic Fungi: Present Taxonomic Concepts in Thermophilic Moulds in Biotechnology (ed. Johri, B, N., Satyanarayana, T. & Olsen, J.) 43–83 (Springer, 1999).
Takahashi, S. et al. Characterization and improvement of substrate-binding affinity of d-aspartate oxidase of the thermophilic fungus Thermomyces dupontii. Appl Microbiol Biotechnol, https://doi.org/10.1007/s00253-019-09787-y (2019).
Collins, C. M. et al. Molecular cloning and expression analysis of two distinct β-glucosidase genes, bg1 and aven1, with very different biological roles from the thermophilic, saprophytic fungus Talaromyces emersonii. Mycol Res 111, 840–849, https://doi.org/10.1016/j.mycres.2007.05.007 (2007).
Houbraken, J., Spierenburg, H. & Frisvad, J. C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek 101, 403–421, https://doi.org/10.1007/s10482-011-9647-1 (2012).
Martínez, P. M., Appeldoorn, M. M., Gruppen, H. & Kabel, M. A. The two Rasamsonia emersonii α-glucuronidases, ReGH67 and ReGH115, show a different mode-of-action towards glucuronoxylan and glucuronoxylo-oligosaccharides. Biotechnol for Biofuels 9, 105, https://doi.org/10.1186/s13068-016-0519-9 (2016).
Ozawa, K. et al. Identification and characterization of thermostable glucose dehydrogenases from thermophilic filamentous fungi. Appl Microbiol Biotechnol 101, 173–183, https://doi.org/10.1007/s00253-016-7754-7 (2017).
Pollegioni, L. et al. Yeast d-Amino Acid Oxidase: Structural Basis of its Catalytic Properties. J Mol Biol 324, 535–546, https://doi.org/10.1016/S0022-2836(02)01062-8 (2002).
Sacchi, S. et al. Engineering the Substrate Specificity of d-Amino-acid Oxidase. J biol chem 277, 27510–27516, https://doi.org/10.1074/jbc.M203946200 (2002).
Takahashi, S., Abe, K., Shibata, K. & Kera, Y. Functions and metabolism of D-amino acids in microorganisms in The handbook of microbial metabolism of amino acids. (ed. D’Mello, J. P. F.) 315–331 (CABI, 2017).
Pollegioni, L., Piubelli, L., Sacchi, S., Pilone, M. S. & Molla, G. Physiological functions of D-amino acid oxidases: from yeast to humans. Cell Mol Life Sci 64, 1373–1394, https://doi.org/10.1007/s00018-007-6558-4 (2007).
Pollegioni, L., Caldinelli, L., Molla, G., Sacchi, S. & Pilone, M. S. Catalytic Properties of d-Amino Acid Oxidase in Cephalosporin C Bioconversion: A Comparison between Proteins from Different Sources. Biotechnol Prog 20, 467–473, https://doi.org/10.1021/bp034206q (2004).
Todone, F. et al. Active Site Plasticity in d-Amino Acid Oxidase: A Crystallographic Analysis. Biochemistry 36, 5853–5860, https://doi.org/10.1021/bi9630570 (1997).
Porter, D. J., Voet, J. G. & Bright, H. J. Mechanistic Features of the d-Amino Acid Oxidase Reaction Studied by Double Stopped Flow Spectrophotometry. J biol chem 252, 4464–4473 (1977).
Komarova, N. V., Golubev, I. V., Khoronenkova, S. V., Chubar, T. A. & Tishkov, V. I. Engineering of Substrate Specificity of D-Amino Acid Oxidase from the Yeast Trigonopsis variabilis: Directed Mutagenesis of Phe258 Residue. Biochemistry (Mosc) 77, 1181–1189, https://doi.org/10.1134/s0006297912100100 (2012).
Setoyama, C. et al. Engineering the Substrate Specificity of Porcine Kidney d-Amino Acid Oxidase by Mutagenesis of the “Active-Site Lid”. J Biochem 139, 873–879, https://doi.org/10.1093/jb/mvj094 (2006).
Katane, M. et al. Role of the active site residues arginine-216 and arginine-237 in the substrate specificity of mammalian d-aspartate oxidase. Amino acids 40, 467–476, https://doi.org/10.1007/s00726-010-0658-4 (2011).
Sacchi, S., Caldinelli, L., Cappelletti, P., Pollegioni, L. & Molla, G. Structure-function relationships in human D-amino acid oxidase. Amino acids 43, 1833–1850, https://doi.org/10.1007/s00726-012-1345-4 (2012).
Subramanian, K. et al. Modulating D-amino acid oxidase (DAAO) substrate specificity through facilitated solvent access. PLoS One 13, e0198990, https://doi.org/10.1371/journal.pone.0198990 (2018).
Wong, K. S., Fong, W. P. & Tsang, P. W. A single Phe54Tyr substitution improves the catalytic activity and thermostability of Trigonopsis variabilis d-amino acid oxidase. N biotechnol 27, 78–84, https://doi.org/10.1016/j.nbt.2009.11.002 (2010).
Wang, S. J., Yu, C. Y., Lee, C. K., Chern, M. K. & Kuan, I. C. Subunit fusion of two yeast d-amino acid oxidases enhances their thermostability and resistance to H2O2. Biotechnol Lett 30, 1415–1422, https://doi.org/10.1007/s10529-008-9694-5 (2008).
Dib, I., Slavica, A., Riethorst, W. & Nidetzky, B. Thermal Inactivation of d-Amino Acid Oxidase From Trigonopsis variabilis Occurs Via Three Parallel Paths of Irreversible Denaturation. Biotechnol bioeng 94, 645–654, https://doi.org/10.1002/bit.20854 (2006).
Piubelli, L., Caldinelli, L., Molla, G., Pilone, M. S. & Pollegioni, L. Conversion of the dimeric d-amino acid oxidase from Rhodotorula gracilis to a monomeric form. A rational mutagenesis approach. FEBS Lett 526, 43–48, https://doi.org/10.1016/S0014-5793(02)03111-3 (2002).
Pollegioni, L. et al. Engineering the Properties of D-Amino Acid Oxidases by a Rational and a Directed Evolution Approach. Curr Protein Pept Sci 8, 600–618, https://doi.org/10.2174/138920307783018677 (2007).
Betancor, L. et al. Use of Physicochemical Tools to Determine the Choice of Optimal Enzyme: Stabilization of d-Amino Acid Oxidase. Biotechnol Prog 19, 784–788, https://doi.org/10.1021/bp025761f (2003).
Slavica, A., Dib, I. & Nidetzky, B. Single-Site Oxidation, Cysteine 108 to Cysteine Sulfinic Acid, in d-Amino Acid Oxidase from Trigonopsis variabilis and Its Structural and Functional Consequences. Appl Environ Microbiol 71, 8061–8068, https://doi.org/10.1128/aem.71.12.8061-8068.2005 (2005).
Yu, J. H. et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41, 973–981, https://doi.org/10.1016/j.fgb.2004.08.001 (2004).
Langarica-Fuentes, A., Handley, P. S., Houlden, A., Fox, G. & Robson, G. D. An investigation of the biodiversity of thermophilic and thermotolerant fungal species in composts using culture-based and molecular techniques. Fungal Ecology 11, 132–144, https://doi.org/10.1016/j.funeco.2014.05.007 (2014).
Tedeschi, G., Pollegioni, L. & Negri, A. Assays of d-Amino Acid Oxidases. Methods Mol Biol 794, 381–395, https://doi.org/10.1007/978-1-61779-331-8_26 (2012).
Yamada, R. et al. Purification and properties of d-aspartate oxidase from Cryptococcus humicolus UJ1. Biochim biophys acta 1294, 153–158, https://doi.org/10.1016/0167-4838(96)00012-x (1996).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26, 283–291, https://doi.org/10.1107/S0021889892009944 (1993).
Acknowledgements
This study was supported by the Grant-in-Aid for Scientific Research (C) (23580106) to S. Takahashi from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Contributions
Y.S. and S.T. conceived the experiments. Y.S., T.F. and S.T. conducted the experiments. Y.S., T.F., K.A., Y.K. and S.T. analyzed the results and wrote the manuscripts.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shimekake, Y., Furuichi, T., Abe, K. et al. A novel thermostable d-amino acid oxidase of the thermophilic fungus Rasamsonia emersonii strain YA. Sci Rep 9, 11948 (2019). https://doi.org/10.1038/s41598-019-48480-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48480-y
This article is cited by
-
Modeling the role of charged residues in thermophilic proteins by rotamer and dynamic cross correlation analysis
Journal of Molecular Modeling (2023)
-
A novel hyperthermophilic methylglyoxal synthase: molecular dynamic analysis on the regional fluctuations
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.