Abstract
Whether the effect of migration-selection-drift equilibrium on population structure is governed by spatial or environmental differences is usually elucidated by isolation-by-distance (IBD), isolation-by-environment (IBE), and isolation-by-resistance (IBR) tests. The population structure of Ammopiptanthus mongolicus, a broad-leaved evergreen psammophyte in eastern Central Asia, was previously thought to follow an isolation by distance pattern. However, recent studies have emphasized the effects of environmental factors on its growth and distribution, suggesting an important influence of local adaptation on the genetic structure of the species. Using inter-simple sequence repeat (ISSR) markers, we verified the previously inferred low intra-population variation and high inter-population differentiation. However, in contrast to previous studies, the results of partial Mantel tests and a maximum likelihood population effects mixed model (MLPE) suggested that local climate differences, rather than geographic distances or resistance distances, are the main factor affecting population differentiation. Further analysis with removal of multicollinear climatic variables and univariate MLPE found that summer and winter precipitation were crucial for shaping the current population genetic structure. Since local precipitation is related to the regeneration, colonization, and overwintering survival of A. mongolicus, its influence on demographic change may explain its effect on the population genetic structure. In addition, precipitation is related to terrain despite westward decreases, which explains the independence of genetic difference and geographic distance. The identified role of IBE suggests that collecting germplasm resources from genetically differentiated populations could be a more effective strategy to preserve the overall genetic diversity of the species than the establishment of corridors to enhance gene flow among populations.
Similar content being viewed by others
Introduction
Random genetic drift, environment-leading selection, genetic draft (hitchhiking), and background selection may affect the genetic diversity of organisms1. Geographic distance and environmental difference are two key factors affecting genetic structure between populations2. The former is related to the interplay of genetic drift and movement, while the latter is usually related to the adaptability to environmental pressure3. In changeable environments, selection determines genetic diversity of adapted genes, and the genetic diversity of neutral genes would be also reduced by random associations with genetic backgrounds of different fitness (i.e. genetic draft)4. Due to the combined effects of genetic drift and variation-reducting selection, the distribution of genetic variation among populations may be uneven and the genetic diversity may be lower than expected predicted by census population size5,6. Such decline in genetic diversity may in turn limit the adaptability of populations to environmental change. Gene rescue by gene flow can reduce the threat of local extinction7, although the influx of genes may be still constrained by environmental selection8.
The population structure of a species distributed across an environmental gradient may be affected by both geographic and environmental factors simultaneously. Autocorrelations of geographic and environmental distances can be additive for genetic differentiation among populations but often confound each other3,9,10. The additive effect of such autocorrelations is due not only to the proportional alignment of environmental differences with geographic distance but also to the promotion of divergence by ecological barriers to gene flow1,3. That is, when environmental conditions differ, the reduced establishment success of immigrants may accelerate the genetic fixation rate due to the decreased chance of outcrossing, thereby enhancing genetic isolation. This phenomenon not only appears in adaptive loci but could also extend to the whole genome via genetic draft caused by selective sweeps1. In this situation, the synergistic effects of environmental-driven and draft selection will lead to positive correlations of both neutral and adaptive loci (instead of adaptive loci only) with environmental differences3.
The impact of geographic distance or environmental difference on genetic differences among populations of a species reveals differences in resilience in adapting to heterogeneous environments11. Spatial resilience focuses on the patterns and processes of connectivity among locations. The local system resilience may be affected by the geographic distance and environmental heterogeneity among localities. Populations with isolation-by-distance (IBD) reveal positive correlations between genetic distances and geographic distances among populations, in which the genetic diversity turnover relies on the genetic rescue from neighboring populations. By contrast, isolation-by-environment (IBE) indicates populations harboring different genotypes. Variations in genetic composition of IBE populations are sensitive to environmental changes. Populations that are already adapted to alternative environments could harbor higher resilience and potential to adapt to environmental change. Therefore, the resilience of IBE populations is determined by the degree of environmental differences and the adaptability of the population. In addition, terrain and environmental variation may impede the direction and success of dispersal, i.e. isolation-by-resistance (IBR)12,13. IBR represents the ecological process or physiological limitation of organisms to dispersal14. Lower terrain and environmental resistance and unimpeded habitat connectivity enhance the resilience for species persistence. Since populations affected by IBE potentially have more restricted niche tolerance than a population of a comparable species with only IBD, the mechanism structuring populations must be considered when formulating conservation policies.
An understanding of local adaptation and dispersal limitation can support the development of more appropriate management strategies. For example, when evaluating “single-large or several-small” (SLOSS) strategies for planning a protected area, the several-small strategy and/or collection of germplasm resources from different populations for ex situ conservation should be adopted for species with a signature of local adaptation, whereas a single-large strategy may be appropriate for species with IBD or low genetic structure15. In other words, the test of IBD, IBE, and IBR can help to understand the process of population genetic differentiation, which will provide a reference for habitat conservation and management of endangered species.
Each plant occupies its own niche, and spatial and resource competition and environmental adaptation determine plants’ distributions16. External changes on the landscape and environment in combination with adaptability can also affect their population structure2,17, especially in desert areas where the environment is poor. The xeric plant Ammopiptanthus mongolicus (Maxim. ex Kom.) Cheng f. (Leguminosae) is the only broad-leaved evergreen shrub in the deserts of eastern Central Asia. A. mongolicus is listed as a second-grade vulnerable (VU) plant in the Red List of Threatened Species of China (the Red Book)18. Understanding the population genetic structure not only increases the understanding of its demographic dynamics, but also provides information for conservation (e.g., determining management units)2,19. Previous research using inter-simple sequence repeat (ISSR) markers concluded that the genetic differentiation of A. mongolicus was related to geographic distance, i.e. IBD20. Codominant marker evidence (isozymes) indicated that this species is an outcrossed but self-compatible entomophilous plant21. The small pollination range of insects and the gravity propagation of seeds were suggested as limiting factors for long-distance gene flow in A. mongolicus20. Ammopiptanthus mongolicus grows in rocky, gravelly, sandy soils of dry valleys, basins, and rocky dunes with a soil depth of less than 30 cm22. Most of the desert psammophytes exhibit a spatial distribution strongly associated with scattered fertile soils (i.e., fertile island hypothesis)23,24. In contrast, A. mongolicus can grow in heterogenous microhabitats25. Despite the environmental versatility of A. mongolicus at a fine spatial scale, recent studies have shown its global distribution is limited by local climatic conditions (e.g., temperature and humidity), soil organic matter and total nitrogen26. Given that environmental variation can critically influence distribution and population sizes of A. mongolicus, the isolation-by environment is expected to exert a relevant influence on the genetic structuring of the species. However, previous genetic studies have failed to consider the impact of the local environment on the genetic differentiation of A. mongolicus populations20,27.
In this study, we explored the effects of geographic distance and local climate on population structure by performing population genetic analyses. We used a multilocus marker, the ISSR, to verify whether A. mongolicus populations are genetically differentiated as inferred by Ge et al.20. We further explored the factors that hinder gene flow among genetically differentiated populations. In addition, since this species has a wide-ranging latitudinal distribution with a varying altitudinal distribution (ranging from ~1000 to 2000 m above sea level), we hypothesized that geographic distance, differences in local climate, and resistance of gene flow to altitudinal and climatic differences drive population genetic differentiation, i.e. isolation-by-distance (IBD), isolation-by-environment (IBE), and isolation-by-resistance (IBR). Accordingly, by outlining the genetic structure and identifying the influencing factors of A. mongolicus, conservation suggestions for this endangered Tertiary relict are provided.
Results
Low intra-population genetic variation and high inter-population differentiation
From a total of 200 samples collected from 10 populations (Fig. 1 and Table 1), 105 sharp and clear bands (loci) of the ISSR marker were recorded, of which 71 loci were polymorphic. The genetic diversity estimated from these 71 among-population polymorphic loci revealed that the percentage of within-population polymorphic loci (%P) ranged from 9.86 (EJNYG) to 29.58% (ALSY and WH). The overall expected heterozygosity (HE) was below 0.12 and the Shannon index (I) was below 0.16 in all studied populations, with two distant populations (WLTH and ALSY) exhibiting the largest values (Table 2). Although the genetic diversity was slightly higher in the whole species than within populations, it was still low, especially HE (I = 0.452 ± 0.024, HE = 0.296 ± 0.019, total populations). The high genetic diversity of the total population relative to each single population suggests high differentiation among the populations.
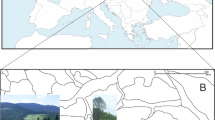
Sampling sites. (a) The map shows the relative locations of the distribution of A. mongolicus in the inland of temperate Asia; (b) the detailed sampling sites in this study and the topographic variation of the distribution. The current altitude layer is publicly available from WorldClim version 2.067 (www.worldclim.org), and the map was generated with the package raster72 (http://www.rspatial.org/) in R58.
AMOVA was used to assess the population genetic structure and revealed that 76.58% of the genetic variation was partitioned among populations, while the remaining 23.42% was attributed to differences between individuals within populations (Table 3). The inference of high genetic differentiation was also confirmed by both the neighbor-joining (NJ) tree (Fig. 2) and discriminant analysis of principal components (DAPC, Fig. 3). Our results supported the existence of ten genetic clusters, indicating that each population had its own genetic signature. In DAPC, seven principal components (PCs) were retained according to the 1000-run K-means algorithm assessed by the Bayesian information criterion (BIC), and the optimal number of clusters was 10, which corresponds to the number of sampled populations. Figure 3 shows both the component and scatter plots; all samples were clearly assigned to their own populations, except one sample in population ALSYSHT with roughly half of its genetic component from HLS and a few samples of HLS with small proportions of genetic admixture with ALSYSHT (the component plot of Fig. 3).
STRUCTURE analysis showed that the optimal grouping number (K) of genetic components was two based on the logarithmic probability change rate of successive K-value data. When K = 2, the populations ALSZNRG, EJNYG, ALYSHT, and HLS clustered together, while the remaining populations formed another group (Fig. 4). Except for two ALYSHT samples, only weak genetic admixture was detected between these two groups (Table 3). The second best K was three, with the WH and WLT populations forming the third group (Fig. 8). The grouping pattern of STRUCTURE was consistent with that of the NJ tree (Fig. 2). When K = 10, almost every population had its own unique genetic components, except for a composite component in ALZBURG similar to a part of EJNYG (Fig. 4). Although there were slight differences, the STRUCTURE, DAPC and NJ tree analyses yielded congruent inferences of obvious genetic differentiation.
IBE explains the population structure of A. mongolicus
Models of IBD, IBE, and IBR were tested to explain the genetic differentiation patterns among populations of A. mogolicus. The results of the partial Mantel test suggested that the population genetic structure could be explained by IBE (r = 0.609, p = 0.001) instead of IBD (r = 0.137, p = 0.212), IBRclim (r = −0.031, p = 0.254), or IBRalt (r = 0.179, p = 0.250) (Table 4 and Fig. 5). However, there was a marginally significant correlation between geographic distance and environmental difference (Mantel test, r = 0.368, p = 0.073), implying that the farther the geographic distance, the greater the environmental difference. Model selections for the maximum likelihood population effect mixed effect (MLPE) revealed that the IBE was the first-ranked model explaining the population genetic structure according to the ranking of the AIC and BIC (Tables 4 and 5), consistent with the inference of the partial Mantel test. The second ranked model was IBRclim, with only a small difference in AIC compared to the IBE model (ΔAIC = 0.25, Tables 4 and 5). Both IBE and IBRclim attribute population genetic structure to the climatic effect; the former explains the impact of climate differences on the survival and reproduction of colonizers, while the latter emphasizes the facilitation or inhibition of the migration (gene flow) process of organisms by climate differences. However, despite small ΔAIC between IBE and IBRclim, the effect size of the fixed effect (climatic composite resistance distance) is small (fixed estimate = 0.020) in IBRclim, suggesting that the environmental resistance during migration contributes less than the selective pressure after colonization.
Linear regression lines showing the correlations among genetic, geographic, and environmental distances. (a) The test of isolation-by-distance (IBD); (b) the test of isolation-by-environment (IBE); (b) the test of isolation-by-resistance in climate (IBRclim); (d) the test of isolation-by-resistance in altitude (IBRalt); (e) the test of correlation between geographic and environmental distance. Among these linear relationships, only the climatic distance was significantly correlated with genetic distance (i.e. IBE), as supported by the Mantel test, partial Mantel test, and MLPE (Table 4).
The Mantel test has been criticized for high Type 1 error due to multicollinearity9,28. Therefore, we removed the bioclimatic factors with multicollinearity and conducted the partial Mantel test again using each retained single factor (Fig. 6) to further explore which bioclimatic variable is the key factor affecting population genetic differentiation. Four bioclimatic factors, bio3, bio4, bio6, and bio18, were retained; only bio18 (precipitation of the warmest quarter) was positively correlated with the genetic distance (r = 0.553, p = 0.007, Fig. 7). Since some bioclimatic factors were removed due to collinearity with bio18, we tested environmental distances based on these individual factors (bio12, bio13, and bio16) for correlations with the genetic distance among populations, which confirmed their positive correlations with genetic distance (bio12: r = 0.563, p = 0.005; bio13: r = 0.554, p = 0.003; bio16: r = 0.567, p = 0.004, by partial Mantel test, conditioning on geographic distance). Bio12 (annual precipitation), bio13 (precipitation of the wettest month), bio16 (precipitation of the wettest quarter), and bio18 (precipitation of the warmest quarter) are all bioclimatic dimensions related to precipitation. According to the monthly precipitation records (Fig. 8a), the annual precipitation mostly accumulates from June to September. The regional precipitation not only decreases westward but is also obviously related to topography (Fig. 8b).
Flow chart of the experimental design to further filter the environmental factors affecting population genetic structure. (a) Bioclimatic layers were extracted from the open database WorldClim version 2.067 (www.worldclim.org); (b) factors (layers) with multicollinearity were removed using variance inflation factor (VIF) analysis, and factors with VIF > 10 were discarded; (c) the remaining bioclimatic factors (bio3, bio4, bio6, and bio18) were correlated with the genetic distance by the partial Mantel test, and the key bioclimatic factor (bio18) was identified; (d) factors related to the key bioclimatic factor were identified (bio12, bio13, bio16, and bio18).
Plots of linear regressions showing the correlations between genetic distance and differences in each single bioclimatic factor. (a) Genetic distance vs. bio3 (partial Mantel test: r = −0.361, p = 0.934, conditioning on geographic distance); (b) genetic distance vs. bio4 (partial Mantel test: r = 0.287, p = 0.127, conditioning on geographic distance); (c) genetic distance vs. bio6 (partial Mantel test: r = −0.095, p = 0.645, conditioning on geographic distance); (d) genetic distance vs. bio18 (partial Mantel test: r = 0.553, p = 0.007, conditioning on geographic distance).
Univariate MLPE regression was also conducted to test the IBE model with each of the 19 bioclimate distances as the fixed variable and the population effect as the random variable. The ranked AIC revealed that both models with bio17 and bio19 as the fixed variable had the smallest AIC values and significantly better fits than the other models (ΔAIC > 5, Table 6). Bio17 and bio19 are the precipitation of the driest and coldest quarters, respectively. In our study area, the coldest and driest seasons are the same, resulting in the same estimates in bio17- and bio19-univariate MLPE. Although the most crucial climatic factor affecting the genetic distance differed between the partial Mantel test (summer precipitation) and MLPE (winter precipitation), both analyses suggest that the regional precipitation difference is the key factor affecting the genetic structure of A. mongolicus.
Discussion
IBE is the best model on population structure
The population genetic structure of A. mongolicus was previously suggested to fit the IBD model20, which implies an inverse proportion of effective dispersal to geographical distance29,30,31. Over the past decade, accumulating studies have indicated that geographic distance or geographic barriers may not be the only factor affecting gene flow. Environmental differences may be the key factor underlying effective migration9,10,31. In this study, we suggest that the adaptability of A. mongolicus to local climate affects its seed germination and colonization. The effect of selection pressure on population differentiation is usually faster than that of drift and could occur at a small geographic scale32,33,34. Notably, environmental differences were marginally correlated with geographic distance. We therefore suggest that the previous inference of IBD20 could be due to the intercorrelation between geographic and environmental differences. The increasing number of open databases is now helping to clarify ecological and evolutionary phenomena. A meta-analysis showed that 74.3% of phylogeographic studies (52 of 70 studies) revealed significant IBE patterns, including 37.1% (27 studies) revealing spatial autocorrelation (i.e. covariates with IBD)9. Similarly, from 106 IBE studies, Shafer and Wolf10 reported effect sizes of 0.34 (95% CI 0.24–0.42) and 0.26 (95% CI 0.13–0.37) for a mixed-effect model with and without controlling spatial autocorrelation, respectively, suggesting that spatial autocorrelation reduces IBE correlations for environmental variables. These studies indicated the relevance of environmental autocorrelation for the spatial effect (i.e. IBD). That is, the previous inference that the population differentiation of A. mongolicus aligns with geographic distance20 probably reflects differential adaptation to the local climate. Differential adaptability to heterogeneous environments provides a better explanation than IBD in A. mongolicus, i.e. divergent selection is more important than neutral processes.
Genetic draft explains low genetic diversity
The low estimates of genetic diversity are consistent with the previous estimation by Ge et al.20, which included populations located farther south but no populations in Alashan (ALSY, ALSZNRG, ALSYSHT, and ALSZCHE). The genetic diversity of A. mongolicus was also lower than that of other desert species estimated by ISSR, e.g. Achillea fragrantissima in Egypt35, Citrullus colocynthis in India36, and Lasiurus sindicus in India37. Although the factors affecting the genetic diversity of species vary, the selection pressure of local precipitation with genetic draft may be the limiting factor affecting the genetic variation of desert plant populations, such as A. mongolicus in this case.
Environmental heterogeneity would reduce the chance of dispersal38,39, and the constraint of the range of distribution may lead to deleterious erosion of genetic diversity due to increased inbreeding and genetic draft39. Precipitation is an important limiting factor for the reproductive success of desert plants. The growth pattern of A. mongolicus is similar to that of desert deciduous plants or summer annuals, with blossoming and germination during the high-rainfall season40. Rapid blooming is advantageous for plant reproductive success in the desert40. However, pollinators tend to visit flowers of the same or adjacent plants instead of distant flowers in the short blooming season, which may reduce the outcrossing rate of A. mongolicus (inbreeding coefficient FIS > 0 in all loci21).
In addition, rainfall restrictions in deserts may also result in strong selection on A. mongolicus. With a selective sweep, genetic variation of adjacent genes decreases along with adaptive loci, which will even expands to most genome regions. Compared with other plants that may also be affected by genetic draft, such as Dactylis glomerata L. in the plateau of Central Asia and Western China41, A. mongolicus exhibits extremely low intra-population genetic variation, suggesting that regional environmental pressures (especially precipitation) in the desert have a more severe impact on this broad-leaved green plant. Rainfall-induced declines in outcrossing opportunities and strong selective sweeps could explain the low genetic variation of A. mongolicus, which may also be resistant to the rescue effect of gene flow among populations.
Differential local precipitation is the key to population differentiation
Summer rainfall almost completely determines the annual precipitation in the distribution of A. mongolicus. In general, the annual precipitation tends to decline in a southeast-to-northwest direction across the Asian continent42, but fluctuations in terrain (e.g. the Hetao Plain, Helan Mountains, and Mongolian Highlands) make the local climate more complicated. Such local differential precipitation may have long been the selective pressure not only for the breeding and dispersion of A. mongolicus but also for the water supply in the dry season.
Several studies have indicated that water is the key factor affecting the seed germination43 and seedling growth44 of A. mongolicus. In summer (July and August), the legume of A. mongolicus is ripe and dehiscent, and seeds fall off, quickly absorb water and germinate43. In a manipulation experiment, 85% of seedlings wilted in a 5-day drought treatment44, indicating that the demand for water is a limiting factor for the regeneration of A. mongolicus. Due to the lack of defoliation in winter, supplementing evapotranspiration with some precipitation may also affect A. mongolicus survival in winter. Although the local precipitation is small and varies little in winter (the cumulative precipitation ranges from 0 to 6.06 mm in Dec~Feb), such differences may cause local adaptation. Differential adaptability to precipitation among populations might accelerate the process of population differentiation by stalling maladaptive immigration.
As described above, differential local precipitation might also affect pollinators’ species and visiting frequency45. Differences in precipitation could vary the ratio of bee pollinators to fly pollinators; the former require dry soil for nesting, whereas the latter require moist environments for larval growth and metamorphosis46. Changes in the abundance of pollinators could affect the success of pollination and seed yield, even though the connectivity of the plant and pollinator relationship may not be disturbed by precipitation47. In addition, precipitation can also affect rhizobium symbionts48 and pathogen infectivity49, thereby affecting plant health and population regeneration. The presence of some endosymbiotic fungi (dark septate endophytes) can facilitate the growth of A. mongolicus under drought conditions50. We have not explored differences in soil microorganisms and endophytes among different populations, but distributional differences of these endophytes may also cause differential adaptability to precipitation among populations. The local adaptation caused by differential precipitation may lead to divergent directions of genetic draft, which may explain apparent population differentiation in A. mongolicus.
Concluding remarks
In conclusion, the selective pressure of the environmental gradient (differences in precipitation) is strong for A. mongolicus and likely explains the low genetic variation within populations and high population differentiation. Most individuals carry not only locally adapted genes but also homogenized genomic variation, which decreases successful emigration to populations with different environments, i.e. selection against maladapted dispersers12,31. A. mongolicus is the only evergreen shrub in the desert of Northwest China and is an important wintering place for several small animals, i.e. an umbrella species. Given the low genetic variation within populations and maladapted gene flow among populations, every population is a unique evolutionarily significant unit and should be considered as a unique management unit for conservation. The high dependence of adaptability on precipitation is not propitious for effective gene flow among populations. Therefore, the establishment of ecological corridors51,52,53 may not be an appropriate strategy for conservation. Germplasms from different populations should be actively preserved to maintain the complete gene pool and increase the evolutionary resilience9 of A. mongolicus in the face of increasingly severe climate change.
Materials and Methods
Species studied and sampling
The genus Ammopiptanthus was suggested to have originated from the broad-leaved evergreen Tethyan flora54, as supported by molecular dating indicating that the genus Ammopiptanthus split from its sister taxa in the early Miocene (chloroplast DNA matK sequences:19.6 Mya; nuclear ITS sequences: 21.8 Mya)55. Ammopiptanthus mongolicus is discontinuous distributed in western Inner Mongolia, northern Ningxia and Northern Gansu in China, ranging from 36°27′N–42°01′N, 102°36′E–108° 49′E26. The sampling area of this study was the core distribution of A. mongolicus in Inner Mongolia, China. We chose 10 populations covering the main distribution range of A. mongolicus (Table 1). Fresh leaves were sampled from 20 individuals per population, and each sampled plant was distant from other plants by at least 20 meters. A total of 200 individuals were sampled. The sampled leaves were placed immediately in a liquid nitrogen tank and stored in a −20 °C refrigerator after carrying to the laboratory.
Molecular techniques
Genomic DNA was extracted using a commercial kit DNAquick Plant System (TIANGEN Biotech Co., Ltd., Beijing, China). DNA quality was checked by agarose gel electrophoresis and by the DNA absorbance ratio (OD260/OD280: 1.7~1.9) in a WD-9403C UV Viewing Cabinet (BEIJING LIUYI Biotechnology Co., Ltd., Beijing, China). ISSR amplification was performed using fifteen primers (Table 7) with the following PCR procedure: denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at the proper temperature for 1 min (Table 7), and extension at 72 °C for 1 min, with a final 10-min extension at 72 °C. PCR was conducted in an MJMini personal thermal cycler (Bio-Rad, Hercules, USA) and T100™ thermal cycler (Bio-Rad, Hercules, USA). All PCR products were checked by 1.5% agarose gel electrophoresis, and the appearance of bands was read. Ghost bands were excluded by comparison with a negative control in which water was used as the template with the same ISSR protocol. The ISSR experiments were repeated twice to ensure that the peak signals affirming the bands (loci) were not PCR errors. Only loci that were consistently present or absent in all preliminary tests were read in the formal experiment.
Genetic diversity and population genetic structure
The genetic diversity was estimated by the indices of percentage of polymorphic loci (%P), average number of different alleles per locus (NA), effective number of alleles per locus (NE), Shannon’s information index (I), expected heterozygosity (HE), and unbiased heterozygosity (UHE) using GenAlEx v. 6.556. The contributions of genetic variation between and within populations were assessed by analysis of molecular variance (AMOVA). The significance of genetic differentiation between populations was estimated by ΦST under 999 permutations. We also conducted DAPC to determine if the spatially structured population was also genetically structured using the package adegenet57 in R58. The best clustering number (k) was inferred by the k-means algorithm with 106 simulations evaluated by BIC (the elbow in the BIC curve and the smallest BIC). The optimal number of PCs retained for DAPC was evaluated by a-score optimization. A component plot and scatter plot were drawn to illustrate the population clustering pattern of A. mongolicus. Patterns of genetic admixture were assessed by Bayesian clustering analysis, a population model-based approach based on Hardy-Weinberg and linkage equilibria59, with the assistance of STRUCTURE 2.3.460. We estimated the posterior probability of the grouping number (K = 1–20) by 10 independent runs using 106 steps of Markov chain Monte Carlo (MCMC) replicates after 10% burn-in for each run to evaluate consistency. The best grouping number was evaluated by ΔK61 in STRUCTURE HARVESTER ver. 0.6.9462. In addition, to understand the relationships between each of the lineages, we transformed the number of differences in ISSR loci between individuals into a triangular matrix and then constructed an NJ tree using MEGA663.
Testing IBD, IBE, and IBR
To test the effects of geographic distance and environmental differences on genetic structure, the partial Mantel test was conducted using the R package vegan64. We calculated the pairwise genetic distances among populations using FST/(1 − FST)65. Euclidean distances of geographic distance were calculated using the R package fossil66. We also collected 19 standard bioclimatic variables of 10 sampling sites as environmental data from WorldClim version 2.067. We considered the 19 bioclimatic variables as different environmental space vectors and used the Canberra distance to calculate the distance between populations in this vector space. To test IBD, the genetic distance was used as the response, the geographic distance as the predictor, and the environmental distance as the condition factor. To test IBE, the roles of environmental distance and geographic distance were interchanged. In addition, to test whether these environmental factors impeded gene flow, the climatic composite resistance surface was transformed from raster layers of the bioclimatic variables using Circuitscape 4.068. We also transformed the altitudinal layer into an altitudinal resistance surface. These two resistance surfaces were used to test IBR, namely IBRclim and IBRalt, respectively. The Mantel statistic was based on Spearman’s rank correlation with 9999 permutations.
Mantel and partial Mantel tests have been strongly criticized for inflated type-I error, potential collinearity between environmental variables when building an environmental dissimilarity matrix, low power, etc.9,28,69. Therefore, we fit linear mixed-effects models using the MLPE parameterization, which has been found to perform better than other regression-based statistical approaches70, to account for the non-independence of values within pairwise distance matrices and to distinguish the effects of multiple independent variables. Mixed-effects models were fit by maximum likelihood to test the effects of fixed factors (geographic, bioclimatic, and two resistance distances) with the random effect of populations. AIC and BIC were used as the objective criteria to evaluate model fit from four models of single fixed factor, six combinations of double fixed factors, four combinations of triple fixed factors, and the full model (the combinations of all fixed factors, Table 5).
Since IBE was suggested as the first-ranked model by both the partial Mantel test and model selection for MLPE (see Results), we further identified the most crucial environmental factors affecting genetic distance using two strategies. First, we re-executed the partial Mantel test by calculating the distance of each environmental factor between populations. To avoid unnecessary weighting due to intercorrelations among bioclimatic variables, we used variance inflation factor (VIF) analysis to reduce multicollinearity71. We discarded variables with high VIF values (VIF > 10) and then calculated the distances of the retained bioclimatic variables among populations to test the IBE hypothesis (Fig. 6). This remaining factor is the most likely environmental factor affecting the population genetic structure of A. mongolicus. Testing one variable by a Mantel (or partial Mantel) test has been suggested to be more credible than testing multiple variables28. Second, we performed model selection to evaluate 19 models with every single bioclimatic distance as the fixed factor in MLPE. These single-bioclimate-distance IBE models were ranked by AIC, and the model with the lowest AIC was suggested as the best one for the prediction of population genetic structure.
Data Aavailability
All genetic and environmental data used in this study are available in the Supplementary Data.
References
Nosil, P., Funk, D. J. & Ortiz-Barrientos, D. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402, https://doi.org/10.1111/j.1365-294X.2008.03946.x (2009).
Manel, S., Schwartz, M. K., Luikart, G. & Taberlet, P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197, https://doi.org/10.1016/s0169-5347(03)00008-9 (2003).
Orsini, L., Vanoverbeke, J., Swillen, I., Mergeay, J. & De Meester, L. Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol. Ecol. 22, 5983–5999, https://doi.org/10.1111/mec.12561 (2013).
Neher, R. A. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 44, 195–215, https://doi.org/10.1146/annurev-ecolsys-110512-135920 (2013).
Leffler, E. M. et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10, e1001388, https://doi.org/10.1371/journal.pbio.1001388 (2012).
Gillespie, J. H. Is the population size of a species relevant to its evolution? Evolution 55, 2161–2169, https://doi.org/10.1111/j.0014-3820.2001.tb00732.x (2001).
Frankham, R. Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 24, 2610–2618, https://doi.org/10.1111/mec.13139 (2015).
Fitzpatrick, S. W. et al. Gene flow from an adaptively divergent source causes rescue through genetic and demographic factors in two wild populations of Trinidadian guppies. Evol. Appl. 9, 879–891, https://doi.org/10.1111/eva.12356 (2016).
Sexton, J. P., Hangartner, S. B. & Hoffmann, A. A. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68, 1–15, https://doi.org/10.1111/evo.12258 (2014).
Shafer, A. B. & Wolf, J. B. Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecol. Lett. 16, 940–950, https://doi.org/10.1111/ele.12120 (2013).
Savolainen, O., Pyhäjärvi, T. & Knürr, T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 38, 595–619, https://doi.org/10.1146/annurev.ecolsys.38.091206.095646 (2007).
McRae, B. H. Isolation by resistance. Evolution 60, 1551–1561, https://doi.org/10.1111/j.0014-3820.2006.tb00500.x (2007).
McRae, B. H. & Beier, P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. USA 104, 19885–19890, https://doi.org/10.1073/pnas.0706568104 (2007).
Peterman, W. E., Connette, G. M., Semlitsch, R. D. & Eggert, L. S. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol. Ecol. 23, 2402–2413, https://doi.org/10.1111/mec.12747 (2014).
Diniz-Filho, J. A. F. & De Campos Telles, M. P. Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conserv. Biol. 16, 924–935, https://doi.org/10.1046/j.1523-1739.2002.00295.x (2002).
Silvertown, J. Plant coexistence and the niche. Trends Ecol. Evol. 19, 605–611, https://doi.org/10.1016/j.tree.2004.09.003 (2004).
Cowling, R. M., Rundel, P. W., Lamont, B. B., Kalin Arroyo, M. & Arianoutsou, M. Plant diversity in mediterranean-climate regions. Trends Ecol. Evol. 11, 362–366, https://doi.org/10.1016/0169-5347(96)10044-6 (1996).
Fu, L. G. China Plant Red Data Book. 368–371 (Science Press, 1992).
Palsbøll, P. J., Bérubé, M. & Allendorf, F. W. Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16, https://doi.org/10.1016/j.tree.2006.09.003 (2007).
Ge, X. J., Yu, Y., Yuan, Y. M., Huang, H. W. & Yan, C. Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of northwest China as revealed by ISSR analysis. Ann. Bot. 95, 843–851, https://doi.org/10.1093/aob/mci089 (2005).
Chen, G.-Q., Crawford, D., Huang, H. & Ge, X.-J. Genetic structure and mating system of Ammopiptanthus mongolicus (Leguminosae), an endangered shrub in north-western China. Plant Species Biol. 24, 179–188, https://doi.org/10.1111/j.1442-1984.2009.00253.x (2009).
Liu, G.-H. Study on the endangered reasons of Ammopiptanthus mongolicus in the desert of Alashan. Bull. Bot. Res. 18, 341–345 (1998).
Schlesinger, W. H. et al. Biological feedbacks in global desertification. Science 247, 1043–1048, https://doi.org/10.1126/science.247.4946.1043 (1990).
Garner, W. & Steinberger, Y. A proposed mechanism for the formation of ‘Fertile Islands’ in the desert ecosystem. J. Arid Environ. 16, 257–262, https://doi.org/10.1016/S0140-1963(18)30941-8 (1989).
Jia, X. H., Li, X. R., Zhang, J. G. & Zhang, Z. S. Analysis of spatial variability of the fractal dimension of soil particle size in Ammopiptanthus mongolicus’ desert habitat. Environ. Geol. 58, 953–962, https://doi.org/10.1007/s00254-008-1575-7 (2008).
Liu, M., Wu, S., Pan, B. & Wang, D. Geographical distribution and habitat characteristic of Ammpopiptanthus Cheng f. (Fabaceae) in China. Arid Land Geogr. 2, 380–387 (2017).
Su, Z., Pan, B., Zhang, M. & Shi, W. Conserv. Genet. and geographic patterns of genetic variation of endangered shrub Ammopiptanthus (Fabaceae) in northwestern China. Conserv. Genet. 17, 485–496, https://doi.org/10.1007/s10592-015-0798-x (2015).
Guillot, G., Rousset, F. & Harmon, L. Dismantling the Mantel tests. Methods Ecol. Evol. 4, 336–344, https://doi.org/10.1111/2041-210x.12018 (2013).
Aitken, S. N. & Whitlock, M. C. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44, 367–388, https://doi.org/10.1146/annurev-ecolsys-110512-135747 (2013).
Bolnick, D. I. & Otto, S. P. The magnitude of local adaptation under genotype-dependent dispersal. Ecol. Evol. 3, 4722–4735, https://doi.org/10.1002/ece3.850 (2013).
Wang, I. J. & Bradburd, G. S. Isolation by environment. Mol. Ecol. 23, 5649–5662, https://doi.org/10.1111/mec.12938 (2014).
Huang, Q. Y., Beharav, A., Youchun, U. C., Kirzhner, V. & Nevo, E. Mosaic microecological differential stress causes adaptive microsatellite divergence in wild barley, Hordeum spontaneum, at Neve Yaar, Israel. Genome 45, 1216–1229, https://doi.org/10.1139/G02-073 (2002).
Owuor, E. D., Fahima, T., Beiles, A., Korol, A. & Nevo, E. Population genetic response to microsite ecological stress in wild barley, Hordeum spontaneum. Mol. Ecol. 6, 1177–1187, https://doi.org/10.1046/j.1365-294X.1997.00296.x (1997).
Li, Y. C., Fahima, T., Beiles, A., Korol, A. B. & Nevo, E. Microclimatic stress and adaptive DNA differentiation in wild emmer wheat, Triticum dicoccoides. Theor. Appl. Genet. 98, 873–883, https://doi.org/10.1007/s001220051146 (1999).
Badr, A. et al. Genetic diversity of Achillea fragrantissima in Egypt inferred from phenotypic variations and ISSR markers associated with traits of plant size and seed yield. Plant Genet. Resour. 15, 239–247, https://doi.org/10.1017/s1479262115000568 (2016).
Verma, K. S., Ul Haq, S., Kachhwaha, S. & Kothari, S. L. RAPD and ISSR marker assessment of genetic diversity in Citrullus colocynthis (L.) Schrad: a unique source of germplasm highly adapted to drought and high-temperature stress. 3 Biotech. 7, 288, https://doi.org/10.1007/s13205-017-0918-z (2017).
Sharma, R., Rajora, M. P., Dadheech, R., Bhatt, R. K. & Kalia, R. K. Genetic diversity in sewan grass (Lasiurus sindicus Henr.) in the hot arid ecosystem of thar desert of Rajasthan, India. J. Environ. Biol. 38, 419–426, https://doi.org/10.22438/jeb/38/3/MS-265 (2017).
Lee, C. R. & Mitchell-Olds, T. Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Mol. Ecol. 20, 4631–4642, https://doi.org/10.1111/j.1365-294X.2011.05310.x (2011).
Young, A., Boyle, T. & Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 11, 413–418, https://doi.org/10.1016/0169-5347(96)10045-8 (1996).
Mulroy, T. W. & Rundel, P. W. Annual plants: Adaptations to desert environments. BioScience 27, 109–114, https://doi.org/10.2307/1297607 (1977).
Zhang, C. et al. AFLP-based genetic diversity of wild orchardgrass germplasm collections from Central Asia and Western China, and the relation to environmental factors. Plos One 13, e0195273, https://doi.org/10.1371/journal.pone.0195273 (2018).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978, https://doi.org/10.1002/joc.1276 (2005).
Zhao, X.-Y. et al. Germination responses to moisture in seed germination and seedling emergence of Ammopiptanthus mongolicus. J. Inner Mongol. Agric. Univ. 30, 57–61 (2009).
Liu, M., Shi, J. & Lu, C. Identification of stress-responsive genes in Ammopiptanthus mongolicus using ESTs generated from cold- and drought-stressed seedlings. BMC Plant Biol. 13, 88, https://doi.org/10.1186/1471-2229-13-88 (2013).
Rech, A. R. et al. The macroecology of animal versus wind pollination: ecological factors are more important than historical climate stability. Plant Ecol. Divers. 9, 253–262, https://doi.org/10.1080/17550874.2016.1207722 (2016).
Devoto, M., Medan, D. & Montaldo, N. H. Patterns of interaction between plants and pollinators along an environmental gradient. Oikos 109, 461–472, https://doi.org/10.1111/j.0030-1299.2005.13712.x (2005).
Lance, R. F., Bailey, P., Lindsay, D. L. & Cobb, N. S. Precipitation and the robustness of a plant and flower-visiting insect network in a xeric ecosystem. J. Arid Environ. 144, 48–59, https://doi.org/10.1016/j.jaridenv.2017.03.015 (2017).
Rathi, S. et al. Selection of Bradyrhizobium or Ensifer symbionts by the native Indian caesalpinioid legume Chamaecrista pumila depends on soil pH and other edaphic and climatic factors. FEMS Microb. Ecol. 94, fiy180–fiy180, https://doi.org/10.1093/femsec/fiy180 (2018).
Anderson, P. K. et al. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544, https://doi.org/10.1016/j.tree.2004.07.021 (2004).
Li, X. et al. Dark septate endophytes isolated from a xerophyte plant promote the growth of Ammopiptanthus mongolicus under drought condition. Sci. Rep. 8, 7896, https://doi.org/10.1038/s41598-018-26183-0 (2018).
Su, Z. et al. Genetic diversity and structure of an endangered desert shrub and the implications for conservation. AoB Plants 9, plx016, https://doi.org/10.1093/aobpla/plx016 (2017).
Berry, K. H., Weigand, J. F., Gowan, T. A. & Mack, J. S. Bidirectional recovery patterns of Mojave Desert vegetation in an aqueduct pipeline corridor after 36 years: I. Perennial shrubs and grasses. J. Arid Environ. 124, 413–425, https://doi.org/10.1016/j.jaridenv.2015.03.004 (2016).
Townsend, P. A. & Levey, D. J. An experimental test of whether habitat corridors affect pollen transfer. Ecology 86, 466–475, https://doi.org/10.1890/03-0607 (2005).
Sun, H. Tethys retreat and Himalayas-Hengduanshan Mountains uplift and their significance on the origin and development of the Sino-Himalayan elements and alpine flora. Acta Bot. Yunnanica 24, 273–288 (2002).
Xie, L. & Yang, Y. Miocene origin of the characteristic broad-leaved evergreen shrub Ammopiptanthus (Leguminosae) in the desert flora of eastern Central Asia. International J. Plant Sci. 173, 944–955, https://doi.org/10.1086/667232 (2012).
Peakall, R. & Smouse, P. E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539, https://doi.org/10.1093/bioinformatics/bts460 (2012).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405, https://doi.org/10.1093/bioinformatics/btn129 (2008).
R Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2015).
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
Hubisz, M. J., Falush, D., Stephens, M. & Pritchard, J. K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332, https://doi.org/10.1111/j.1755-0998.2009.02591.x (2009).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620, https://doi.org/10.1111/j.1365-294X.2005.02553.x (2005).
Earl, D. A. & Vonholdt, B. M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361, https://doi.org/10.1007/s12686-011-9548-7 (2012).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729, https://doi.org/10.1093/molbev/mst197 (2013).
Dixon, P. & Palmer, M. W. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930, https://doi.org/10.1658/1100-9233 (2003).
Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228 (1997).
Vavrek, M. J. fossil: palaeoecological and palaeogeographical analysis tools. Palaeontol. Electronica 14 (2011).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315, https://doi.org/10.1002/joc.5086 (2017).
McRae, B. H., Shah, V. B. & Mohapatra, T. K. Circuitscape 4 User Guide. The Nature Conservancy. http://www.circuitscape.org (2013).
Legendre, P., Fortin, M.-J., Borcard, D. & Peres-Neto, P. Should the Mantel test be used in spatial analysis? Methods Ecol. Evol. 6, 1239–1247, https://doi.org/10.1111/2041-210x.12425 (2015).
Shirk, A. J., Landguth, E. L. & Cushman, S. A. A comparison of regression methods for model selection in individual-based landscape genetic analysis. Mol. Ecol. Resour. 18, 55–67, https://doi.org/10.1111/1755-0998.12709 (2018).
Mansfield, E. R. & Helms, B. P. Detecting Multicollinearity. Am. Stat. 36, 158–160, https://doi.org/10.1080/00031305.1982.10482818 (1982).
Raster: Geographic Data Analysis and Modeling v. 2.8–4 (2018).
Acknowledgements
We appreciate Bing-Hong Huang’s assistance in data analyses. This research was financially supported by the National Natural Science Foundation of China (NSFC31760120) and the Ministry of Science and Technology in Taiwan (MOST 105-2628-B-003 -002 -MY3 & MOST 105-2628-B-003-001-MY3) and was also subsidized by the National Taiwan Normal University (NTNU), Taiwan.
Author information
Authors and Affiliations
Contributions
R.H.G. conceived and designed the experiments. S.J. and R.H.G. collected plant materials. S.J. performed the laboratory experiments. W.Z., Y.Z.Y. and Y.J.L. assisted in experiments and general affairs. S.J., M.X.L. and P.C.L. analyzed the data. S.J., R.H.G. and P.C.L. wrote the paper. All authors participated in the discussion, critically reviewed the manuscript, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, S., Luo, MX., Gao, RH. et al. Isolation-by-environment as a driver of genetic differentiation among populations of the only broad-leaved evergreen shrub Ammopiptanthus mongolicus in Asian temperate deserts. Sci Rep 9, 12008 (2019). https://doi.org/10.1038/s41598-019-48472-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48472-y
This article is cited by
-
Conservation genetics and potential geographic distribution modeling of Corybas taliensis, a small ‘sky Island’ orchid species in China
BMC Plant Biology (2024)
-
Population structure and genome-wide evolutionary signatures reveal putative climate-driven habitat change and local adaptation in the large yellow croaker
Marine Life Science & Technology (2023)
-
Gene-ecological zonation and population genetic structure of Tectona grandis L.f. in India revealed by genome-wide SSR markers
Tree Genetics & Genomes (2021)
-
Evidence of Morphological Divergence and Reproductive Isolation in a Narrow Elevation Gradient
Evolutionary Biology (2021)
-
Contrasting effects of local environment and grazing pressure on the genetic diversity and structure of Artemisia frigida
Conservation Genetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.