Abstract
Type I interferon (IFN-I) pathway plays a central role in the systemic lupus erythematosus (SLE) pathogenesis. Recent data suggest that SLE is associated with variants in IFN-I genes, such as tyrosine kinase 2 (TYK2), which is crucial in anti-viral immunity. Here, five TYK2 single nucleotide polymorphisms (SNPs) were genotyped in 368 childhood-onset SLE Mexican patients and 516 sex-matched healthy controls. Allele frequencies were also estimated in four indigenous groups. SLE protection was associated with TYK2 risk infection variants affecting residually its catalytic domain, rs12720356 (OR = 0.308; p = 0.041) and rs34536443 (OR = 0.370; p = 0.034), but not with rs2304256, rs12720270, and rs280500. This association was replicated in a 506 adult-onset SLE patients sample (OR = 0.250; p = 0.005, and OR = 0.277; p = 0.008, respectively). The minor alleles of both associated SNPs had a lower frequency in Mestizos than in Spaniards and were absent or rare in indigenous, suggesting that the presence of these alleles in the Mexican Mestizo population was derived from the Spaniards. For the first time, we report genetic variants with a protective effect in childhood- and adult-onset SLE Mexican population. Our results suggest that the frequency of IFN-I alleles associated with SLE, may have been shaped in populations exposed to infectious diseases for long periods, and this could be an explanation why Native American ancestry is associated with a higher SLE prevalence and an earlier onset.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) (OMIM # 152700) is an autoimmune disease characterized by a broad spectrum of clinical manifestations and the production of autoantibodies against several nuclear antigens. Over the last decade, genome-wide association studies (GWAS) have identified up to 90 risk loci for SLE in various ethnic populations1,2. SLE susceptibility is greater in some ethnic groups, including Africans and Hispanics with an Amerindian origin, which exhibit a higher occurrence of lupus nephritis compared to Caucasian individuals3,4. Previous studies also demonstrate that Hispanics have an earlier age of SLE onset and are more likely to develop severe clinical manifestations5,6,7.
A haplotype located on IFN-regulatory factor 5 (IRF5) shows one of the strongest associations with SLE (OR = 10.46), and was identified in the Mexican Mestizo population. This haplotype occurs at the highest frequency in Mestizos worldwide, and it was still higher in a Mexican indigenous group8. Several studies demonstrate that the type I interferon (IFN-I) pathway plays a central role in SLE pathogenesis, in agreement with the concurrence of elevated IFN-α serum levels in several SLE patients9,10. Supporting this relationship, association studies in different populations indicate that IFN-I signaling genes contribute to SLE. Recent data also suggest that SLE is associated with variants in the tyrosine kinase 2 gene (TYK2), which is essential for IFN-I signaling and plays an important role in anti-viral immunity. However, association studies show inconsistent effect directions among populations. Sigurdsson et al. identified two TYK2 single-nucleotide polymorphisms (SNPs)—rs2304256: Val362Phe and rs12720356: Ile684Ser—that are associated with decreased SLE susceptibility in patients of Nordic ancestry11. Graham et al. reported that SLE susceptibility is significantly associated with the rs12720270 TYK2 SNP in UK families. In the Han Chinese population, a gene–gene interaction between TYK2 and IRF5 is associated with SLE susceptibility, suggesting that the combined effect of variants located in IFN genes may also play a crucial role in SLE pathogenesis12. These varied observations may reflect differences that rely among populations13.
Considering the strong association of IFN-pathway genes with SLE in Mexican patients, here we investigated the contribution of TYK2 variants to the development of childhood- and adult-onset SLE in the Mexican population. We also determined whether the gene–gene interaction of TYK2 with the risk haplotype of IRF5 was associated with this disease.
Results
Comparison of the allele and genotype frequencies of the TYK2 variants among populations
The allele and genotype frequencies of all five analyzed SNPs were in Hardy–Weinberg equilibrium. We compared these allele frequencies with those reported in the 1000 Genomes Project Database, and with those determined in the Mexican indigenous population in this study. The allele frequencies of the genotyped SNPs in the healthy control group were similar to those previously reported among individuals of Mexican ancestry from Los Angeles, CA, USA (MXL). Only polymorphisms rs12720356 and rs280500 significantly differed in frequency between the healthy control group and the Spanish population from the 1000 Genomes Project Database (Iberians, IBS) (Table 1).
The variants rs12720356 and rs34536443 were nearly absent in the indigenous population, with frequencies of 0.003 and 0.001, respectively. These frequencies were significantly lower than those observed among the Mexican Mestizo healthy controls (Table 1, and Supplemental Table 1), suggesting that the presence of these alleles in the Mexican Mestizo population was derived from the Spaniards.
Association of TYK2 SNPs with childhood- and adult-onset SLE in the Mexican population
Association analysis revealed a protective OR for the C allele of rs12720356 (OR = 0.308; p = 0.041) and for the C allele of rs34536443 (OR = 0.370; p = 0.034), which remained significant after correction for gender and ancestry. The polymorphisms rs2304256, rs12720270, and rs280500 were not associated with childhood-onset SLE (Table 2). The protective association of both rs12720356 and rs34536443 variants was replicated in an adult-onset SLE Mexican patients sample (OR = 0.250; p = 0.005, and OR = 0.277; p = 0.008, respectively) (Supplementary Table 2).
LD structure of the TYK2 SNPs in the Mexican population
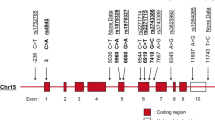
Haplotype construction revealed five different allele combinations with a frequency of >1% in the Mexican population, two of which were associated with childhood-onset SLE. The CAGAG haplotype, contained the derived alleles rs12720356C and rs2304256A and the three wildtype alleles rs12720270G, rs280500A, and rs34536443G, was associated with protection against childhood-onset SLE (OR = 0.359, p = 0.048). On the other hand, the ACGGG haplotype included only the risk allele of rs280500, and was associated with SLE risk (OR = 1.63, p = 0.034). No other haplotype showed a significant association with SLE (Table 3). LD patterns revealed that only TYK2 rs2304256 and rs12720270 variants were in LD (r2 = 0.83) (Fig. 1).
Gene-gene interactions between TYK2 and IRF5
The genotypes of three IRF5 SNPs, that form the previously identified SLE risk haplotype in the Mexican population, were used for gene–gene interaction analysis between IRF5 and TYK2. Epistasis analysis generated a three-way epistatic model between the TYK2 rs280500 polymorphism and the two IRF5 rs2004640 and rs2070197 polymorphisms (Table 4). However, this interaction did not reach statistical significance (TA = 0.617, CV = 8/10, p = 0.184; Fig. 2 and Table 4).
Multifactor dimensionality reduction models of the TYK2 and IRF5 interaction. Dark gray columns indicate controls, and light gray columns indicate cases, with the numbers of controls and cases noted at the bottom of each column. (A) The best single-locus model for IRF5 rs2070197/C. (B) The best two-locus model, including the TYK2 variant rs280500/G and IRF5 variant rs2070197/C. (C) The three-locus model, showing interaction between the TYK2 variant rs280500/G and the IRF5 variants rs2004640/G and rs2070197/C.
Discussion
The Mexican Mestizo population has a highly heterogeneous genetic structure that mainly comprises Amerindian (56%), European (41%), and a small proportion of African (3%) ancestries7. We previously reported a substantial Amerindian contribution to the higher frequency of some SLE risk variants in the Mestizo population—particularly variants in innate immune response genes, such as IRF5, which is part of the IFN signaling pathway, and exhibits altered expression in SLE patients14. In the present study, we evaluated five variants—rs280500, rs12720270, rs2304256, rs12720356, and the rare functional variant rs34536443—located in TYK2, which is also involved in IFN-I and III signaling15. Comparing the frequencies of these TYK2 variants between Mexican Mestizos and IBS revealed significant differences only in rs12720356 and rs280500, which both showed a higher frequency in IBS (0.02 vs. 0.07 and 0.05 vs. 0.2, respectively) (Table 1). To advance our understanding, we further determined the allele frequencies of the TYK2 variants in four representative Mexican Amerindian groups: Nahuatl, Maya, Tarahumara, and Zapoteco. The minor allele frequencies (MAFs) of these five SNPs were lower in Amerindians than in the Mestizo and IBS populations. Furthermore, the rs12720356 and rs34536443 variants were nearly absent in the indigenous population, with frequencies of 0.003 and 0.001, respectively, and notably both were monomorphic in the Tarahumaras and Zapotecos (Supplemental Table 1). These observations suggest that these alleles in the Mestizo population were derived from IBS.
We also analyzed the contributions of the five TYK2 SNPs with regard to susceptibility to childhood-onset SLE. In contrast to the observations in Caucasian and Asian populations, the rs2304256, rs280500, and rs12720270 variants were not associated with childhood-onset SLE in the Mexican population (Table 2)12,13,16. These discrepancies may be explained by genetic differences in allele frequencies, LD patterns, or gene–gene interactions among populations. In fact, the LD pattern observed for rs2304256 and rs12720270 SNPs among Asian populations and in our present Mexican population (China r2 = 0.95, Japan r2 = 0.85, and Mexico r2 = 0.83) substantially differs from LD patterns reported in Caucasian populations (Sweden r2 = 0.40, Finland r2 = 0.49, and UK r2 = 0.20)17. However, we did not identify the gene–gene interactions between TYK2 and IRF5 (Table 4 and Fig. 2) that were previously reported in a Chinese population12.
Our analysis revealed that the rs12720356 and rs34536443 variants conferred protection against both childhood- (OR = 0.308, p = 0.041; OR = 0.370, p = 0.034, respectively) and adult-onset SLE (OR = 0.250; p = 0.005, and OR = 0.277; p = 0.008, respectively) in Mexican population. These findings are in agreement with previous reports that the rs12720356 polymorphism is significantly associated with protection against SLE in individuals of European ancestry11,16. The rs34536443 variant has also been described as protective against other autoimmune diseases, including type 1 diabetes, rheumatoid arthritis, and multiple sclerosis, in other Caucasian populations11,18.
Interestingly, rs12720356 and rs34536443 are missense variants. The rs12720356 variant causes an Ile → Ser amino acid change at position 684, which is a pseudo-kinase region JH2 of TYK2. The rs34536443 variant causes a Pro → Ala amino acid change at position 1104, which is located in the C-terminal JH1 catalytic domain of TYK2. Previous reports state that both non-synonymous variants are catalytically impaired but exhibit residual functionality in response to IFN-α/β and other pro-inflammatory cytokines19. In contrast to in vitro experiments, in vivo studies in Tyk2 kinase-inactive (Tyk2K923E) mice suggest that the impaired kinase activity impacts IFN-I signaling through activation of STAT1-4 factors, and alters the viral defense20. Thus, these non-synonymous variants might imbalance TYK2 function in the IFN-I pathway, decreasing STAT1-4 phosphorylation, and thus decreasing IFN-I signaling, ultimately counteracting the autoimmune response.
Overall, the presently available data suggest that although most immune genes have been under positive selection in the indigenous population, due to several epidemic episodes after the Spanish conquest, variants within TYK2, failing to respond to infections, may have been under negative selection. This may be partly explained by the restricted function of TYK2 in IFN-α signaling. Even though Tyk2-deficient mice respond normally to IL-6 and IL-10 against infections, they require higher IFN-α concentrations to respond in the absence of Tyk221. Nevertheless, they cannot induce IL-12 signaling through STAT4 phosphorylation. This is an effect similar to that found in the B10.Q/J mouse strain, which is characterized by a single missense TYK2 mutation (E775K) and is strongly sensitive to Toxoplasma gondii infection but resistant to experimental collagen-induced arthritis22.
Thus, we hypothesize that the scarcity or absence of these SLE-protective alleles in the present-day indigenous populations, may also indicate that these alleles, failing to respond to infections, might had been purged from populations like Native Americans, exposed to endemic infectious diseases for long periods, or negatively selected during the severe epidemics brought by Spaniards during America’s colonization, leading to selection for individuals who could contend against infections. This hypothesis is in line with recent findings, where homozygosity for TYK2 P1104A confers a predisposition for tuberculosis by a preferential impairment of IL-23-dependent IFN-γ immunity, which could explain the protective effect to SLE, observed in this study. In addition, this observation could be also supported by the fact that the 1104A allele has decreased from 9% to 4.2% over the past 4000 years in Europeans, suggesting that it has been negative selected23. Altogether, is in agreement with several studies suggesting that genes involved in the immune response are targets of natural selection in humans, and that certain autoimmune diseases seem to be mediated by these alleles24,25.
In conclusion, for the first time, we report two TYK2 genetic variants with a protective effect in the Mexican childhood- and adult-onset SLE population. These rare variants, rs12720356 and rs34536443, have a lower frequency in Mestizos than in Spaniards and were absent or rare in indigenous, suggesting that the presence of these alleles in the Mexican population was derived from the Spaniards. Thus, the Mexican Mestizos may have inherited higher frequencies of SLE risk alleles from the indigenous population, while protective variants may have been subject to negative selection.
Our results suggest that frequency of IFN-I alleles associated with SLE, may have been shaped in populations exposed to infectious diseases for long periods, and this could be an explanation why Native American ancestry is associated with a higher SLE prevalence and an earlier onset.
Several TYK2 polymorphisms are associated with autoimmune and inflammatory diseases, but studies in different populations show inconsistent results regarding the direction of the effect on SLE susceptibility. This is likely due to the different genetic backgrounds of the studied populations. Studies of variants in genes related to IFN-I signaling in autoimmune diseases are relevant, since such investigations may elucidate key factors in the development of such diseases in certain groups, including the Mexican population. This study brings insights in the genetic complexity of admixed populations such as the Mexican, and highlights the relevance of dissecting variants that may contribute to autoimmune diseases, such as SLE.
Methods
Population sample
We performed a case-control association study including 368 patients with childhood-onset SLE and 516 sex-matched healthy blood bank donors, all from Mexico City. All patients fulfilled the American College of Rheumatology (ACR) criteria for SLE diagnosis, as previously described26,27. As a replication of associated SNPs with childhood-onset SLE, an independent cohort of 967 individuals from Mexico City consisting of 506 adult-onset SLE patients fulfilling the ACR criteria for SLE diagnosis, and 461 healthy controls, was included. To evaluate the allele frequencies in Native Americans, we also included 300 indigenous individuals belonging to four of the most representative Mexican ethnic groups (Maya, Nahuatl, Tarahumara, and Zapoteco), all of whom were from the previously described Metabolic Analysis in an Indigenous Sample (MAIS) cohort28. This study was conducted in accordance with the Declaration of Helsinki. All participants gave their signed informed consent, and the study was approved by the Instituto Nacional de Medicina Genómica local ethics and research committees. Informed consent from a parent and/or legal guardian was obtained for the children study participation, and all included children assented.
Genotyping
Genomic DNA was isolated from whole blood samples using a Puregene Blood Core kit (Qiagen, Valencia, California, USA). We genotyped five TYK2 SNPs—including four previously described as associated with SLE (rs280500, rs12720270, rs2304256, and rs12720356) and the rare functional variant rs34536443—using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA). The results were analyzed using SDS 2.3 software (Applied Biosystems). All SNPs had a call rate of over 98%. To validate the SNP genotyping accuracy, we directly sequenced samples from a randomly selected subset of 15 patients and 15 controls using an automated ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Life Technologies, CA, USA). All sequences showed 100% reproducibility. Ancestry was determined in the discovery cohort using 96 ancestry informative markers (AIMs) contained in a GoldenGate Genotyping Assay (Illumina, San Diego, CA), as previously described29. Allele frequencies from the continental populations were obtained from the 1000 Genomes Project Database30.
Statistical analysis
Data from this case-control association study were analyzed by logistic regression using PLINK (v.1.07)31. All associations were evaluated under an additive model with adjustment for gender and ancestry. We determined the allele frequencies for cases and controls, the odds ratio (OR), and the 95% confidence interval (95% CI). A P value of <0.05 after Bonferroni multiple testing correction was considered to indicate statistical significance. Haploview (v1.0) was used to calculate linkage disequilibrium (LD) through pairwise r2 values, to determine allele and haplotype frequencies, and to construct haplotypes32. Gene–gene interactions between TYK2 and IRF5 were determined using Multifactor Dimensionality Reduction (MDR) software (v.3.0.2.5)33.
Data Availability
All data generated or analyzed during this study are included in this manuscript.
References
Teruel, M. & Alarcon-Riquelme, M. E. The genetic basis of systemic lupus erythematosus: What are the risk factors and what have we learned. J Autoimmun 74, 161–175, https://doi.org/10.1016/j.jaut.2016.08.001 (2016).
Goulielmos, G. N. et al. The genetics and molecular pathogenesis of systemic lupus erythematosus (SLE) in populations of different ancestry. Gene 668, 59–72, https://doi.org/10.1016/j.gene.2018.05.041 (2018).
Lau, C. S., Yin, G. & Mok, M. Y. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus 15, 715–719 (2006).
Sanchez, E. et al. Impact of genetic ancestry and sociodemographic status on the clinical expression of systemic lupus erythematosus in American Indian-European populations. Arthritis Rheum 64, 3687–3694, https://doi.org/10.1002/art.34650 (2012).
Calvo-Alen, J. et al. Clinical, immunogenetic and outcome features of Hispanic systemic lupus erythematosus patients of different ethnic ancestry. Lupus 12, 377–385 (2003).
Alarcon-Riquelme, M. E. et al. Genome-Wide Association Study in an Amerindian Ancestry Population Reveals Novel Systemic Lupus Erythematosus Risk Loci and the Role of European Admixture. Arthritis Rheumatol 68, 932–943, https://doi.org/10.1002/art.39504 (2016).
Johnson, N. A. et al. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet 7, e1002410, https://doi.org/10.1371/journal.pgen.1002410 (2011).
Aitman, T. J. et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439, 851–855, https://doi.org/10.1038/nature04489 (2006).
Pascual, V., Farkas, L. & Banchereau, J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol 18, 676–682, https://doi.org/10.1016/j.coi.2006.09.014 (2006).
Crow, M. K. Type I interferon in the pathogenesis of lupus. J Immunol 192, 5459–5468, https://doi.org/10.4049/jimmunol.1002795 (2014).
Sigurdsson, S. et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet 76, 528–537, https://doi.org/10.1086/428480 (2005).
Tang, L. et al. Genetic association and interaction between the IRF5 and TYK2 genes and systemic lupus erythematosus in the Han Chinese population. Inflamm Res 64, 817–824, https://doi.org/10.1007/s00011-015-0865-2 (2015).
Cunninghame Graham, D. S., Akil, M. & Vyse, T. J. Association of polymorphisms across the tyrosine kinase gene, TYK2 in UK SLE families. Rheumatology (Oxford) 46, 927–930, https://doi.org/10.1093/rheumatology/kel449 (2007).
Reddy, M. V. et al. Genetic association of IRF5 with SLE in Mexicans: higher frequency of the risk haplotype and its homozygozity than Europeans. Hum Genet 121, 721–727, https://doi.org/10.1007/s00439-007-0367-6 (2007).
Velazquez, L., Fellous, M., Stark, G. R. & Pellegrini, S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70, 313–322 (1992).
Diogo, D. et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS One 10, e0122271, https://doi.org/10.1371/journal.pone.0122271 (2015).
Jarvinen, T. M. et al. Tyrosine kinase 2 and interferon regulatory factor 5 polymorphisms are associated with discoid and subacute cutaneous lupus erythematosus. Exp Dermatol 19, 123–131, https://doi.org/10.1111/j.1600-0625.2009.00982.x (2010).
Mero, I. L. et al. A rare variant of the TYK2 gene is confirmed to be associated with multiple sclerosis. Eur J Hum Genet 18, 502–504, https://doi.org/10.1038/ejhg.2009.195 (2010).
Li, Z. et al. Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J Immunol 190, 2335–2344, https://doi.org/10.4049/jimmunol.1203118 (2013).
Prchal-Murphy, M. et al. TYK2 kinase activity is required for functional type I interferon responses in vivo. PLoS One 7, e39141, https://doi.org/10.1371/journal.pone.0039141 (2012).
Shimoda, K. et al. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity 13, 561–571 (2000).
Shaw, M. H. et al. A natural mutation in the Tyk2 pseudokinase domain underlies altered susceptibility of B10.Q/J mice to infection and autoimmunity. Proc Natl Acad Sci USA 100, 11594–11599, https://doi.org/10.1073/pnas.1930781100 (2003).
Boisson-Dupuis, S. et al. Tuberculosis and impaired IL-23-dependent IFN-gamma immunity in humans homozygous for a common TYK2 missense variant. Sci Immunol 3, https://doi.org/10.1126/sciimmunol.aau8714 (2018).
Quach, H. et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell 167, 643–656 e617, https://doi.org/10.1016/j.cell.2016.09.024 (2016).
Nedelec, Y. et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell 167, 657–669 e621, https://doi.org/10.1016/j.cell.2016.09.025 (2016).
Velazquez-Cruz, R. et al. Association of PDCD1 polymorphisms with childhood-onset systemic lupus erythematosus. Eur J Hum Genet 15, 336–341, https://doi.org/10.1038/sj.ejhg.5201767 (2007).
Tan, E. M. et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 25, 1271–1277 (1982).
Contreras-Cubas, C. et al. Heterogenous Distribution of MTHFR Gene Variants among Mestizos and Diverse Amerindian Groups from Mexico. PLoS One 11, e0163248, https://doi.org/10.1371/journal.pone.0163248 (2016).
Seldin, M. F. et al. Amerindian ancestry in Argentina is associated with increased risk for systemic lupus erythematosus. Genes Immun 9, 389–393, https://doi.org/10.1038/gene.2008.25 (2008).
Genomes Project, C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65, https://doi.org/10.1038/nature11632 (2012).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, https://doi.org/10.1086/519795 (2007).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265, https://doi.org/10.1093/bioinformatics/bth457 (2005).
Hahn, L. W., Ritchie, M. D. & Moore, J. H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19, 376–382 (2003).
Acknowledgements
This study was supported by grants from the Consejo Nacional de Ciencia y Tecnología (CONACYT) from Mexico (CB 2010-135155 to LO, CB 2014-243394 to CCC, and Salud 2014-202307 to HGO).
Author information
Authors and Affiliations
Contributions
C.C.C., H.G.-O. and L.O. were involved in the study conception and design, data interpretation, and writing of the manuscript. V.B. participated in the sample and clinical information collection. C.C.C., H.G.-O., F.B.O. and P.B. carried out genotyping and statistical analyses. R.V.C., F.B.O., P.B., A.M.H. and V.B. participated in study design coordination, data interpretation, and critical editing of the manuscript. R.E.B.C., J.R.B., M.A.L.H., Y.S. and O.M.M. participated in the replication cohort sample collection. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Contreras-Cubas, C., García-Ortiz, H., Velázquez-Cruz, R. et al. Catalytically Impaired TYK2 Variants are Protective Against Childhood- and Adult-Onset Systemic Lupus Erythematosus in Mexicans. Sci Rep 9, 12165 (2019). https://doi.org/10.1038/s41598-019-48451-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48451-3
This article is cited by
-
Genome-wide association study identifies Sjögren’s risk loci with functional implications in immune and glandular cells
Nature Communications (2022)
-
Systemic Lupus Erythematosus in Children and Young People
Current Rheumatology Reports (2021)
-
Janus Kinase Inhibition and SLE: Is this a Plausible Treatment Option for SLE?
Current Treatment Options in Rheumatology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.