Abstract
The family Brassicaceae includes plants that are non-host for arbuscular mycorrhizal fungi (AMF) such as the model plant Arabidopsis thaliana (arabidopsis) and the economically important crop plant Brassica napus (rapeseed). It is well known that Trichoderma species have the ability to colonize the rhizosphere of Brassicaceae plants, promoting growth and development as well as stimulating systemic defenses. The aim of the present work is to ascertain that Brassicaceae plants increase productivity when AMF and Trichoderma are combinedly applied, and how such an effect can be ruled. This simultaneous application of a Trichoderma harzianum biocontrol strain and an AMF formulation produces a significant increase in the colonization by Trichoderma and the presence of AMF in arabidopsis and rapeseed roots, such colonization accompanied by improved productivity in both Brassicaceae species. Expression profiling of defense-related marker genes suggests that the phytohormone salicylic acid plays a key role in the modulation of the root colonization process when both fungi are jointly applied.
Similar content being viewed by others
Introduction
Arbuscular mycorrhizal fungi (AMF) are able to establish symbiotic relationships with the majority of terrestrial plants, including species of great economic interest in agriculture1. As a result, the plant benefits from improved water and nutrient uptake, while the fungus receives a place to develop the mycorrhizal structures and the uptake of sugars produced during the photosynthetic process2,3,4. AMF are also able to improve systemic plant responses to environmental5,6 and biotic7,8 stresses. However, the loss of essential genes for symbiosis during evolution9 has led to the inability of several flowering plant lineages, including most plants of the Brassicaceae family, to form AMF symbiotic relationships10,11. Brassica is the most important genus of the Brassicaceae because it includes crop plants such as Brassica oleracea (broccoli, cabbage, cauliflower), B. rapa (turnip), B. nigra (black mustard) and B. napus (rapeseed), as well as the model plant Arabidopsis thaliana (arabidopsis)12.
Trichoderma is a genus of soil-borne filamentous fungi widely used as a source of biocontrol agents in agriculture owing to effective antagonisitic mechanisms such as mycoparasitism, antibiosis or competition against plant pathogens and nematodes13,14. Some strains are able to induce plant defenses15 and stimulate plant growth and development16,17 by establishing a molecular dialogue with the roots18,19. Mutualistic beneficial interactions between Trichoderma spp. and plants such as arabidopsis, tomato, cucumber, pea or canola20,21,22 and even woody plants23 have been observed.
Although Trichoderma and AMF are commonly applied as beneficial microorganisms in agriculture, little is known about the molecular changes occurred in the plants when these fungi are used together. It has been reported that the combined application of Trichoderma and AMF has positive effects in the nutritional composition of several plant species including marigold, tomato, cucumber or melon24,25,26,27. Nevertheless, negative effects for plants have been also described when these two fungi were applied together28,29.
A complex regulatory network mediated by phytohormones, involving the salycilic acid (SA), jasmonic acid (JA) and ethylene (ET) pathways, plays a key role during the AMF or Trichoderma root colonization process30,31,32,33. During the first hours of plant-microbe interaction a SA-dependent response known as systemic acquired resistance (SAR) takes place in plant roots32. In addition, beneficial microorganisms activate the mechanism known as induced acquired resistance (ISR), mediated by JA and ET34. These signaling pathways can interact antagonistically or synergistically and are effective at fighting against a broad range of attackers and colonizers35, including the symbiotic interactions with beneficial microbes34. In order to reach an effective association with the plant, AMF and Trichoderma establish a molecular dialogue with their host that bypasses plant defenses allowing the root system to be colonized during the first hours of interaction36,37. This process requires microorganisms to produce and secrete small-sized molecules recognizable by plant cell receptors19,38. As far as Trichoderma is concerned, response mediated by SA is expected at the root level during the first hours of colonization14,37, whereas in the case of AMF, a response mediated by JA takes place39.
The objectives of this study have been to analyze: i) the effect of the combined application of T. harzianum T34 and an AMF formulation on fungal root colonization of two non-host (arabidopsis and rapeseed) and one host (Solanum lycopersicum, tomato) AMF plants, and on arabidopsis and rapeseed silique production; ii) the plant transcriptomic changes derived from the combined use of these two fungal inocula; and iii) the role of SA and JA on fungal root colonization when T34 and AMF are applied together.
Results
Arabidopsis and rapeseed production
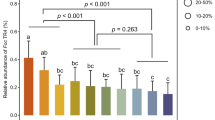
The number of siliques was used to determine the productive capacity of the different Brassicaceae plants used. The production of siliques was significantly greater in arabidopsis and rapeseed plants inoculated with T. harzianum T34 in contrast to the untreated control plants (Fig. 1), whereas production was lower in plants only challenged with AMF. However, the highest number of siliques was recorded for arabidopsis and rapeseed plants treated with AMF plus T34.
Number of siliques produced by arabidopsis (a) and rapeseed (b) plants. Arabidopsis (ARA) and rapeseed (BN) inoculated with T. harzianum T34 (-T34) and AMF (-AMF). Each value represents the average of 45 plants from 3 independent experiments (15 plants each), with their standard deviation. Two-way analysis of variance (ANOVA) was performed, followed by Sidak’s multiple comparison test, indicating significant differences as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Root colonization
Using a quantitative real-time PCR (qPCR) analysis, the colonization of T. harzianum T34 and AMF when applied together or separately was evaluated in arabidopsis, rapeseed and tomato roots. The T34 and/or AMF colonization rates are shown in Table 1. For single inoculations, DNA of T34 was detected in arabidopsis, rapeseed and tomato roots, whereas DNA of AMF was only detected in tomato roots. The combined application of AMF and T34 led to significantly increased levels of T. harzianum in both arabidopsis and rapeseed roots and an opposite effect in tomato roots. After the combined inoculation of T34 and AMF significantly increased levels of AMF were measured in tomato roots, as well as the presence of AMF was detected in roots of the two Brassicaceae plants.
To determine the AMF species in these plants, DNA extracted from root samples was amplified and sequenced using genus-specific primers, and a sequence analysis showed that Rhizophagus fasciculatus and Rhizophagus irregularis were the only fungal species present in roots of arabidopsis and rapeseed plants, respectively (data not shown).
Effect of T34 and AMF combined application in local defense of arabidopsis and tomato plants
To explore whether the response of host and non-host mycorrhizal plants to combined or separated applications of AMF and T34 involved different local defense responses, we analyzed by qPCR the expression levels of defense-related phytohormone marker genes genes at two time points, in roots untreated or previously inoculated with T34, AMF and AMF plus T34. In the case of Arabidopsis, roots of 3-week-old plants (Fig. 2), just one week after fungal inoculation, and roots of 5-week-old-plants (Fig. 3) were analyzed; in the case of tomato, roots of 4-week-old plants (Fig. 4), just one week after fungal inoculation, and roots of 7-week-old-plants (Fig. 5) were analyzed. Compared to the arabidopsis control plants, the application of T34 alone increased the expression levels of PR-1 and CALS5, both SA-responsive markers, and ICS1, involved in SA biosynthesis just one week after fungal inoculation (Figs 2 and 3). By contrast, the expression levels of VSP2 and PDF1.5 genes, both JA-responsive markers, were reduced at both time points, whereas expression of LOX1, involved in the JA biosynthetic pathway, were reduced only in 5-week-old-plants (Figs 2 and 3). Although the inoculation of AMF alone also increased the expression levels of the two marker genes of the SA-responsive pathway at both time points, and ICS1, one week after T34 application, (Figs 2 and 3) it was accompanied by increased expression levels of VSP2 and a reduction of LOX1 levels in roots of 7-week-old-plants (Figs 2 and 3). The combined application of AMF and T34 caused significant changes in the expression of the six genes analyzed, increasing the transcript levels of all of them, except for those of VSP2, and PDF1.5 one week after fungal application (Figs 2 and 3). These profiles are significant not only in comparison with the control condition, but also with those from arabidopsis plants treated with single applications of T34 or AMF. Changes in SA- and JA-signaling defense pathways in arabidopsis roots should affect fungal colonization rates when AMF and T34 are applied together.
Real time reverse transcription polymerase chain reaction (qRT-PCR) analysis of some defense genes in roots of 3 week-old A. thaliana plants inoculated with T. harzianum T34 and/or AMF. Genes of the isochorismate synthase 1 (ICS1), pathogenesis-related protein 1 (PR-1), callose synthase 5 (CALS5), lipoxygenase 1 (LOX1), vegetative storage protein (VSP2) and plant defensin 1.5 (PDF1.5). Values correspond to relative measurements against arabidopsis grown in the absence of the fungus (2−ΔΔCt = 1). The arabidopsis actin gene was used as an internal reference. Bars represent standard deviations of the means of three root pools of five plants each one collected from three independent experiments. Two-way analysis of variance (ANOVA) was performed, followed by Sidak’s multiple comparison test, indicating significant differences as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Real time reverse transcription polymerase chain reaction (qRT-PCR) analysis of some defense genes in roots of 5 week-old A. thaliana plants inoculated with T. harzianum T34 and/or AMF. Genes of the isochorismate synthase 1 (ICS1), pathogenesis-related protein 1 (PR-1), callose synthase 5 (CALS5), lipoxygenase 1 (LOX1), vegetative storage protein (VSP2) and plant defensin 1.5 (PDF1.5). Values correspond to relative measurements against arabidopsis grown in the absence of the fungus (2−ΔΔCt = 1). The arabidopsis actin gene was used as an internal reference. Bars represent standard deviations of the means of three root pools of five plants each one collected from three independent experiments. Two-way analysis of variance (ANOVA) was performed, followed by Sidak’s multiple comparison test, indicating significant differences as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Real time reverse transcription polymerase chain reaction (qRT-PCR) analysis of expression of some defense genes in roots of 4 weeks-old tomato plants inoculated with T. harzianum T34 and/or AMF. Genes of the isochorismate synthase 1 (ICS1), pathogenesis-related protein 1 (PR-1), lipoxygenase 1 (LOX1) and ethylene signaling protein (EIN2). Values correspond to relative measurements against tomato grown in the absence of the fungus (2−ΔΔCt = 1). The tomato actin gene was used as endogenous reference control. Bars represent standard deviations of the means of three root pools of five plants each one collected from three independent experiments. Two-way analysis of variance (ANOVA) was performed, followed by Sidak’s multiple comparison test, indicating significant differences as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Real time reverse transcription polymerase chain reaction (qRT-PCR) analysis of expression of some defense genes in roots of 7 weeks-old tomato plants inoculated with T. harzianum T34 and/or AMF. Genes of the isochorismate synthase 1 (ICS1), pathogenesis-related protein 1 (PR-1), lipoxygenase 1 (LOX1) and ethylene signaling protein (EIN2). Values correspond to relative measurements against tomato grown in the absence of the fungus (2−ΔΔCt = 1). The tomato actin gene was used as endogenous reference control. Bars represent standard deviations of the means of three root pools of five plants each one collected from three independent experiments. Two-way analysis of variance (ANOVA) was performed, followed by Sidak’s multiple comparison test, indicating significant differences as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Compared with tomato control plants (Figs 4 and 5), the single application of T. harzianum increased the expression levels of ICS1 and PR-1, one week after T34 application and reduced those of ICS1 and EIN2, a major regulator of the ET signaling pathway, in roots of 7-week-old-plants, while the expression of LOX1 did not significantly change. The application of AMF alone caused increased expression of LOX1 but did not modify the PR-1 expression at both time points (Figs 4 and 5). Compared to the control, the combined application of AMF plus T34 showed a similar gene expression profile to that observed in tomato plants treated with T34 which would be indicating the prevalence of Trichoderma over the AMF effect.
Colonization of SA- and JA-impaired arabidopsis mutants
In order to analyze the role played by the SA- and JA-signaling defense pathways in the process of fungal colonization of a non-host AMF plant such as arabidopsis we employed mutants impaired in SA biosynthesis (sid2) and in JA response (coi1-30).
The application of AMF plus T34 caused the death of sid2 plants during the floral development stage (data not shown). Roots were collected 10 days after T34 application in order to perform a comparative analysis of the colonization pattern by T. harzianum in the wild type and the two mutant lines of arabidopsis. When T34 was applied alone, a significant increase of fungal DNA was detected in sid2 mutant roots as compared with that of the Col-0 wild type. The highest levels of fungal DNA was detected in the roots of plants treated with the AMF plus T34 condition (Table 2). With respect to the coi1-30 mutant, the colonization pattern of T34 was similar to that observed in Col-0 roots. As expected for a non-host AMF plant, no fungal DNA was detected in Col-0 roots inoculated with AMF alone, although AMF DNA was detected in the sid2 mutant inoculated only with AMF. In addition, the highest levels of AMF DNA were detected in sid2 plants inoculated with AMF plus T34. The fungal root colonization ability was the same for both the mutant coi1-30 and the Col-0 wild type.
Discussion
It is well documented that the application of beneficial fungi, such as AMF and Trichoderma spp., improves crop productivity and reduces the use of agrochemicals and their negative effects on human health and the environment40,41,42. In the present study we found that T. harzianum T34 increases silique production in Brassicaceae plants of arabidopsis and rapeseed (Fig. 1), while AMF inoculation significantly reduces silique production. Such positive effects are in agreement with those reported for this T. harzianum strain32 and other strains of T. atroviride and T. virens21 in arabidopsis and of T. asperellum in rapeseed42. Negative responses to AMF application were also observed in non-host AMF plants43,44, including Brassicaceae45,46. The increased productivity in response to the combined application of AMF and Trichoderma in the two non-host mycorrhizal plants used in this study (Fig. 1) is in agreement with previous findings reported in marigold24, melon27, cucumber26 and black lentil47; although such positive effects are also dependent on the fungal strains used29. It has also been described that the combined application of AMF and organic matter may help to facilitate plant nutrient uptake48, although this effect has been associated to edaphic characteristics and the presence of other soil microorganisms49.
The higher number of siliques recorded for arabidopsis and rapeseed after the combined inoculation of AMF plus T34 could be related to the changes detected in the ability of T34 to colonize the roots of both plants, which was enhanced by the presence of AMF We were not able to observe the formation of vesicles nor arbuscules after the joint application of T. harzianum and AMFs although, interestingly, AMF DNA was detected in the roots of the two non-host mycorrhizal plants only when AMF and T34 were applied together, indicating that T. harzianum facilitates the presence of AMF in arabidopsis and rapeseed roots (Table 1). Even though some authors have reported the development of rudimentary AM phenotypes in non-mycorhizal plants11, without arbuscule formation10, it is clear that arabidopsis is not a true AMF host11. The beneficial effects on a host plant is not always accompanied by the formation of arbuscules or vesicles50 and this fact may explain the increased productivity observed in arabidopsis and rapeseed plants from the treatment T34 plus AMF (Fig. 1).
Recently it has been suggested that plants belonging to Brassicaceae have lost the ability to form AMF symbiotic relationships during evolution, probably due to the loss of AMF symbiosis-related genes9 and/or the ability of plants to recognize AMF effectors that play a crucial role in establishing the symbiotic association51. Trichoderma is also able to produce several secreted proteins capable of being recognized by plant receptors, which allow Trichoderma-plant associations to take place19. It has been described that during the establishment of a symbiotic association there is a transient suppression of plant defenses that facilitates Trichoderma penetration19,32,37 and that may allow the access of AMF to the Brassicaceae roots when both fungi are applied together. Moreover, it has been described that a combination of Trichoderma and AMF favours the germination of Trichoderma conidia52 that subsequently can parasitize the AMF53,54. The increased levels of T34 DNA detected in arabidopsis and rapeseed roots after T34 plus AMF application are in agreement with these two reports.
In contrast to our observations in Brassicaceae plants, the combined application of AMF and T34 in tomato was associated with increased and reduced DNA levels of AMF and T34, respectively (Table 1). These results are in line with increased AMF colonization of melon plants after the combined inoculation of T. harzianum with Septoglomus constrictum (formerly G. constrictum), G. claroideum or R. irregularis27. However, a T. harzianum root colonization decrease was also reported by these authors when this fungus was inoculated together with F. mosseae, G. claroideum or S. constrictum. The decreased levels of T. harzianum detected in tomato roots in response to the AMF plus T34 application could be due to fungal competition for space or nutrients, as previously reported between AMF and Trichoderma24,28,55, or to the AMF-induced plant defense against Trichoderma56.
Plant interactions with beneficial or pathogenic fungi triggers a complex phytohormone networking that leads to local and systemic defense responses, in which the SA- and JA/ET-dependent pathways are mainly involved34,35. As biotrophic fungi, Trichoderma and AMF are sensitive to SA-dependent defenses which prevent the fungus from entering into the vascular system32,57. During the early stages of the interaction, the establishment of a symbiotic relation requires the suppression of SA-dependent defense responses in the plant, which are compensated by an increase of JA-/ET-regulated defenses7,37. The alternation of SA- and JA-dependent defense responses leads to an undulating defensive response in the plant once the fungal colonizer has achieved an effective root colonization57. In the present work, we have chosen two time points. The first one week after T34 application and the second at flowering, when AMF establishment is more efficient58. Increased levels of SA-response genes, such as PR-1 and CALS5, accompanied by decreased expression levels of the JA/ET-marker, were detected in both cases (Figs 2–5). These results indicate that a local SA response is needed to avoid massive fungal root colonization. Similar increases in PR-1 expression have been reported in arabidopsis and tomato plants grown under in vivo conditions at longer time points after Trichoderma was applied14,37.
Different defense responses were observed in non-host and host mycorrhizal plants challenged with AMF (Figs 2–5). In the non-host plant, arabidopsis, a local SA- and JA-mediated response was observed. This result is consistent with the defense responses observed in incompatible associations between AMF and non-host plants and in the pea myc- mutant plant, which is unable to form AMF symbiosis59. The increased expression of LOX1 observed in tomato roots colonized by AMF agrees with the enhancement of JA biosynthesis previously described in mycorrhized tomato roots7,39. In any case, it is difficult to propose a model for hormonal crosstalk in plants occurring under multi-fungal interactions60. We have observed in arabidopsis roots that marker genes of the JA and SA pathways overlap in the combined application of AMF and T34.
The differences observed in the colonization pattern among host and non-host AMF plants seems to be related to the expression of SA-related genes, as assessed by the use of the arabidopsis mutants impaired in SA biosynthesis (sid2) and JA response (coi1-30), respectively (Table 2). The fungal DNA levels detected in the insensitive JA mutant agreed with those observed for the wild type, indicating that the lack of responses mediated by JA does not seem necessary for the mycorrhization process. However, in the case of the SA-deficient mutant, inoculation with AMF alone leads to root colonization even in the absence of T34, suggesting that this hormone plays a crucial role in fungal colonization, at least in arabidopsis.
In conclusion: i) SA seems to be the key phytohormone to prevent mycorrhization in Brassicaceae plants; ii) the simultaneous application of Trichoderma and AMF produces a significant increase of the Trichoderma root colonization levels and favours the presence of the AMF in Brasicaceae roots; and iii) the combined application of AMF and T. harzianum leads to a significant increase in the number of siliques in Brassicaceae plants. This finding may have a significant impact from an agronomic point of view, since the combined application of both types of fungi can produce a significant increase in the productivity of this important family of plants.
Methods
Plant material
The Arabidopsis thaliana ecotype Col-0, its SA-impaired mutant sid2 and its JA-impaired mutant coi1-30, as well as Solanum lycopersicum cv. Marmande and Brassica napus cv. Jura were the plants used in this study. The SA-impaired mutant sid2 SALK_088254 was kindly provided by Dra. P. García-Agustín (University Jaume I, Castellón, Spain), and the JA-impared mutant coi1-30 was kindly provided by Dr. Roberto Solano (CNB-CSIC, Madrid, Spain). Seeds from arabidopsis, tomato and rapeseed plants were surface-sterilized as previously described61.
Fungal cultures
Trichoderma harzianum CECT 2413 (Spanish Type Culture Collection, Valencia, Spain) (referred to as strain T34), was used throughout this study. Strain T34 was routinely grown on potato-dextrose-agar (PDA, Sigma-Aldrich, Madrid, Spain) in the dark at 28 °C and the spores were stored at −80 °C in a 20% glycerol solution. Spores were harvested from 7-day-old PDA plates as previously described57, and a final concentration was determined using a haemocytometer and adjusted to 2 × 107 spore mL−1.
The mycorrhizal formulation Miratext-02, provided by Mirat Fertilizantes (Salamanca, Spain), was used as the AMF inoculum, which contained at least 1 × 106 spore kg−1 in an inert substrate and included five different AMF species: [Glomus microagregatum, Funneliformis mosseae (formerly G. mosseae), Claroideoglomus claroideum (formerly G. claroideum), Rhizophagus irregularis (formerly G. intraradices) and R. fasciculatus (formerly G. fasciculatum)].
Fungal inoculation and plant growth conditions
Seeds were plated on Murashige and Skoog (MS) (Duchefa, Haarlem, The Netherlands) solid medium (agar 1%) with sucrose (1%) and plates mantained in a growth chamber at 22 °C, 40% relative humidity (RH) and a 16 h light/8 h dark photoperiod at 80–100 μE m−2 s−1, for 7 (rapeseed), 10 (arabidopsis) and 16 (tomato) days. Seedlings were individually transferred to 0.2 (for arabidopsis) and 5 (for rapeseed and tomato) L-pots, containing a mixture of peat/vermiculite (3:1), maintained under greenhouse conditions as previously described57 and watered as needed. For each type of plant, the four considered treatments were: untreated (control) and plants treated with AMF, T34, and AMF plus T34. The inoculation of AMF was done by burying 1 g of Miratext-02 (1000 spore g−1) per pot at 5 cm below the substrate surface just before transplanting the seedlings. Plants treated with T. harzianum T34 were root inoculated with 1 mL of a conidial suspension containing 2 × 107 spore mL−1 one week after the seedlings have been transplanted. Twenty plants were used for each type of plant and treatment, and each assay was repeated three times.
The siliques from 15 arabidopsis and rapeseed plants per condition were collected at the end of the life cycle and counted (11 weeks for arabidopsis and 19 weeks for rapeseed). The analyses of fungal-root colonization and the expression of defense genes were performed with roots of a total of 60 plants of arabidopsis, rapeseed and tomato. For each type of plant, roots from five plants per each treatment were pooled and root pools from three independent assays were considered. Roots were collected one week after T34 inoculation and during the formation of the floral primordia in 5- (arabidopsis), 10- (rapeseed) and 7- (tomato) week-old plants, except in the case of the arabidopsis mutants since sid2 mutant was unable to reach reproductive growth in the presence of T3432. In this last case, roots were collected 10 days after fungal inoculation when the plants were 4 weeks old. All root material was washed with water until there was no remaining substrate, immediately frozen with liquid nitrogen and pulverized with a mortar.
Quantification of fungal root colonization
The quantification of T34 and AMF DNA in the roots of rapeseed, arabidopsis and tomato plants was performed by qPCR as previously described62,63, with some modifications. DNA was extracted from roots of the untreated (control) and AMF, T34, and AMF plus T34 inoculated plants. A mix was prepared in a 10-μL volume using 5 μL of Brilliant SYBR Green QPCR Master Mix (Roche, Penzberg, Germany), 10 ng of DNA, the forward and reverse primers at a final concentration of 100 nM, and nuclease-free PCR-grade water to adjust the final volume. The 18S rRNA gene of AMF64 and the Actin genes of Trichoderma, arabidopsis, rapeseed and tomato were used; their corresponding primer pairs are indicated in Table 3. Amplifications were performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, USA) programmed for 40 cycles under the following conditions: denaturation, 95 °C for 15 s; annealing, 60 °C for 1 min; extension, 72 °C for 1 min. Each PCR was performed in triplicate by using the DNA extracted from 3 root pools of 5 plants each one for each treatment and plant type. Cycle threshold values served to calculate the amount of fungal DNA using standard curves. Values of Trichoderma or AMF DNA were referred to the amount of arabidopsis, rapeseed or tomato DNA in every corresponding sample.
The primers used for the quantification of the DNA of each species are sufficiently specific, without showing any of them, cross reaction with any of the other fungal and/or plant species used.
Gene expression studies
Roots of three groups of 5 arabidopsis or tomato plants per assayed condition were separately collected and used for RNA extraction using the TRI reagent (Ambion, Austin, USA), following the manufacturer’s instructions. The cDNA synthesis was performed as previously described61. Gene expression was analyzed by qPCR. PCR mixtures and amplification conditions were as previously described65. The primers used are given in Table 3, and the Actin gene was used as the arabidopsis and tomato endogenous control. Data are expressed using 2−ΔΔCT method66.
Statistical analysis
The statistical analysis of the data was carried out with the Statistix 8.0 software. When a comparison of means was performed against a control, the Student’s t-test was employed at P < 0.05; significant differences are denoted using an asterisk. Combined effects of strain T34 and AMF were analyzed by two-way ANOVA followed by Sidak’s multiple comparison test, indicating significant differences as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
References
Baum, C., El-Tohamy, W. & Gruda, N. Increasing the productivity and product quality of vegetable crops using arbuscular mycorrhizal fungi: a review. Sci. Hortic. 187, 131–141 (2015).
Parniske, M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775 (2008).
Veresoglou, S. D., Chen, B. & Rillig, M. C. Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol. Biochem. 46, 53–62 (2012).
Roth, R. & Paszkowski, U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 39, 50–56 (2017).
Latef, A. A. H. A. et al. Arbuscular mycorrhizal symbiosis and abiotic stress in plants: a review. J. Plant Biol. 59, 407–426 (2016).
Lenoir, I., Fontaine, J. & Sahraoui, A. L. H. Arbuscular mycorrhizal fungal responses to abiotic stresses: a review. Phytochemistry. 123, 4–15 (2016).
Pozo, M. J. & Azcón-Aguilar, C. Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398 (2007).
Song, Y. Y. et al. Priming of anti-herbivore defense in tomato by arbuscular mycorrhizal fungus and involvement of the jasmonate pathway. J. Chem. Ecol. 39, 1036–1044 (2013).
Delaux, P. M. Comparative phylogenomics of symbiotic associations. New Phytol. 213, 89–94 (2017).
Veiga, R. S. et al. Arbuscular mycorrhizal fungi reduce growth and infect roots of the non-host plant Arabidopsis thaliana. Plant Cell Environ. 36, 1926–1937 (2013).
Cosme, M., Fernández, I., Van der Heijden, M. G. A. & Pieterse, C. M. J. Non-mycorrhizal plants: the exceptions that prove the rule. Trends Plant Sci. 23, 577–587 (2018).
Huang, C. H. et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 33, 394–412 (2015).
Lorito, M., Woo, S. L., Harman, G. E. & Monte, E. Translational research on Trichoderma: from omics to the field. Annu. Rev. Phytopathol. 48, 395–417 (2010).
Medeiros, H. A. et al. Tomato progeny inherit resistance to the nematode Meloidogyne javanica linked to plant growth induced by the biocontrol fungus Trichoderma atroviride. Sci. Rep. 7, 40216, https://doi.org/10.1038/srep40216 (2017).
Yedidia, I., Benhamou, N. & Chet, I. Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65, 1061–1070 (1999).
Shoresh, M. & Harman, G. E. Differential expression of maize chitinases in the presence or absence of Trichoderma harzianum strain T22 and indications of a novel exo-endo-heterodimeric chitinase activity. BMC Plant Biol. 10, 136 (2010).
Hermosa, R., Viterbo, A., Chet, I. & Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158, 17–25 (2012).
Hermosa, R. et al. The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 16, 69–80 (2013).
Mendoza-Mendoza, A. et al. Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85 (2018).
Vinale, F. et al. Trichoderma-plant pathogen interactions. Soil. Biol. Biochem. 40, 1–10 (2008).
Contreras-Cornejo, H. A., Macías-Rodríguez, L., Cortés-Penagos, C. & López-Bucio, J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592 (2009).
Samolski, I., Rincón, A. M., Pinzón, L. M., Viterbo, A. & Monte, E. The quid74 gene from Trichoderma harzianum has a role in root architecture and plant biofertilization. Microbiology 158, 129–138 (2012).
Carrero-Carrón, I. et al. Interactions between Trichoderma harzianum and defoliating Verticillium dahlia in resistant and susceptible wild olive clones. Plant Pathol. 67, 1758–1767 (2018).
Calvet, C., Pera, J. & Barea, J. M. Growth response of marigold (Tagetes erecta L.) to inoculation with Glomus mosseae, Trichoderma aureoviride and Pythium ultimum in a peat perlite mixture. Plant Soil 148, 1–6 (1993).
Datnoff, L. E., Nemec, S. & Pernezny, K. Biological control of Fusarium crown and root rot of tomato in Florida using Trichoderma harzianum and Glomus intraradices. Biol. Control 5, 427–431 (1995).
Chandanie, W. A., Kubota, M. & Hyakumachi, M. Interactions between the arbuscular mycorrhizal fungus Glomus mosseae and plant growth-promoting fungi and their significance for enhancing plant growth and suppressing damping-off of cucumber (Cucumis sativus L.). Appl. Soil Ecol. 41, 336–341 (2009).
Martínez‐Medina, A., Pascual, J. A., Lloret, E. & Roldán, A. Interactions between arbuscular mycorrhizal fungi and Trichoderma harzianum and their effects on Fusarium wilt in melon plants grown in seedling nurseries. J. Sci. Food Agric. 89, 1843–1850 (2009).
Green, H., Larsen, J., Olsson, P. A., Jensen, D. F. & Jakobsen, I. Suppression of the biocontrol agent Trichoderma harzianum by mycelium of the arbuscular mycorrhizal fungus Glomus intraradices in root-free soil. J. Appl. Environ. Microbiol. 65, 1428–1434 (1999).
Martínez, A., Obertello, M., Pardo, A., Ocampo, J. A. & Godeas, A. Interactions between Trichoderma pseudokoningii strains and the arbuscular mycorrhizal fungi Glomus mosseae and Gigaspora rosea. Mycorrhiza 14, 79–84 (2004).
Martínez-Medina, A., Pascual, J. A., Pérez-Alfocea, F., Albacete, A. & Roldán, A. Trichoderma harzianum and Glomus intraradices modify the hormone disruption induced by Fusarium oxysporum infection in melon plants. Phytopathology 100, 682–688 (2010).
Martínez-Medina, A., Roldán, A. & Pascual, J. A. Interaction between arbuscular mycorrhizal fungi and Trichoderma harzianum under conventional and low input fertilization field condition in melon crops: growth response and Fusarium wilt biocontrol. Appl. Soil Ecol. 47, 98–105 (2011).
Alonso‐Ramírez, A. et al. Salicylic acid prevents Trichoderma harzianum from entering the vascular system of roots. Mol. Plant Pathol. 15, 823–831 (2014).
Jogaiah, S., Abdelrahman, M., Tran, L. S. P. & Ito, S. I. Different mechanisms of Trichoderma virens‐mediated resistance in tomato against Fusarium wilt involve the jasmonic and salicylic acid pathways. Mol. Plant Pathol. 19, 870–882 (2018).
Pieterse, C. M. et al. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375 (2014).
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S. & Van Wees, S. C. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 (2009).
Bonfante, P. & Requena, N. Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr. Opin. Plant. Biol. 14, 451–457 (2011).
Morán-Diez, E. et al. Transcriptomic response of Arabidopsis thaliana after 24 h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 169, 614–620 (2012).
Maillet, F. et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–64 (2011).
Hause, B., Maier, W., Miersch, O., Kramell, R. & Strack, D. Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol. 130, 1213–1220 (2002).
Rooney, D. C. et al. Mycorrhizas and biomass crops: opportunities for future sustainable development. Trends Plant Sci. 14, 542–549 (2009).
Woo, S. L. et al. Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126 (2014).
Kowalska, J. Organically grown Brassica napus - use of border strips and Trichoderma. Soil Plant Sci. 64, 529–536 (2014).
Barto, K., Friese, C. & Cipollini, D. Arbuscular mycorrhizal fungi protect a native plant from allelopathic effects of an invader. J. Chem. Ecol. 36, 351–360 (2010).
Veiga, R. S., Howard, K. & van der Heijden, M. G. No evidence for allelopathic effects of arbuscular mycorrhizal fungi on the non-host plant Stellaria media. Plant Soil 360, 319–331 (2012).
Lambers, H. & Teste, F. P. Interactions between arbuscular mycorrhizal and non‐mycorrhizal plants: do non‐mycorrhizal species at both extremes of nutrient availability play the same game? Plant Cell Environ. 36, 1911–1915 (2013).
Rinaudo, V., Bàrberi, P., Giovannetti, M. & van der Heijden, M. G. Mycorrhizal fungi suppress aggressive agricultural weeds. Plant Soil 333, 7–20 (2010).
Sharma, N., Yadav, K. & Aggarwal, A. Synergistic effect of arbuscular mycorrhizae and Trichoderma sp. on growth, nutrient uptake and yield of Phaseolus mungo L. cultivars. J. Trop. Plant Physiol. 8, 23–31 (2016).
Hammer, E. C. et al. A mycorrhizal fungus grows on biochar and captures phosphorus from its surfaces. Soil Biol. Biochem. 77, 252–260 (2014).
Akhter, A., Hage-Ahmed, K., Soja, G. & Steinkellner, S. Compost and biochar alter mycorrhization, tomato root exudation, and development of Fusarium oxysporum f. sp. lycopersici. Front. Plant Sci. 6, 529, https://doi.org/10.3389/fpls.2015.00529 (2015).
Van der Heijden, M. G. et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72 (1998).
Kamel, L., Keller‐Pearson, M., Roux, C. & Ané, J. M. Biology and evolution of arbuscular mycorrhizal symbiosis in the light of genomics. New Phytol. 213, 531–536 (2017).
Filion, M., St‐Arnaud, M. & Fortin, J. A. Direct interaction between the arbuscular mycorrhizal fungus Glomus intraradices and different rhizosphere microorganisms. New Phytol. 141, 525–533 (1999).
De Jaeger, N., Declerck, S. & De la Providencia., I. E. Mycoparasitism of arbuscular mycorrhizal fungi: a pathway for the entry of saprotrophic fungi into roots. FEMS Microbiol. Ecol. 73, 312–322 (2011).
Lace, B. et al. Gate crashing arbuscular mycorrhizas: in vivo imaging shows the extensive colonization of both symbionts by Trichoderma atroviride. Environ. Microbiol. Rep. 7, 64–77 (2015).
Rasmussen, S. R. et al. Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci. Rep. 6, 29733, https://doi.org/10.1038/srep29733 (2016).
Zhu, H. et al. The possible involvement of salicylic acid and hydrogen peroxide in the systemic promotion of phenolic biosynthesis in clover roots colonized by arbuscular mycorrhizal fungus. J. Plant Physiol. 178, 27–34 (2015).
Rubio, M. B. et al. Identifying Trichoderma parareesei beneficial qualities for plants. Appl. Environ. Microbiol. 80, 1864–1873 (2014).
Chialva, M. et al. Understanding changes in tomato cell walls in roots and fruits: the contribution of arbuscular mucorrhizal colonization. Int. J. Mol. Sci. 20, 415 (2019).
García‐Garrido, J. M. & Ocampo, J. A. Regulation of the plant defense response in arbuscular mycorrhizal symbiosis. J. Exp. Bot. 53, 1377–1386 (2002).
Vos, I. A., Moritz, L., Pieterse, C. M. & Van Wees, S. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 6, 639, https://doi.org/10.3389/fpls.2015.00639 (2015).
Rubio, M. B. et al. The combination of Trichoderma harzianum and chemical fertilization leads to the deregulation of phytohormone networking, preventing the adaptive responses of tomato plants to salt stress. Front. Plant Sci. 8, 294, https://doi.org/10.3389/fpls.2017.00294 (2017).
Morán-Diez, E. et al. The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum-plant beneficial interaction. Mol. Plant Microbe Interact. 22, 1021–1031 (2009).
Pérez, E. et al. The importance of chorismate mutase in the biocontrol potential of Trichoderma parareesei. Front. Microbiol. 6, 1181, https://doi.org/10.3389/fmicb.2015.01181 (2015).
Lee, J., Lee, S. & Young, J. P. W. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol 65, 339–349 (2008).
Montero-Barrientos, M. et al. Transgenic expression of the Trichoderma harzianum hsp70 gene increases Arabidopsis resistance to heat and other abiotic stresses. J. Plant Physiol. 167, 659–665 (2010).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Yang, H. et al. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 538, 113–122 (2014).
Berger, S., Mitchell‐Olds, T. & Stotz, H. U. Local and differential control of vegetative storage protein expression in response to herbivore damage in Arabidopsis thaliana. Physiol. Plant. 114, 85–91 (2002).
Zimmerli, L., Stein, M., Lipka, V., Schulze‐Lefert, P. & Somerville, S. Host and non‐host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 40, 633–646 (2004).
Berruti, A., Lumini, E., Balestrini, R. & Bianciotto, V. Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes. Front. Microbiol. 6, 1559, https://doi.org/10.3389/fmicb.2015.01559 (2016).
Bostock, R. M. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu. Rev. Phytopathl. 4, 3545–3580 (2005).
Acknowledgements
This work was supported by the grants AGL2015-70671-C2-1-R from the Spanish Ministry of Economy and Competitiveness, and SA230U13 and SA009U16 from the Regional Government of Castile and Leon (Spain).
Author information
Authors and Affiliations
Contributions
J.P. performed the experiments; J.P., E.M. and C.N. conceived and designed the experiments; J.P., R.H., E.M. and C.N. analyze the data; J.P., R.H., E.M. and C.N. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Poveda, J., Hermosa, R., Monte, E. et al. Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity. Sci Rep 9, 11650 (2019). https://doi.org/10.1038/s41598-019-48269-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48269-z
This article is cited by
-
IPD3, a master regulator of arbuscular mycorrhizal symbiosis, affects genes for immunity and metabolism of non-host Arabidopsis when restored long after its evolutionary loss
Plant Molecular Biology (2024)
-
Recent advances in the use of Trichoderma-containing multicomponent microbial inoculants for pathogen control and plant growth promotion
World Journal of Microbiology and Biotechnology (2024)
-
Trichoderma: a multipurpose, plant-beneficial microorganism for eco-sustainable agriculture
Nature Reviews Microbiology (2023)
-
A potential role of salicylic acid in the evolutionary behavior of Trichoderma as a plant pathogen: from Marchantia polymorpha to Arabidopsis thaliana
Planta (2023)
-
Hyphosphere microbiome of arbuscular mycorrhizal fungi: a realm of unknowns
Biology and Fertility of Soils (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.