Abstract
Hidradenitis suppurativa (HS) is a chronic inflammatory disorder characterized by painful nodules, sinus tracts, and scars occurring predominantly in intertriginous regions. The prevalence of HS is currently 0.053–4%, with a predominance in African-American women and has been linked to low socioeconomic status. The majority of the reported literature is retrospective, population based, epidemiologic studies. In this regard, there is a need to establish a repository of biospecimens, which represent appropriate gender and racial demographics amongst HS patients. These efforts will diminish knowledge gaps in understanding the disease pathophysiology. Hence, we sought to outline a step-by-step protocol detailing how we established our HS biobank to facilitate the formation of other HS tissue banks. Equipping researchers with carefully detailed processes for collection of HS specimens would accelerate the accumulation of well-organized human biological material. Over time, the scientific community will have access to a broad range of HS tissue biospecimens, ultimately leading to more rigorous basic and translational research. Moreover, an improved understanding of the pathophysiology is necessary for the discovery of novel therapies for this debilitating disease. We aim to provide high impact translational research methodology for cutaneous biology research and foster multidisciplinary collaboration and advancement of our understanding of cutaneous diseases.
Similar content being viewed by others
Introduction
Hidradenitis suppurativa (HS) is a chronic inflammatory disorder characterized by painful nodules, sinus tracts and scars that occur predominantly in intertriginous regions1,2. The wide range of reported prevalence is 0.05–4%, but may be an underestimation since the diagnosis is often delayed or missed altogether1,2,3. HS occurs more frequently in women (female to male ratio of 3:1) and patients of African descent1,4,5,6. Patients suffering from HS consistently report an impaired quality of life and possess among the poorest scores reported when using the dermatology quality of life index (DLQI)7,8. The underlying etiology of HS is presently unknown and current therapy is often inadequate or ineffective1,2,3,9. Consequently, there is a need for better understanding of the pathogenesis of HS and development of novel therapies.
To this end, we propose a HS-specific biobanking protocol to establish a standardized process for consistent and ethical data collection and analysis for future clinical and basic science studies. Biobanks are well-organized collections of human biological material which are stored for research and can help drive translational research10,11,12. However, differences in biospecimen collection and management amongst biorepositories and institutions in this age of national and international collaboration can lead to variability in quality and molecular integrity that ultimately interferes with results and reproducibility13,14. Though there are existing biobanking protocols for many other conditions15,16, there is no central source to describe best practices for ethical issues, biospecimen collection, processing techniques and biorepository development (storage) for HS biobank development. Biobanking is particularly important for complex and largely understudied diseases like HS10,12. This protocol was successfully instituted at the Johns Hopkins University School of Medicine (JHUSOM) and has already contributed to translational studies17.
The purpose of this manuscript is to present a detailed, step-by-step protocol for collecting HS specimens and building an HS biobank. During the collection of biospecimens, careful clinical phenotyping, patient histories and in selected cases, ultrasound imaging, should be accompanied by photography. Figure 1 provides an overview to our approach to specimen collection for translational studies in HS, emphasizing the role of collecting clinical phenotypic data for correlation with basic laboratory findings. There are a number of unique challenges to collecting biospecimens for translational research in HS. Herein, we acknowledge and present our approaches to address them.
Methods
All samples are collected, stored and used in agreement with the ethical and research guidelines set by biobanking privacy laws18 and the Institutional Review Boards (IRB) of JHUSOM and University of California, San Francisco. The methods for specimen collection for genetic studies were approved at Columbia University Irving Medical Center. Informed consent was obtained from all participants and/or their legal guardian(s). These methods can be applied elsewhere after institutional IRB approval, further evolving new technologies and knowledge.
Overview of Biospecimen Collection
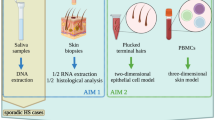
Biospecimen categories and applications- detailed in Figs 2 and 3
-
i.
Skin biopsies and surgically resected tissue samples can be collected from diseased tissue (“lesional”), areas adjacent to the diseased tissue but without gross evidence of disease (“peri-lesional”), normal-appearing tissue of an HS patient (“unaffected”), as well as healthy individuals (“normal”). Precise descriptions of the morphology, disease severity and exact location of lesions biopsied enable experimental comparisons of morphologically similar lesions from different individuals. Where possible, the lesional, peri-lesional and unaffected skin should be collected from the same anatomical region, as there are region-specific differences in epidermal appendages and the cutaneous microbiome19. Photographic documentation before and after sample collection can aid in future clinical-pathological correlations. Punch biopsies (4 mm or larger in size) and excisional biopsies should be deep enough to include subcutaneous fat. Surgically resected tissue tends to provide larger and deeper tissue sections than biopsies, providing opportunities for more assays and increasing the tissue repository for future evaluation. Tissue sections can be utilized for histology, immunohistochemistry (IHC), and immunofluorescence (IF). Tissue proteins can be obtained for Western Blotting (WB), Enzyme-linked immunosorbent assay (ELISA), zymography and tissue proteomics20. Whole tissues can be processed for keratinocytes, fibroblasts and immune cells. Nucleic acids can also be extracted for transcriptomic, genetic analysis and microbiome analysis21,22,23.
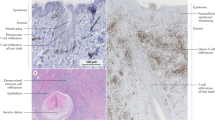
Figure 2 Figure 3 Examples of analysis techniques for collected biospecimens. (a) Hematoxylin & Eosin staining of HS lesional skin from fresh-frozen tissue. (b) Fontana-Masson staining of HS skin from paraffin-embedded tissue. Scale bar: 100 μm. (c) Immunofluorescence of HS skin for keratin 14 (red), MPO-myeloperoxidase (green), and DAPI (blue). (d) HS skin was digested and primary fibroblasts (first panel) and primary epidermal cells- keratinocytes and melanocytes- (second panel) were cultured. Peripheral blood was collected from HS patients and monocytes/macrophages (third panel) and neutrophils (fourth panel) were isolated and cultured. Scale bar: 50 μm. (e) HS skin was digested into single cell suspension and stained for flow cytometry analysis. (f) Protein was extracted from peripheral blood cells and used to perform Western blot. Uncropped Western Blot in Supplemental Fig. S1. (g) RNA was extracted from homogenized healthy control and HS skin and was quantified with a spectrophotometer. The 260/280 absorbance ratio of ~2 represents the purity and quality of the RNA. (h) Heatmap representing the microbiome of healthy control and HS skin developed by next-generation sequencing.
-
ii.
Saliva can yield high quality human RNA/DNA to assess biomarkers or genetic mutations and polymorphisms in HS patients. Direct24,25,26 or oral swab27 collection of saliva may also be utilized to understand changes in the oral microbiome that may be linked to systemic disease manifestations. Bacterial RNA/DNA extraction methods have minimal if any effect on saliva microbiome profiles, making saliva an ideal biospecimen for this analysis28. In addition, saliva can be used for proteomic analysis29,30.
-
iii.
Blood cells, such as peripheral blood mononuclear cells (PBMCs) and neutrophils, can be isolated to provide insight into the systemic response and contributions to HS31,32. These cells may be used for flow cytometry17,32,33,34, proteomics35, transcriptomic and genetic analysis as well as functional analyses36,37, including cell culture/co-culture experiments38,39.
-
iv.
Serum can also aid in understanding the systemic effects of HS. Inflammatory and other biomarkers may be identified using either large scale proteomic platforms or individual analyses, and can be related to onset, specific disease stages, and/or disease progression40. Likewise, RNA and DNA can also be extracted for transcriptomic and genetic analysis, respectively22,41.
-
v.
Skin swabs can be used to catalog the composition of and shifts in the HS skin microbiome. Skin cells, chemicals and microbes collected on skin swabs can be used to analyze molecular and microbial characteristics via mass spectrometry and microbial 16 S rRNA amplicon sequences42, as well as high-throughput next-generation sequencing23,43,44. Details have been outlined in Figs. 2 and 3.
Outpatient versus inpatient biospecimen collection
Biospecimens may be collected during outpatient clinic visits or in the operating room (OR). The advantages and disadvantages of each mode of collection have been outlined in Table 1. Biospecimens collected during outpatient visits allows for concurrent clinical correlations. Collection of HS tissue resections in the OR provides abundant tissue for analysis; however, the clinical context may not be fully assessed if the patient was not evaluated clinically by the investigators prior to surgery. When collecting surgically resected tissue in the OR, it is important to bring specific supplies to the site of tissue collection (Fig. 4).
Recommendations for Tissue Collection- Outlined in Table 2
Recommendation 1. Consents and protocols
Institution-specific IRB-approved protocols and consents provide the opportunity for prospective biobanking and tissue analysis, retrospective analysis of archived tissue and multi-institutional collaborations. During clinical encounters, patients should be informed about ongoing HS research studies and offered the opportunity to participate. If interested, informed consent should be obtained, allowing for biospecimen collection, analysis of collected material, permission to contact for future studies and for clinical chart reviews. When tissue is collected in the OR, consent may be obtained immediately prior to surgery concurrent with informed consent for the surgical procedure. Alternatively, informed consent for the study can be obtained in advance of surgery. In the authors’ experience, the latter process allows the patient more time for questions and consideration of the implications of their participation.
Recommendation 2. Tissue collection and processing
OR: Tissue should be placed on prepared aluminum foil pieces once resected in the OR. After gross appearance of the tissue is observed and recorded, including photographs if possible (Fig. 5), the tissue should be partitioned as follows:
-
Five 0.3–0.5 cm sections should be placed into individual 2 mL cryogenic vials.
-
Three 0.5–1 cm sections should be wrapped in foil for optimal cutting tissue compound (OCT) embedding, paraffin and RNA extraction. Timely processing prevents RNA degradation.
-
One larger tissue section (approximately 2.5 cm) should be placed in a 50 mL conical tube with sterile Phosphate-buffered saline (PBS), kept at room temperature and either processed or placed on ice within 30 minutes. If isolating immune cells, this tissue section must be processed on the same day it is collected.
-
Remaining sections should be wrapped in foil to be stored for other analyses.
All cryogenic tubes containing tissue sections and foil wrapped tissue sections should be placed in a container with liquid nitrogen for transport back to the lab. If immediate snap freezing is not feasible, tissues should be frozen within 30 minutes of resection. Each patient should have a representative tissue section placed in formalin and sent for routine histopathologic examination and archiving by the surgical team.
Outpatient clinic
The biospecimen noted above can be collected during outpatient visits with appropriate staffing and supplies. Smaller tissue samples can be obtained using punch biopsies or small elliptical excisions and can be treated in the aforementioned manner. Table 1 outlines helpful considerations when comparing biospecimen collection in outpatient or OR settings
Initial tissue processing: The specimens should be quickly returned to the laboratory bench, paying special attention to the liquid nitrogen level in order to prevent spills. The bench should be lined with disposable underpads. The larger tissue sections and the individual cryogenic vials should be placed in a storage box with a de-identified label (e.g. HS_001) and stored at −80 °C for future use.
Samples can be processed as fresh frozen or as formalin fixed paraffin embedded tissue (FFPE). Both incur advantages and disadvantages, outlined in Fig. 6. For the tissue designated for OCT (fresh frozen tissue), ‘peel-away’, disposable and variously sized histology cryomolds can be used. The cryomold should be slowly filled with enough OCT media to cover the tissue section, paying special attention to avoid bubbles. Forceps should be used to place the tissue section in an orientation such that the epidermis and dermis can be visualized on a cross-sectional cut (Fig. 6). The cryomold should be immersed, not fully, into liquid nitrogen (or slurry of methanol and dry ice) for 30 seconds or until nearly frozen. At this point, it can be transferred to a foam cooler of dry ice. Individual samples can be wrapped in labeled foil and sealed in a plastic bag. These samples can be stored at −80 °C or sectioned with a cryostat immediately. If continuing with the cryostat sectioning, samples may be cut into 3–8 μm sections and placed on labeled plus-plus glass slides. Cryostat sectioned slides should immediately be placed at −20 °C or −80 °C for permanent storage.
Tissue designated for FFPE should be placed in labeled cassettes and submerged in 10% formalin. The container should be stored at 4 °C for at least 24 hours for proper fixation before paraffin embedding.
Tissue designated for RNA should be placed in a 50 mL conical tube with 10–15 mL of TRIzol® or RNAlater®, and immediately stored at −80 °C or future use. Figure 4 outlines specific supplies and resources.
Recommendation 3. Preparation of tissue for protein analysis
Homogenization of whole tissue
Designated tissue sections can be homogenized and utilized to assess proteins, using experiments such as WB, ELISA and proteomics. Several 3–5 mm tissue sections should be placed in a mortar with enough liquid nitrogen to flash freeze the tissue. Using a pestle, the flash frozen tissue should be ground into powder-like substance to which lysis buffer should be added and processed accordingly45. The lysate can be stored at −80 °C until further use45.
Recommendation 4. Preparation of tissue for nucleic acid analysis
DNA and RNA extraction
The recommended protocol above yields a frozen specimen from which you can extract nucleic acids using purification protocols or manufactured kits43,46,47,48,49,50,51,52. The specimen can be stored at −80 °C until required.
Recommendation 5. Preparation of tissue for cell culture
Extraction of primary cells from tissue
Fresh tissue placed in sterile PBS can be used to isolate primary cells. Tissue samples can be digested using various methods53,54,55,56,57 to derive single cell suspensions. This allows for the isolation of cell types of interest via subsequent extracellular and/or intracellular staining and flow cytometry.
For fibroblast and keratinocyte isolation specifically, tissue can be separated into the dermis (fibroblasts) and epidermis (keratinocytes and melanocytes) as previously described58. Confluent cultured HS fibroblasts, keratinocytes, and melanocytes are pictured in Fig. 3d. Primary cultures of human fibroblasts and keratinocytes can also be used to create induced pluripotent stem cells (iPSC), which provide a pathway to model and treat human diseases59,60,61.
Extraction of immune cells from tissue
As described in the previous section, skin tissue samples can be digested to derive single cell suspensions, which can be stained and sorted using flow cytometry for specific immune cell selection53,54,55,56,57. Specific to T cells, skin tissue samples can be minced accordingly, and subsequently cultured. Alternatively, explants can be cultured directly in 24-well plate wells with a small decrease in number of T cells isolated62. We encourage our readers to consider that recent studies suggest HS is a complex immune-mediated process and this method that focuses on a single immune cell type, the T cell, is limited in scope.
Recommendation 6. Processing of saliva samples
DNA sample preparation
Saliva can be self-collected in specialized tubes that allow for ambient storage for up to five years. DNA extraction reagents and protocols are provided by kit manufacturers. Extracted DNA can be stored at −20 °C (−80 °C for storage beyond 6 months) in aliquots. Each kit typically yields 100 μg of DNA, ensuring adequate amounts for multiple genetic analyses63. Saliva can also be used for proteomic29, transcriptomic64 and microbiome analysis28,65.
Recommendation 7. Blood processing for plasma, serum, nucleic acid and immune cell isolation
Peripheral blood collection and separation
Specific tubes which can be used are indicated in Fig. 1. To evaluate peripheral blood soluble mediators, plasma and serum can be separated from the blood by centrifugation33, removed as a clear layer, divided into aliquots (minimizing freeze thaw cycles) and stored at −80 °C for subsequent analyses. During serum collection, clotted blood will be discarded after centrifugation. Although serum provides a higher sensitivity for biomarker detection, experimental question and design determine the use of serum and/or plasma66,67,68.
Isolation of immune cells
Following plasma isolation, remaining blood is separated by density gradient centrifugation to collect PBMCs17,69,70. Cells not being analyzed immediately can be frozen in aliquots. Polymorphonuclear cells (PMN; neutrophils), can also be isolated in a similar manner33,but must be used within 5–10 hours after isolation due to short lifespan.
Flow cytometry preparation
If possible, a portion of the PBMC can be immunostained for extracellular and intracellular flow cytometric analyses71.
Selection of desired cell preparations
Immunomagnetic separation to isolate specific cell subpopulations can also be performed from PBMCs. Positive or negative selection may be performed to obtain desired cell populations, depending on the investigator’s preference72.
RNA extraction
RNA can be isolated from PBMC or specific cell populations using TRIzol®. Alternatively, RNA may be isolated from whole blood by standardized kits (e.g. QIAamp RNA Blood Mini Kit, Qiagen, MD, USA). Upon isolation, RNA purity and concentration should be determined (Fig. 3g). RNA should be aliquoted for long term storage at −80 °C.
DNA extraction
Within a day of receiving the sample, peripheral blood collected into a 6 mL tube (with EDTA) can be shipped to the processing location at room temperature, within a 1-week window. DNA can be extracted from PBMCs using standardized kits (e.g. PUREGENE DNA isolation kit, Qiagen, MD, USA)73, and aliquoted for long term storage at −20 °C. However, ideally neutrophils should be isolated on the day of blood collection.
Recommendation 8. Skin swab collections for microbiome studies
Study participants should prepare skin using an unscented, non-antibacterial soap for hygiene for 7 days; avoiding all bathing or washing 24 hours prior to sampling, topical antiseptics for 7 days and systemic antibiotics 6 months prior to sampling. Samples should be collected from selected body sites with no prior cleaning or preparation of the skin surface and site of swab should be documented. Swabs should be obtained from a 4 cm2 area using cotton-tipped applicators soaked in enzymatic lysis buffer (20 mM Tris pH 8, 2 mM EDTA, and 1.2% Triton X-100). Negative controls of ambient air swabs should be collected after each sampling. All samples should be stored at −80 °C until further processing74. Skin swabs can also be used for transcriptomic74 and epigenetic75 analysis.
Recommendation 9. Overview of sample storage and barcoding76 – Fig. 1
Limitations and Discussion
Recommendations have been outlined in Fig. 1 and Table 2. One important limitation to note is that variation in important inflammatory cytokines can be associated with the methods of tissue collection, time to freezing, patient age, the amount of cutaneous ultraviolet light exposure around the time of tissue collection, as well as the biospecimen storage duration77,78,79. Whilst validation studies provide some reassurance to the stability of serum and saliva samples25,78, the existing literature on cutaneous biorepository samples is less robust77. Hence, any case control matching would require thorough demographic matching to ensure the most accurate and reliable results from future studies. Obstacles may be encountered during the development of an HS biobank and we have outlined some common troubleshooting tips in Supplementary Table S1.
Many major gaps in our understanding of HS exist80. The majority of the reported literature are retrospective, population based, epidemiologic studies. The published basic science research reports are usually studies with small sample sizes and with limited clinicopathological correlations. Certainly, the lack of an animal model limits a deeper understanding of cellular and pathologic mechanisms81,82. Therefore, there is a need to establish a translational approach to studying this devastating disease.
The availability of biobanked specimens from clinically well-characterized HS patients with appropriate gender and racial representation will help address these knowledge gaps. When collecting biospecimens, a thorough clinical assessment is needed to improve our understanding of the correlations between the clinical phenotype and the generated molecular data. Thus, detailed descriptions of phenotypes, exposures and treatment outcomes should be included in the clinical evaluation of these patients. Additionally, standardization of disease severity measures and nomenclature are imperative for the generalizability of data collected.
The public’s use of the Internet for obtaining and exchanging health-related information has created opportunities to rapidly engage patients and efficiently collect data through Internet surveys. Additionally, the standardization of electronic health records and changes in policy that give patients access to their own records also provides an opportunity to receive patient-supplied clinical records to complement survey data.
Biobanking serves as a major driver of translational research. Although there are multiple methods for performing various scientific analyses, we are proposing a general roadmap and preferable methods for those conducting HS research and studying clinical manifestations in order to render reproducible and reliable results and conclusions. It is our hope that this protocol will promote the development of other institution-specific biobanks for laboratory studies and collaborative efforts to improve our understanding of HS. These protocols can be updated as new technologies drive advanced studies.
References
Jemec, G. B. Hidradenitis suppurativa. N. Engl. J. Med. 366, 158–164 (2012).
Vekic, D. A., Frew, J. & Cains, G. D. Hidradenitis suppurativa, a review of pathogenesis, associations and management. Part 1. Australas. J. Dermatol. 59, 266–277 (2018).
Jemec, G. B. & Kimball, A. B. Hidradenitis suppurativa: epidemiology and scope of the problem. J. Am. Acad. Dermatol. 73, S4–S7 (2015).
Reeder, V. J., Mahan, M. G. & Hamzavi, I. H. Ethnicity and hidradenitis suppurativa. J. Invest. Dermatol. 134, 2842–2843 (2014).
Vaidya, T., Vangipuram, R. & Alikhan, A. Examining the race-specific prevalence of hidradenitis suppurativa at a large academic center; results from a retrospective chart review. Dermatol. Online J. 23, 12 https://escholarship.org/uc/item/9xc0n0z1 (2017).
Vlassova, N., Kuhn, D. & Okoye, G. A. Hidradenitis suppurativa disproportionately affects African Americans: a single-center retrospective analysis. Acta. Derm. Venereol. 95, 990–991 (2015).
Vekic, D. A. & Cains, G. D. Hidradenitis suppurativa, a review of pathogenesis, associations and management. Part 2. Australas. J. Dermatol. 59, 261–266 (2018).
Matusiak, L., Bieniek, A. & Szepietowski, J. C. Hidradenitis suppurativa markedly decreases quality of life and professional activity. J. Am. Acad. Dermatol. 62, 706–708 (2010).
Kimball, A. B. & Jemec, G. B. E. Hidradenitis Suppurativa: A Disease Primer (ADIS, 2017).
Hewitt, R. E. Biobanking: the foundation of personalized medicine. Curr. Opin. Oncol. 23, 112–119 (2011).
Kauffmann, F. & Cambon-Thomsen, A. Tracing biological collections: between books and clinical trials. JAMA 299, 2318–2318 (2008).
Guerin, J. S. et al. Molecular medicine Ireland guidelines for standardized biobanking. Biopreserv. Biobank 8, 3–63 (2010).
Vaught, J. & Lockhart, N. C. The evolution of biobanking best practices. Clin. Chim. Acta. 413, 1569–1575 (2012).
Dakappagari, N., Zhang, H., Stephen, L., Amaravadi, L. & Khan, M. U. Recommendations for clinical biomarker specimen preservation and stability assessments. Bioanalysis 9, 643–653 (2017).
Moore, H. M., Compton, C. C., Alper, J. & Vaught, J. B. International approaches to advancing biospecimen science. Cancer Epidemiol. Biomarkers Prev. 20, 729–732 (2011).
Biospecimen Research Database: National Cancer Institute, Biorepositories and Biospecimen Research Branch; brd.nci.nih.gov/brd/ (2018).
Byrd, A. S. et al. Collagen deposition in chronic Hidradenitis Suppurativa: Potential role for CD163(+) macrophages. Br. J. Dermatol. 179, 792–794 (2018).
Harrell, H. L. & Rothstein, M. A. Biobanking research and privacy laws in the United States. J. Law Med. Ethics 44, 106–127 (2016).
Perez Perez, G. I. et al. Body site is a more determinant factor than human population diversity in the healthy skin microbiome. PLOS ONE 11, e0151990, https://doi.org/10.1371/journal.pone.0151990 (2016).
Berglund, S. R. et al. Optimized methodology for sequential extraction of RNA and protein from small human skin biopsies. J. Invest. Dermatol. 127, 349–353 (2007).
Peirson, S. N. & Butler, J. N. RNA extraction from mammalian tissues. Methods Mol. Biol. 362, 315–327 (2007).
Radpour, R. et al. Simultaneous isolation of DNA, RNA, and proteins for genetic, epigenetic, transcriptomic, and proteomic analysis. J. Proteome Res. 8, 5264–5274 (2009).
Di Bella, J. M., Bao, Y., Gloor, G. B., Burton, J. P. & Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods. 95, 401–414 (2013).
Goode, M. R., Cheong S. Y., Li, N., Ray, W. C. & Bartlett, C. W. Collection and extraction of saliva DNA for next generation sequencing. J. Vis. Exp. 90, https://doi.org/10.3791/5169 (2014).
Pramanik, R. et al. Effects of the UK biobank collection protocol on potential biomarkers in saliva. Int. J. Epidemiol. 41, 1786–1797 (2012).
Henson, B. S. & Wong, D. T. Collection, storage, and processing of saliva samples for downstream molecular applications. Methods Mol. Biol. 666, 21–30 (2010).
Kim, D. et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 5, https://doi.org/10.1186/s40168-017-0267-5 (2017).
Lim, Y., Totsika, M., Morrison, M. & Punyadeera, C. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci. Rep. 7, 8523, https://doi.org/10.1038/s41598-017-07885-3 (2017).
Vitorino, R., Guedes, S., Manadas, B., Ferreira, R. & Amado, F. Toward a standardized saliva proteome analysis methodology. J. Proteomics 75, 5140–5165 (2012).
Cuevas-Cordoba, B. & Santiago-Garcia, J. Saliva: a fluid of study for OMICS. OMICS 18, 87–97 (2014).
Oh, H., Siano, B. & Diamond, S. Neutrophil isolation protocol. J. Vis. Exp. 17, https://doi.org/10.3791/745 (2008).
Baker, G. J., Castro, M. G. & Lowenstein, P. R. Isolation and flow cytometric analysis of glioma-infiltrating peripheral blood mononuclear cells. J. Vis. Exp. 105, https://doi.org/10.3791/53676 (2015).
Williams, D. W., Anastos, K., Morgello, S. & Berman, J. W. JAM-A and ALCAM are therapeutic targets to inhibit diapedesis across the BBB of CD14+CD16+ monocytes in HIV-infected individuals. J. Leukoc. Biol. 97, 401–412 (2015).
Calderon, T. M. et al. Dopamine Increases CD14(+)CD16(+) Monocyte Transmigration across the Blood Brain Barrier: Implications for Substance Abuse and HIV Neuropathogenesis. J. Neuroimmune Pharmacol. 12, 353–370 (2017).
Koncarevic, S. et al. In-depth profiling of the peripheral blood mononuclear cells proteome for clinical blood proteomics. Int. J. Proteomics 2014, 129259, https://doi.org/10.1155/2014/129259 (2014).
Petukhova, L. et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117 (2010).
Xing, L. et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat. Med. 20, 1043–1049 (2014).
Carmona-Rivera, C., Zhao, W., Yalavarthi, S. & Kaplan, M. J. Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Ann. Rheum. Dis. 74, 1417–1424 (2015).
Carmona-Rivera, C. et al. A role for muscarinic receptors in neutrophil extracellular trap formation and levamisole-induced autoimmunity. JCI Insight 2, e89780, https://doi.org/10.1172/jci.insight.89780 (2017).
Amin, B., Maurer, A., Voelter, W., Melms, A. & Kalbacher, H. New poteintial serum biomarkers in multiple sclerosis identified by proteomic strategies. Curr. Med. Chem. 21, 1544–1556 (2014).
Ge, Q. et al. Profiling circulating microRNAs in maternal serum and plasma. Mol. Med. Rep. 12, 3323–3330 (2015).
Bouslimani, A. et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. USA 112, 2120 (2015).
Castelino, M. et al. Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol 17, 4 (2017).
Yin, Y. et al. Application of High-Throughput Next-Generation Sequencing for HLA Typing on Buccal Extracted DNA: Results from over 10,000 Donor Recruitment Samples. PLOS ONE 11, e0165810, https://doi.org/10.1371/journal.pone.0165810 (2016).
Carmona-Rivera, C., Simeonov, D. R., Cardillo, N. D., Gahl, W. A. & Cadilla, C. L. A divalent interaction between HPS1 and HPS4 is required for the formation of the biogenesis of lysosome-related organelle complex-3 (BLOC-3). Biochim. Biophys. Acta. 1833, 468–478 (2013).
Bonin, S. & Stanta, G. Nucleic acid extraction methods from fixed and paraffin-embedded tissues in cancer diagnostics. Expert Rev. Mol. Diagn. 13, 271–282 (2013).
Kocjan, B. J., Hosnjak, L. & Poljak, M. Commercially available kits for manual and automatic extraction of nucleic acids from formalin-fixed, paraffin-embedded (FFPE) tissues. Acta Dermatovenerol. Alp. Pannonica Adriat. 24, 47–53 (2015).
Tan, S. C. & Yiap, B. C. DNA, RNA, and protein extraction: the past and the present. J. Biomed. Biotechnol. 2009, 574398, https://doi.org/10.1155/2009/574398 (2009).
Sheng, H. F. & Zhou, H. W. Methods, challenges and opportunities for big data analyses of microbiome. Nan Fang Yi Ke Da Xue Xue Bao 35, 931–934 (2015).
Zhang, C. et al. Identification of low abundance microbiome in clinical samples using whole genome sequencing. Genome Biol. 16, https://doi.org/10.1186/s13059-015-0821-z (2015).
Dou, Y., Gold, H. D., Luquette, L. J. & Park, P. J. Detecting Somatic Mutations in Normal Cells. Trends Genet. 34, 545–557 (2018).
Salk, J. J., Schmitt, M. W. & Loeb, L. A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 19, 269–285 (2018).
Hotz, C. et al. Intrinsic Defect in Keratinocyte Function Leads to Inflammation in Hidradenitis Suppurativa. J. Invest. Dermatol. 136, 1768–1780 (2016).
Campanelli, A. P. et al. CD4+CD25+ T cells in skin lesions of patients with cutaneous leishmaniasis exhibit phenotypic and functional characteristics of natural regulatory T cells. J. Infect. Dis. 193, 1313–1322 (2006).
Zaba, L. C. et al. Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell-polarizing myeloid dendritic cells. J. Invest. Dermatol. 129, 79–88 (2009).
Hyder, L. A. et al. TREM-1 as a potential therapeutic target in psoriasis. J. Invest. Dermatol. 133, 1742–1751 (2013).
Kok, A. et al. Early initiation of combined antiretroviral therapy preserves immune function in the gut of HIV-infected patients. Mucosal. Immunol. 8, 127–140 (2015).
Kim, D. et al. To Control Site-Specific Skin Gene Expression, Autocrine Mimics Paracrine Canonical Wnt Signaling and Is Activated Ectopically in Skin Disease. Am. J. Pathol. 186, 1140–1150 (2016).
Rodriguez-Piza, I. et al. Reprogramming of human fibroblasts to induced pluripotent stem cells under xeno-free conditions. Stem Cells 28, 36–44 (2010).
Howden, S. E. et al. Simultaneous Reprogramming and Gene Correction of Patient Fibroblasts. Stem Cell Reports 5, 1109–1118 (2015).
Aasen, T. & Izpisua Belmonte, J. C. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat. Protoc. 5, 371–382 (2010).
Clark, R. A. et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J. Invest. Dermatol. 126, 1059–1070 (2006).
James, G., McMullin, M. F., Duncombe, A. S., Clarke, M. & Anderson, L. A. The MOSAICC study: Assessing feasibility for biological sample collection in epidemiology studies and comparison of DNA yields from saliva and whole blood samples. Ann. Hum. Genet. 82, 114–118 (2018).
Chiang, S. H. et al. RNAPro*SAL: a device for rapid and standardized collection of saliva RNA and proteins. BioTechniques 58, 69–76 (2015).
Karched, M., Bhardwaj, R. G., Pauline, E. M., George, S. & Asikainen, S. Effect of preparation method and storage period on the stability of saliva DNA. Arch. Oral. Biol. 81, 21–25 (2017).
Liu, L. et al. Differences in metabolite profile between blood plasma and serum. Anal. Biochem. 406, 105–112 (2010).
Yu, Z. et al. Differences between human plasma and serum metabolite profiles. PLOS ONE 6, e21230, https://doi.org/10.1371/journal.pone.0021230 (2011).
Nishiumi, S., Suzuki, M., Kobayashi, T. & Yoshida, M. Differences in metabolite profiles caused by pre-analytical blood processing procedures. J. Biosci. Bioeng. 125, 613–618 (2018).
Byrd, A. S., O’Brien, X. M., Johnson, C. M., Lavigne, L. M. & Reichner, J. S. An extracellular matrix-based mechanism of rapid neutrophil extracellular trap formation in response to Candida albicans. J. Immunol. 190, 4136–4148 (2013).
Byrd, A. S. et al. NETosis in Neonates: Evidence of a Reactive Oxygen Species-Independent Pathway in Response to Fungal Challenge. J. Infect. Dis. 213, 634–639 (2016).
Williams, D. W. et al. CCR2 on CD14(+)CD16(+) monocytes is a biomarker of HIV-associated neurocognitive disorders. Neurol. Neuroimmunol. Neuroinflamm. 1, e36, https://doi.org/10.1212/NXI.0000000000000036 (2014).
Williams, D. W. et al. Mechanisms of HIV entry into the CNS: increased sensitivity of HIV infected CD14+CD16+ monocytes to CCL2 and key roles of CCR2, JAM-A, and ALCAM in diapedesis. PLOS ONE 8, e69270, https://doi.org/10.1371/journal.pone.0069270 (2013).
Majumdar, G., Vera, S., Elam, M. B. & Raghow, R. A streamlined protocol for extracting RNA and genomic DNA from archived human blood and muscle. Anal. Biochem. 474, 25–27 (2015).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Tierling, S. et al. DNA methylation studies on imprinted loci in a male monozygotic twin pair discordant for Beckwith-Wiedemann syndrome. Clin. Genet. 79, 546–553 (2011).
Jacobs, G., Wolf, A., Krawczak, M. & Lieb, W. Biobanks in the Era of Digital Medicine. Clin. Pharmacol. Ther. 103, 761–762 (2018).
Zhou, J. H., Sahin, A. A. & Myers, J. N. Biobanking in genomic medicine. Arch. Pathol. Lab. Med. 139, 812–818 (2015).
Enroth, S., Hallmans, G., Grankvist, K. & Gyllensten, U. Effects of Long-Term Storage Time and Original Sampling Month on Biobank Plasma Protein Concentrations. EBioMedicine 12, 309–314 (2016).
Shabihkhani, M. et al. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin. Biochem. 47, 258–266 (2014).
Hoffman, L. K. et al. Major gaps in understanding and treatment of hidradenitis suppurativa. Semin. Cutan. Med. Surg. 36, 86–92 (2017).
van der Zee, H. H., Laman, J. D. & Prens, E. P. Can animal skin diseases or current transgenic mice serve as a model for hidradenitis suppurativa? Dermatology 225, 9–13 (2012).
Lu, C. & Fuchs, E. Sweat gland progenitors in development, homeostasis, and wound repair. Cold Spring Harb. Perspect. Med. 4, https://doi.org/10.1101/cshperspect.a015222 (2014).
Acknowledgements
We kindly thank Pierre Coulombe, PhD, Avi Rosenberg, MD, PhD, Marco Delsante, PhD, Sean Zhang, MD, PhD, Ben Larman, PhD, Jelani Zarif, PhD, Karen Brown, PhD, Marc Edwards, PhD, Jonathan Reichner, PhD, Nate Archer, PhD, Janielle Maynard, PhD, Carrie Cox, RN, MS, Ellie Meeder, Valerie Spatafore, Nora Viloria, the JHUSOM Reference Histology and Immunopathology Cores, Fatimah Jackson, PhD (Howard University), and Robert Jackson, PhD (University of Maryland) for their insight and assistance. Also, we are grateful for the Andrew W. Mellon Foundation, and the Office of Faculty Development in the Office of the Provost at Howard University for supporting the 2019 Junior Faculty Writing and Creative Works Summer Academy. Dr. Byrd was a participant in the Johns Hopkins Ethnic Skin Fellowship, which was supported in part by a grant from the Valeant Pharmaceuticals. The company was not involved in the conception of the research idea, conduct of the experiments, and the writing of this manuscript. Supported in part by the Intramural Research Program, NIAMS/NIH, ZIA AR041199, the Johns Hopkins School of Medicine Physician Scientist Training Program (PSTP), the Danby Hidradenitis Suppurativa Foundation Research Grant, and the American Academy of Dermatology Diversity Mentoring Program.
Author information
Authors and Affiliations
Contributions
A.S.B., C.C.-R., D.W.W., M.L.K., R.J.M., L.P., H.B.N., D.K., H.L. and C.A.D. performed and validated the experimental protocols and techniques; A.S.B., Y.D., U.J.O., Q.Q.Q., C.C.-R., D.W.W., L.P., H.B.N., L.A.B., W.D.S., R.A., J.W.F., M.J.K., S.K., L.A.G., L.S.M., A.A., M.A.L. and G.A.O. were involved in the overall design, draft, and/or manuscript preparation; U.J.O. prepared Figures 1 and 3, assisted by A.S.B. and Q.Q.; W.D.S. prepared Figure 2 and Table S1; Y.D., U.J.O., A.S.B., and Q.Q.Q. prepared Figures and Tables; G.A.O., J.A.C., S.M.M., J.M.S., O.A. and K.P.B. surgically resected the analyzed tissue sections and provided clinical and scientific input. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
A.S.B. and G.A.O. are investigators for Eli Lilly. A.S.B. recieved honorarium for speaking at the 77th Annual Society for Investigative Dermatology meeting - AbbVie Sponsored Symposium. J.M.S. is a consultant for Allergen and Co-Founder of LifeSprout, which had no impact or conflicts related to this manuscript. L.S.M. has received grant support from AstraZeneca, MedImmune, Pfizer, Regeneron Pharmaceuticals, Moderna Therapeutics, and Boehringer Ingelheim, and is on the scientific advisory board for Integrated Biotherapeutics, and is a shareholder of Noveome Biotherapeutics, which are all unrelated to the work reported in this manuscript. A.A. received grant funding or honorarium from AbbVie, Galderma, LEO Pharma, Novartis, Sanofi, and Valeant, and hese relationships or funds had no impact or conflicts related to this manuscript. M.A.L. has received consulting honorarium from Abbvie, Incyte, and Xbiotech, but these relationships had no impact on this manuscript. G.A.O. received honorarium from AbbVie for participation on their Hidradenitis Suppurativa Advisory Board. Y.D., U.J.O., Q.Q.Q., C.C.-R., D.W.W., M.L.K., R.J.M., L.P., H.B.N., L.A.B., W.D.S., J.A.C., S.M.M., O.A., K.P.B., D.K., H.L., C.A.D., R.A., J.W.F., M.J.K., S.K., and L.A.G. declare no potential conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41598_2019_48226_MOESM1_ESM.docx
Supplementary Figure S1: Uncropped Western Blots, Supplementary Table S1: Common Troubleshooting Tips in Biospecimen Collection
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Byrd, A.S., Dina, Y., Okoh, U.J. et al. Specimen Collection for Translational Studies in Hidradenitis Suppurativa. Sci Rep 9, 12207 (2019). https://doi.org/10.1038/s41598-019-48226-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48226-w
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.