Abstract
Tumor necrosis factor-α (TNF-α) is a proinflammatory cytokine associated with autoimmune and infectious diseases. Importance of TNF-α in P. falciparum malaria and systemic lupus erythematosus (SLE) have been demonstrated. However, association of functional promoter variants with SLE and malaria is lacking in malaria endemic population. A total of 204 female SLE patients and 224 age and sex matched healthy controls were enrolled in the study. Three hundred fourteen P. falciparum infected patients with different clinical phenotypes were included. TNF-α polymorphisms (G-238A & G-308A) were genotyped by PCR-RFLP. Plasma levels of TNF-α was quantified by ELISA. Heterozygous mutants and minor alleles of TNF-α (G-238A and G-308A) polymorphisms were significantly higher in SLE patients compared to healthy controls and associated with development of lupus nephritis. In addition, both promoter variants were associated with severe P. falciparum malaria. SLE patients demonstrated higher levels of plasma TNF-α compared to healthy controls. TNF-α (G-238A and G-308A) variants were associated with higher plasma TNF-α. In conclusion, TNF-α (G-238A & G-308A) variants are associated with higher plasma TNF-α levels in SLE patients residing in malaria endemic areas and could be a contributing factor in the development of SLE and susceptibility to severe P. falciparum malaria.
Similar content being viewed by others
Introduction
Tumor necrosis factor-alpha (TNF-α) is a pro-inflammatory cytokine produced by wide range of cells such as macrophages, B cells, T cells and mast cells1. TNF-α is primarily produced as a trans-membrane protein that gets released from the membrane by a metalloprotease- TNF alpha converting enzyme (TACE), to form soluble 17 kDa protein2. TNF-α is a pleotropic cytokine with wide range of biological functions: it can initiate host defense against infectious diseases and along with it involved in toxicity and inflammatory processes1. TNF-α exerts its biological effect through specialized types of receptors viz. TNF receptor 1 (TNFR-1) and TNFR-23. Expression of TNF receptors is tissue specific. TNFR1 is normally observed in most tissues but TNFR2 is restricted to cells of the immune system3. TNF-α has both a beneficial and deleterious role and it has been linked with infectious diseases and autoimmune disorders4,5,6,7. The TNF-α gene is located in short arm of chromosome 6 at position 21.3 and spans about 12 kilobase (kb) length8. Till date, 43 single nucleotide polymorphisms (SNPs) at promoter region of TNF-α gene with minor allele frequency data have been reported (https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=7124). Although there are contradictory reports, some SNPs at promoter region of TNF-α have been shown to regulate TNF-α expression and/or soluble TNF-α levels viz. TNF-α G-238A (rs361525), TNF-α G-308A (rs1800629), TNF-α T-857C (rs1799724), and TNF-α T-1031C (rs1799964)9. However, large number of genetic association studies have focused on two common promoter polymorphisms of TNF-α gene (G-238A and G-308A) and these have shown a significant association with SLE as well as P. falciparum infection in different populations10,11.
Malaria infection is believed to be an important selection pressure during human evolution and subjects with possible survival advantage genotypes against lethal malaria are more prevalent in malaria endemic areas12. This was true across the continents where malaria was endemic. Plasmodium falciparum infection is a life-threatening disease with diverse clinical manifestations13,14,15. TNF-α is an important molecule that works like a double-edged sword in malaria infection16. TNF-α can protect an individual against severe infection17 but when production is unregulated it could be damaging to the host. Low levels of TNF-α has been associated with susceptibility to P. falciparum infection. While various reports have demonstrated elevated plasma levels of TNF-α in severe malaria compared to uncomplicated infection18,19. Mortality in P. falciparum infection is also associated with very high plasma levels of TNF-α18,20. These observations collectively indicate the importance of TNF-α in P. falciparum malaria: optimum levels are essential for protection against infection. Recently, numerous studies have been carried out in different populations to established possible link between TNF-α gene polymorphisms and susceptibility/resistance to P. falciparum infection and/or clinical severity21,22,23,24. Most of the reports22,23,24 have included TNF-α promoter polymorphisms, that are believed to affect mRNA expression and alter plasma levels of protein molecule. TNF-α (G-308A) mutants have been associated with susceptibility to P. falciparum infection23, higher levels of parasitaemia22 and severe malaria25. However, an independent study on South-West Nigerian infected patients failed to demonstrate such association24. Another common TNF-α promoter (G-238A) variant is also linked to elevated parasitaemia22 and severe P. falciparum malaria24.
SLE is characterized by production of autoantibodies against self-antigens, formation of immune complexes, deposition of these complexes in tissues leading to organ damage and its failure26. Lupus nephritis remains one of the severe clinical manifestations and contributes to significant morbidity and mortality27,28. About 50–60% of SLE patients present with kidney dysfunction and the rate of renal affection is higher in Asian population29. Although type I interferons have been shown to play an important role in the pathogenesis of lupus nephritis30, there are cumulative evidence to suggest that TNF-α may also have a crucial role in renal dysfunction6,31. This has been demonstrated in the mouse model of SLE (MRL/lpr): elevated levels have been reported in serum and kidney tissue such as glomeruli, vascular smooth muscle, perivascular infiltrating cells and tubular epithelial cells32,33. Furthermore, the severity of proteinuria has been found to correlate with the degree of TNF-α expression in the kidney32. There are several reports of elevated TNF-α in SLE patients34,35,36,37. In a study involving African American, European American and Hispanic American SLE patients, high levels of plasma TNF-α was observed and a positive correlation with IFN-α was demonstrated37. A significant positive correlation between plasma TNF-α, with clinical severity and anti-ds DNA has also been reported in several studies34,35,36. Interestingly, TNF-α expression was found to be high in the renal tissue of patients with lupus nephritis38,39. Defective clearance of apoptotic bodies has been suggested to be an important factor in the pathogenesis in SLE40,41. TNF-α has been shown to induce apoptosis42,43,44. Elevated TNF-α in SLE patients could be one of the reasons for increased apoptosis, elevated production of nuclear debris followed by defective clearance of dead or dying cells. However, anti TNF-α therapy has provided no therapeutic advantage in the treatment of SLE45,46.
Importance of TNF-α in P. falciparum malaria has been widely investigated16,47,48 and it has been demonstrated that TNF-α is an important molecule for parasite clearance49. However, uncontrolled production of this cytokine during P. falciparum infection can lead to clinical complications50,51. In malaria endemic areas, subjects with moderate TNF-α producing genotypes have survival advantage12. The association of TNF-α in mouse models of lupus6,52 and in the clinical manifestations in humans has been documented34,35,36,37. For instance, higher expression of TNF-α has been associated with lupus nephritis38,39. Since TNF-α appears to be important to some aspects of the pathogenesis in both SLE and P. falciparum malaria, we hypothesized a possible relationship between TNF-α promoter variants with predisposition to SLE, notably lupus nephritis, in patients residing in a malaria endemic area.
There are limited studies in the Indian population10,53,54, especially, in the malaria endemic belts, for possible association between TNF-α polymorphisms (G-238A and G-308A) and SLE. A recent study has shown a significant association between heterozygotes and minor allele to SLE10. A study from south of India demonstrated an association between TNF-α promoter haplotype and protection against SLE53. In our study, we have enrolled SLE patients and controls from malaria endemic areas of Eastern India and investigated the association of TNF-α promoter variants with SLE. Furthermore, we have quantified plasma levels of TNF-α to assess the genotype-phenotype relationship. The novelty of our study relies on the enrolment of SLE patients and P. falciparum infected cases from malarial endemic areas. The question we have addressed is whether individuals from malarial endemic areas are vulnerable to the development of SLE if genetically susceptible.
Results
Baseline characteristics
In the present study, we enrolled 428 female subjects (224 healthy controls and 204 SLE patients) and 314 P. falciparum infected patients including 103 uncomplicated malaria (UM), 68 cerebral malaria (CM), eighty four multi organ dysfunctions (MODs) and 59 non-cerebral severe malaria (NCSM) (Table 1).
Prevalence of TNF-α promoter (G-238A & G-308A) polymorphisms
Prevalence of heterozygous (GA) and homozygous mutants (AA) for G-238A polymorphism was 12% and 1% respectively (Table 2). Similarly, GA and AA genotype frequency for G-308A polymorphism was 11% and 2% respectively. Distributions of TNF-α(G-308A) polymorphism in healthy female controls deviated from Hardy-Weinberg equilibrium (HWE) (G-308A: χ2 = 11.35, P value = 0.0007; G-238A: χ2 = 3.5, P value = 0.061).
TNF-α (G-238A and G-308A) polymorphism are associated with SLE
As shown in Table 2, the prevalence of GA and minor allele ‘A’ for TNF-α (G-308A) polymorphism were significantly high in SLE patients compared to healthy controls (GA: P = 0.005, OR = 2.18; A: P = 0.002, OR = 1.99). Similarly, frequency of GA for TNF-α (G-238A) polymorphism were more frequent in SLE patients than healthy female controls (P = 0.008, OR = 2.02). Although minor allele for TNF-α (G-238A) polymorphism was more frequent in SLE patients compared to controls, the difference was not significant after Bonferroni correction (P = 0.032, OR = 1.69).
Furthermore, haplotype analysis (G-308A/G-238A) showed significantly higher prevalence of A-G and A-A in SLE patients compared to healthy controls (A-G: P = 0.049, OR = 1.63; A-A: P = 0.029, OR = 2.57) (Supplementary Table 1).
Distribution of TNF-α (G-238A and G-308A) polymorphism in patients with nephritis
Since the study revealed a significant association between TNF-α promoter (G-238A and G-308A) polymorphisms and SLE, we analyzed the association of these polymorphisms with organ involvement. Lupus nephritis was the most important clinical phenotype observed and often associated with increased mortality in SLE. In our study, 41% of SLE patients had lupus nephritis. We categorized the patients into two broad groups: (1) Patients with lupus nephritis (LN+), and (2) patients without nephritis (LN−). As depicted in Table 3, GA genotype and minor allele (A) of TNF-α (G-238A) polymorphism was more frequent in patients with lupus nephritis (LN+) compared to those patients without renal involvement (LN−) (GA: P = 0.002, OR = 2.89;A: P < 0.001, OR = 2.92). However, distribution of TNF-α (G-308A) polymorphism was comparable among both groups.
Plasma TNF-α level in SLE patient & healthy controls
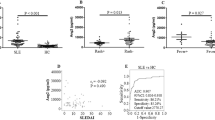
We quantified plasma TNF-α in 90 samples (SLE: 44, HC: 46) by ELISA and mean TNF-α in each group was compared by unpaired ‘t’ test. SLE patients had significantly higher levels of TNF-α compared to healthy controls (P < 0.0001). Plasma levels of TNF-α were compared between LN+ and LN− by student’s t test and results are shown in Fig. 1. The difference in mean TNF-α levels among LN+ and LN− SLE patients was not statistically significant (P = 0.08).
Plasma TNF-α levels in SLE patients and healthy controls. (A) Plasma TNF-α levels was quantified by ELISA in SLE patients (n = 44) and healthy controls (n = 46) and the mean TNF-α were compared by student’s t test. SLE patients displayed significantly higher TNF-α levels compared to healthy controls (P < 0.001). (B) SLE patients were categorized in to two broad group, presence (n = 14) or absence of kidney involvement (n = 30) and mean TNF-α levels was compared. P value less than 0.05 was considered as significant. LN+: lupus nephritis patients; LN−: SLE patients without kidney involvement.
Genotype-phenotype association of TNF-α (G-238A and G-308A) polymorphisms
Several studies have demonstrated a functional relevance of TNF-α promoter polymorphisms (G-238A and G-308A) with expression of TNF -α. We compared plasma levels of TNF-α among different genotypes of TNF-α (G-238A and G-308A). As shown in Fig. 2A,B, for both promoter polymorphism (G-238A and G-308A) the major genotype GG expressed significantly lower levels of plasma TNF-α compared to heterozygous mutant (GA)(P < 0.0001) and homozygous minor genotypes (AA) (G-238A: P = 0.005; G-308A: P = 0.002). Furthermore, we analyzed association of both promoter polymorphism with plasma levels of TNF-α in SLE patients and healthy controls independently (data not shown) and interestingly the observations remained consistent.
Association between TNF-α polymorphisms and levels of plasma TNF-α in SLE patients and control. Plasma TNF-α levels was measured by ELISA, based on availability of plasma samples (SLE: n = 44; HC: n = 46), and correlated with TNF-α genotypes (A) G-238A polymorphism and (B) G-308A polymorphism). Mean plasma TNF-α levels of various genotypes was compared by ANOVA followed by tukey’s multiple comparison post-test. P value less than 0.05 was considered as significant.
Association of TNF-α (G-238A and G-308A) polymorphisms with P. falciparum malaria
Association between TNF-α (G-238A and G-308A) polymorphisms and susceptibility to P. falciparum malaria have been well documented. In the present study, we enrolled 314 P. falciparum infected cases comprising of 103 uncomplicated cases and 211 severe malaria patients and genotyped for TNF-α (G-238A and G-308A) polymorphisms. As shown in Table 4, heterozygous genotype for TNF-α (G-238A) polymorphism and minor allele of G-308A polymorphism were more frequent in SM than UM (GA: P = 0.02, OR = 2.09; A: P = 0.02, OR = 2.05).
Severe malaria patients were further sub-categorized in to CM, MOD and NCSM and distributions of genotypes and allele were compared with UM cases. Results are shown in Table 4. Distributions of heterozygous genotype (GA), minor allele (A) were significantly higher in MOD compared to UM for both TNF-α promoter polymorphisms (G-238A and G-308A). Prevalence of TNF-α (G-308A) heterozygous genotype (GA) was significantly higher in CM cases compared to UM (P = 0.02, OR = 2.65). Comparison of haplotype distribution revealed a significant association of A-A haplotype with predisposition to SM (P = 0.047, OR = 2.33) and MOD (P = 0.011, OR = 3.35) development (Data not shown).
Severe malaria patients displayed higher plasma TNF-α than uncomplicated cases
Severe malaria patients displayed significantly higher plasma TNF-α when compared to uncomplicated P. falciparum infected patients (P = 0.003) (Fig. 3A). Based on various organs involvement, severe malaria patients were further categorized into a) CM [n = 16], b) MOD [n = 21] c) NCSM [n = 18] and compared with uncomplicated cases. Patients with MOD displayed significantly higher levels of plasma TNF-α compared to NCSM (P = 0.004) and UM (P = 0.0002). In addition, a significant difference in mean plasma levels of TNF-α was also observed among CM and UM (P = 0.04) (Fig. 3B).
Plasma TNF-α in different clinical categories of P. falciparum malaria. (A) Plasma TNF-α levels was quantified by ELISA in uncomplicated malaria cases (UM) (n = 12) and severe malaria patients (SM) (n = 55) and the mean TNF-α values were compared by student’s t test. Severe malarial cases displayed significantly higher TNF-α levels compared to uncomplicated malaria (P = 0.0003). (B) Severe malaria cases were further categorized clinically into four sub groups viz. cerebral malaria (CM, n = 16), multi organ dysfunction (MOD, n = 21), non-cerebral severe malaria (NCSM, n = 18) and mean TNF-α levels was compared among them. P value less than 0.05 was considered as significant.
In silico analysis
We observed a significant correlation between TNF-α polymorphisms (G-238A and G-308A) and plasma levels of TNF-α. To validate the above findings, we analyzed functional relevance of these variants in silico. SNPs (rs1800629 and rs361525) were submitted to the FuncPred program and results obtained are shown in Supplementary Table 2. Both the SNPs were found to affect transcription factor binding site (TFBS). However, none of them affect miRNA binding site. SNP with ID rs1800629 was found to have a regulatory potential (RegPot) of 0.0401, which was also an indication of regulatory effects on binding and expression of gene targets.
RegulomeDB database has divided both the SNPs into two distinct categories (Category 1d and Category 4 as shown in Supplementary Table 3). rs1800629 showed RegulomeDB score of 1d and rs361525 which has minimal binding evidence (Category 4). The top ranked SNP rs1800629 had annotation for eQTL + TF binding + any motif + DNase peak and thus very likely to have regulatory functions.
Resampling analysis
As the samples size investigated in the present study was smaller, we performed a resampling analysis and data are shown in Supplementary Table 4. TNF-α (G-308A and G-238A) variants and minor alleles were more frequent in SLE cases and lupus nephritis cases suggesting an important role of TNF-α variants with predisposition to SLE and clinical manifestations.
Discussion
TNF-α is an important cytokine in the pathogenesis as well as control of P. falciparum infection16. Therefore, higher levels observed in malaria infection is a protective phenomenon but very high levels can contribute to severity and mortality. The role of TNF-α in SLE is still conjectural but there are studies implicating it as a contributory factor in the pathogenesis based on experimental and associational studies6,55. It is important to understand the link between TNF-α and SLE in patients residing in malarial endemic areas. In the present study, we observed elevated plasma levels of TNF-α in SLE patients. Furthermore, TNF-α promoter polymorphisms (G-238A and G-308A) were significantly associated with higher plasma levels. These observations provide evidence of a possible role for TNF-α in the pathogenesis of SLE but the precise mechanism(s) is not yet known6,55. TNF-α is a pleotropic cytokine and acts at multiple levels1. In genetically susceptible SLE individuals, malaria might be a trigger for increased production of TNF-α, besides other cytokines, triggering a cascade of events contributing to the development of SLE10,56,57,58.
P. falciparum malaria is predominantly endemic in the Eastern and Northeastern parts of India59. But it remains endemic in most parts of the country. Prevalence of TNF-α promoter polymorphisms (G-238A and G-308A) have been reported in Indian population. Distributions of TNF-α (G-238A and G-308A) genotypes were comparable with previous reports from different parts of the country60,- South-West61, North62 and North-West63 regions. In most of the earlier reports10,60,61,62,63 distribution of TNF-α promoter variants were compatible with HWE in healthy controls. In the present study distribution of TNF-α (G-308A) genotypes deviated from HWE in healthy females (P = 0.0007). The geographical area of Odisha is highly endemic for P. falciparum malaria which contributes to high mortality due to malaria in the country64. Deviation of genotype distribution has been attributed to several factors, and selection pressure remains one of the important causes65. The studied population is endemic to various infectious diseases other than malaria and could be the reason for increased selection pressure on host genome66. Interestingly, two independent studies from South67 and North India68 have reported higher prevalence of heterozygotes (GA) compared to homozygous (GG or AA). They have also shown a deviation of TNF-α G-308A genotypes from HWE. These abnormalities in observations could be due to genotyping methods (ARMS PCR/sequence specific primer PCR) which could give spurious results.
Role of TNF-α promoter variants in SLE have been widely investigated. A recent meta-analysis including 41 published studies worldwide, showed association of minor allele (A) and AA genotype of TNF-α (G-308A) polymorphism with susceptibility to development of SLE11. In the present analysis, we observed higher prevalance of heterozygous (GA) and minor allele (A) in SLE patients compared to healthy females, suggesting a possible role of TNF-α (G-308A) polymorphism in susceptible to SLE. Similar observations have been reported in SLE patients from different geographical areas such as Brazil, Colombo, Mexico, North America, Spain, Taiwan11 and South India10. However, contradictory results have also been reported in Portugese, Thai, Chinese, Italian, African American, Japanese and Argentenian populations11. These discrepancies have been attributed to ethnicity of subjects enrolled for case-control studies and further supported by ethnicity related meta-analysis which revealed significant link between allele ‘A’ with predisposition to SLE in Europeans, Asians and South and North Americans but surprisingly not in African population11. In the present study, TNF-α (G-238A) heterozygous and minor allele (A) were also associated with susceptibility to SLE and it corroborated with other observations. Although the exact mechanism related to TNF-α polymorphism and SLE is yet to be understood, results of the present study and previous reports across the world indicates a strong association of TNF-α promoter variants, higher expression of TNF-α m-RNA and elevated levels of plasma TNF-α in SLE patients from malaria endemic regions.
We analysed the possible association of TNF-α polymorphisms with clinical manifestations of SLE, namely lupus nephritis which is one of the major clinical phenotypes linked to SLE mortality. We observed that heterozygous (GA) and minor allele ‘A’ of TNF-α (G-238A) polymorphism were significantly associated with lupus nephritis. These observations have been corroborated by a recent study on Chinese SLE patients11. However it contradicts an observation from South Indian population10 and a recent meta-analysis11. Furthermore, patients with lupus nephritis had higher levels of plasma TNF-α than those without nephritis.
Functional relevance of TNF-α promoter polymorphisms (G-308A and G-238A) have not been widely investigated. Minor allele for TNF-α (G-308A) polymorphism has been observed to enhance the binding of transcription factors and is associated with increase in mRNA production compared to major allele (G)69. In vitro stimulation of peripheral blood mononuclear cells (PBMC) derived from heterozygous subjects (GA) with lipopolysaccharide, displayed higher TNF-α than those of wild type individuals (GG)70. Furthermore, elevated plasma TNF-α has been associated with mutants for TNF-α (G-308A) polymorphism10. In the present study, we observed higher plasma levels of TNF-α in GA and AA genotypes compared to GG, corroborating earlier observations. Interestingly, other TNF-α promoter polymorphism (G-238A) also revealed similar results: mutants (GA and AA) were associated with higher plasma TNF-α than wild type (GG), which corroborates with an earlier report71. Furthermore, we performed in silico analysis which revealed regulatory effect in binding of transcription factors and enhanced expression of TNF-α gene. Results of the present investigation and earlier reports demonstrate signifcant regulatory role of promoter polymorphisms.
Investigations on possible link between malaria and SLE are limited and contradictory. Epidemiological data have shown lower prevalence of autoimmune diseases in areas where malaria incidence is high72. However, in an earlier observation, we have demonstrated protection against severe malaria and malarial death in complement receptor 1 variants and concluded a possible reason for higher prevalence of CR1 mutants in malaria endemic areas14. We had also observed that CR-1 mutants are susceptible to development of SLE and lupus nephritis since they expressed lower surface CR1 which affects clearance of apoptotic debris73. Furthermore, similar association of FcγRIIb variant (codon 232) with susceptibility to SLE and protection against P. falciparum malaria has been reported74. Lower parasitaemia and minimal clinical severity has been reported in FcγRIIb deficient mice when infected with non-lethal murine plasmodium strain indicating protective nature of the truncated or deficient FcγRIIb against malaria74. This observation has been further supported by higher prevalence of FcγRIIb codon 232 mutant in African and Asian population when compared to other populations across the world where malaria is endemic. The results of the present study and earlier reports collectively demonstrate that certain genotypes are beneficial in protecting humans against P. falciparum malaria and are highly prevalent in malaria endemic areas. Unfortunately, subjects genetically susceptible to SLE and residing in malarial endemic areas have a greater chance to develop SLE compared to those residing in non-endemic areas.
In conclusion, elevated plasma TNF-α is observed in SLE patients and associated with clinical severity. Furthermore, promoter variants of TNF-α gene, associated with higher TNF-α expression, were more prevalent in SLE patients. TNF-α is essential for clearance of malarial parasites49 and people residing in malarial endemic areas often produce optimal levels of TNF-α19 which could be helpful in combating the infection. It could also be one of the contributory factors for inducing SLE in genetically susceptible individuals. Further studies from other malarial endemic areas in the world are important to validate our findings.
Materials and Methods
Subjects
Gender wise analysis has been recommended in numerous earlier reports of genetic association studies73,75,76. SLE is a chronic inflammatory autoimmune disorder and mostly prevalent in females77. In the present study, we enrolled 428 female subjects (224 healthy controls and 204 SLE patients) to investigate possible association of TNF-α polymorphism in SLE. Patients of SLE were diagnosed based on the revised American College of Rheumatology (ACR) classification criteria78 and analyzed based on various clinical manifestations (Table 1). In addition, we enrolled 314 P. falciparum infected patients who reported to or were admitted to Department of Medicine, SCB Medical College, Cuttack, Odisha. Clinical categorization of falciparum infected patients were performed as described earlier14,15,73,79. Healthy females, age matched and residing in the same geographical areas, with no prior history of autoimmune disorders were enrolled as controls (HC). About 5 ml blood was collected from each participant. Plasma was separated and stored at −80 degrees centigrade for later use. The study was approved by the Institutional Human Ethics Committee of Central University of Jharkhand, India and S.C.B. Medical College Cuttack, Odisha, India. Informed written consent was obtained from each patient. The study was conducted in accordance with methods approved by IHECs.
DNA isolation and genotyping of TNF-α (G-238A and G-308A) polymorphisms
Whole genomic DNA was purified from blood samples using Gen Elute Blood Genomic DNA mini Kit (Sigma-Aldrich) according to manufacturer’s instructions. TNF-α promoter polymorphisms (G-238A & G-308A) were genotyped by polymerase chain reaction followed by restriction fragment length polymorphism method as described earlier (Galbraith et al. 1998).
TNF-α quantification
The plasma TNF-α levels in SLE patients, healthy controls and P. falciparum infected cases were quantified by enzyme linked immunosorbent assay (ELISA) kit (eBiosciences) according to manufacturer’s instructions.
Non-coding SNP functional analysis
In order to recognize the effect of SNPs in non-coding regions, tools predicting probable functional effect of SNPs at transcription factor binding sites (TFBS), Intron/exon border consensus sequences (splice sites), Exonic splicing enhancers (ESEs), and miRNA binding were utilized. SNPinfo (FuncPred) and RegulomeDB offer a pool of functional information using series of tools. The SNPs functionality was defined by SNPinfo (FuncPred) (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.php) web server80, which helps in selecting SNPs for genetic association studies. For the current study, two SNPs (rs1800629 and rs361525) were uploaded for batch analysis with the default settings. The output information was a list of SNPs with possible functional effect.
To supplement SNP ranking, SNPs were further analyzed by RegulomeDB (http://regulomedb.org/)81. RegulomeDB categorizes variants into six categories ranging from 1 to 6, where category 1 variants are ‘likely to affect binding and linked to expression of a gene target’, category 2 variants are ‘likely to affect binding’, Category 3 variants are ‘less likely to affect binding’, and Category 4, 5 and 6 variants have ‘minimal binding evidence’. RegulomeDB also allocates a score of 7 for variants with no annotation data available. dbSNP rsIDs were utilized as input for the current study.
Statistical analysis
Genotype and allele distribution among different clinical categories was compared by Fisher’s exact test. P value less than 0.02 was taken as significant (Bonferroni correction for two SNPs 0.05/2 = 0.02). The mean plasma levels of TNF-α in SLE patients and healthy controls was compared by student’s t test and analysis of variance (ANOVA) was employed for study difference in plasma TNF-α in different clinical categories of P. falciparum malaria. The association of TNF-α (G-238A and G-308A) genotypes with plasma TNF-α levels were analyzed by unpaired ‘t’ test or ANOVA followed by an appropriate post-test. Graph Pad Prism 5.01 software was used for these statistical analyses. Haplotype analysis was performed by SNAP Stats online tool. Resampling analysis was performed by bootstrap method in Microsoft excel sheet attached as supplementary file-1.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Kalliolias, G. D. & Ivashkiv, L. B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nature reviews. Rheumatology 12, 49–62, https://doi.org/10.1038/nrrheum.2015.169 (2016).
Black, R. A. Tumor necrosis factor-alpha converting enzyme. The international journal of biochemistry & cell biology 34, 1–5 (2002).
Sedger, L. M. & McDermott, M. F. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants - past, present and future. Cytokine & growth factor reviews 25, 453–472, https://doi.org/10.1016/j.cytogfr.2014.07.016 (2014).
Waters, J. P., Pober, J. S. & Bradley, J. R. Tumour necrosis factor in infectious disease. The Journal of pathology 230, 132–147, https://doi.org/10.1002/path.4187 (2013).
Johnston, B. & Conly, J. Tumour necrosis factor inhibitors and infection: What is there to know for infectious diseases physicians? The Canadian journal of infectious diseases & medical microbiology = Journal canadien des maladies infectieuses et de la microbiologie medicale 17, 209–212 (2006).
Aringer, M. & Smolen, J. S. The role of tumor necrosis factor-alpha in systemic lupus erythematosus. Arthritis research & therapy 10, 202, doi:10.1186/ar2341 (2008).
Brennan, F. M. & McInnes, I. B. Evidence that cytokines play a role in rheumatoid arthritis. The Journal of clinical investigation 118, 3537–3545, https://doi.org/10.1172/JCI36389 (2008).
Shakhov, A. N., Kuprash, D. V., Azizov, M. M., Jongeneel, C. V. & Nedospasov, S. A. Structural analysis of the rabbit TNF locus, containing the genes encoding TNF-beta (lymphotoxin) and TNF-alpha (tumor necrosis factor). Gene 95, 215–221 (1990).
Bayley, J. P., Ottenhoff, T. H. & Verweij, C. L. Is there a future for TNF promoter polymorphisms? Genes and immunity 5, 315–329, https://doi.org/10.1038/sj.gene.6364055 (2004).
Umare, V. D. et al. Impact of TNF-alpha and LTalpha gene polymorphisms on genetic susceptibility in Indian SLE patients. Human immunology 78, 201–208, https://doi.org/10.1016/j.humimm.2016.11.002 (2017).
Yang, Z. C., Xu, F., Tang, M. & Xiong, X. Association Between TNF-alpha Promoter -308 A/G Polymorphism and Systemic Lupus Erythematosus Susceptibility: A Case-Control Study and Meta-Analysis. Scandinavian journal of immunology 85, 197–210, https://doi.org/10.1111/sji.12516 (2017).
Kwiatkowski, D. P. How malaria has affected the human genome and what human genetics can teach us about malaria. American journal of human genetics 77, 171–192, https://doi.org/10.1086/432519 (2005).
Das, B. K. & Panda, A. K. MBL-2 polymorphisms (codon 54 and Y-221X) and low MBL levels are associated with susceptibility to multi organ dysfunction in P. falciparum malaria in Odisha, India. Frontiers in microbiology 6, 778, https://doi.org/10.3389/fmicb.2015.00778 (2015).
Panda, A. K. et al. Complement receptor 1 variants confer protection from severe malaria in Odisha, India. PloS one 7, e49420, https://doi.org/10.1371/journal.pone.0049420PONE-D-12-17272 (2012).
Panda, A. K. et al. Association of ABO blood group with severe falciparum malaria in adults: case control study and meta-analysis. Malaria journal 10, 309, https://doi.org/10.1186/1475-2875-10-309 (2011).
Gimenez, F., Barraud de Lagerie, S., Fernandez, C., Pino, P. & Mazier, D. Tumor necrosis factor alpha in the pathogenesis of cerebral malaria. Cellular and molecular life sciences: CMLS 60, 1623–1635, https://doi.org/10.1007/s00018-003-2347-x (2003).
Cruz, L. N., Wu, Y., Ulrich, H., Craig, A. G. & Garcia, C. R. Tumor necrosis factor reduces Plasmodium falciparum growth and activates calcium signaling in human malaria parasites. Biochim Biophys Acta 1860, 1489–1497, https://doi.org/10.1016/j.bbagen.2016.04.003 (2016).
Kwiatkowski, D. Tumour necrosis factor, fever and fatality in falciparum malaria. Immunology letters 25, 213–216 (1990).
Perera, M. K. et al. Association of high plasma TNF-alpha levels and TNF-alpha/IL-10 ratios with TNF2 allele in severe P. falciparum malaria patients in Sri Lanka. Pathogens and global health 107, 21–29, https://doi.org/10.1179/2047773212Y.0000000069 (2013).
Singh, S., Singh, N. & Handa, R. Tumor necrosis factor-alpha in patients with malaria. Indian journal of malariology 37, 27–33 (2000).
Basu, M. et al. Genetic association of Toll-like-receptor 4 and tumor necrosis factor-alpha polymorphisms with Plasmodium falciparum blood infection levels. Infect Genet l. 10, 686–696, https://doi.org/10.1016/j.meegid.2010.03.008 (2010).
Nguyen, T. N. et al. Association of a functional TNF variant with Plasmodium falciparum parasitaemia in a congolese population. Genes and immunity. 18, 152–157, https://doi.org/10.1038/gene.2017.13 (2017).
Ojurongbe, O. et al. Genetic variants of tumor necrosis factor-alpha -308G/A (rs1800629) but not Toll-interacting proteins or vitamin D receptor genes enhances susceptibility and severity of malaria infection. Immunogenetics. 70, 135–140, https://doi.org/10.1007/s00251-017-1032-4 (2018).
Olaniyan, S. A. et al. Tumour necrosis factor alpha promoter polymorphism, TNF-238 is associated with severe clinical outcome of falciparum malaria in Ibadan southwest Nigeria. Acta tropica. 161, 62–67, https://doi.org/10.1016/j.actatropica.2016.05.006 (2016).
Dunstan, S. J. et al. Variation in human genes encoding adhesion and proinflammatory molecules are associated with severe malaria in the Vietnamese. Genes and immunity. 13, 503–508, https://doi.org/10.1038/gene.2012.25 (2012).
Tsokos, G. C. Systemic lupus erythematosus. The New England journal of medicine. 365, 2110–2121, https://doi.org/10.1056/NEJMra1100359 (2011).
Jaryal, A. & Vikrant, S. Current status of lupus nephritis. Indian J Med Res. 145, 167–178, https://doi.org/10.4103/ijmr.IJMR_163_16 (2017).
Almaani, S., Meara, A. & Rovin, B. H. Update on Lupus Nephritis. Clin J Am Soc Nephrol. 12, 825–835, https://doi.org/10.2215/CJN.05780616 (2017).
Yap, D. Y. & Chan, T. M. Lupus Nephritis in Asia: Clinical Features and Management. Kidney Dis (Basel). 1, 100–109, https://doi.org/10.1159/000430458 (2015).
Crow, M. K. T. I interferon in the pathogenesis of lupus. Journal of immunology. 192, 5459–5468, https://doi.org/10.4049/jimmunol.1002795 (2014).
Iwata, Y., Furuichi, K., Kaneko, S. & Wada, T. The role of cytokine in the lupus nephritis. Journal of biomedicine & biotechnology. 2011, 594809, https://doi.org/10.1155/2011/594809 (2011).
Yokoyama, H., Kreft, B. & Kelley, V. R. Biphasic increase in circulating and renal TNF-alpha in MRL-lpr mice with differing regulatory mechanisms. Kidney international. 47, 122–130 (1995).
Lichtnekert, J. et al. Activated protein C attenuates systemic lupus erythematosus and lupus nephritis in MRL-Fas(lpr) mice. Journal of immunology. 187, 3413–3421, https://doi.org/10.4049/jimmunol.1101125 (2011).
Gabay, C. et al. Circulating levels of tumor necrosis factor soluble receptors in systemic lupus erythematosus are significantly higher than in other rheumatic diseases and correlate with disease activity. The Journal of rheumatology. 24, 303–308 (1997).
Sabry, A. et al. Proinflammatory cytokines (TNF-alpha and IL-6) in Egyptian patients with SLE: its correlation with disease activity. Cytokine 35, 148–153, https://doi.org/10.1016/j.cyto.2006.07.023 (2006).
Studnicka-Benke, A., Steiner, G., Petera, P. & Smolen, J. S. Tumour necrosis factor alpha and its soluble receptors parallel clinical disease and autoimmune activity in systemic lupus erythematosus. British journal of rheumatology. 35, 1067–1074 (1996).
Weckerle, C. E. et al. Large-scale analysis of tumor necrosis factor alpha levels in systemic lupus erythematosus. Arthritis and rheumatism. 64, 2947–2952, https://doi.org/10.1002/art.34483 (2012).
Yamamoto, K. & Loskutoff, D. J. Expression of transforming growth factor-beta and tumor necrosis factor-alpha in the plasma and tissues of mice with lupus nephritis. Lab Invest. 80, 1561–1570 (2000).
Herrera-Esparza, R., Barbosa-Cisneros, O., Villalobos-Hurtado, R. & Avalos-Diaz, E. Renal expression of IL-6 and TNFalpha genes in lupus nephritis. Lupus. 7, 154–158, https://doi.org/10.1191/096120398678919949 (1998).
Shao, W. H. & Cohen, P. L. Disturbances of apoptotic cell clearance in systemic lupus erythematosus. Arthritis research & therapy. 13, 202, https://doi.org/10.1186/ar3206 (2011).
Munoz, L. E., Lauber, K., Schiller, M., Manfredi, A. A. & Herrmann, M. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nature reviews. Rheumatology. 6, 280–289, https://doi.org/10.1038/nrrheum.2010.46 (2010).
Rath, P. C. & Aggarwal, B. B. TNF-induced signaling in apoptosis. Journal of clinical immunology. 19, 350–364 (1999).
Liu, H. et al. TNF-alpha-induced apoptosis of macrophages following inhibition of NF-kappa B: a central role for disruption of mitochondria. Journal of immunology. 172, 1907–1915 (2004).
Aggarwal, S., Gollapudi, S. & Gupta, S. Increased TNF-alpha-induced apoptosis in lymphocytes from aged humans: changes in TNF-alpha receptor expression and activation of caspases. Journal of immunology. 162, 2154–2161 (1999).
Zhu, L. J., Yang, X. & Yu, X. Q. Anti-TNF-alpha therapies in systemic lupus erythematosus. Journal of biomedicine & biotechnology. 2010, 465898, https://doi.org/10.1155/2010/465898 (2010).
Almoallim, H., Al-Ghamdi, Y., Almaghrabi, H. & Alyasi, O. Anti-Tumor Necrosis Factor-alpha Induced Systemic Lupus Erythematosus(). Open Rheumatol J6, 315–319, https://doi.org/10.2174/1874312901206010315 (2012).
de Kossodo, S. & Grau, G. E. Profiles of cytokine production in relation with susceptibility to cerebral malaria. Journal of immunology. 151, 4811–4820 (1993).
de Kossodo, S. & Grau, G. E. Role of cytokines and adhesion molecules in malaria immunopathology. Stem Cells. 11, 41–48, https://doi.org/10.1002/stem.5530110108 (1993).
Clark, I. A. & Cowden, W. B. Roles of TNF in malaria and other parasitic infections. Immunol Ser. 56, 365–407 (1992).
Angulo, I. & Fresno, M. Cytokines in the pathogenesis of and protection against malaria. Clin Diagn Lab Immunol. 9, 1145–1152 (2002).
Iriemenam, N. C. et al. Cytokine profiles and antibody responses to Plasmodium falciparum malaria infection in individuals living in Ibadan, southwest Nigeria. Afr Health Sci. 9, 66–74 (2009).
Xu, Y. et al. Mechanisms of tumor necrosis factor alpha antagonist-induced lupus in a murine model. Arthritis Rheumatol. 67, 225–237, https://doi.org/10.1002/art.38882 (2015).
Rupasree, Y., Naushad, S. M., Rajasekhar, L., Uma, A. & Kutala, V. K. Association of TLR4 (D299G, T399I), TLR9 −1486T>C, TIRAP S180L and TNF-alpha promoter (−1031, −863, −857) polymorphisms with risk for systemic lupus erythematosus among South Indians. Lupus 24, 50–57, https://doi.org/10.1177/0961203314549792 (2015).
Katkam, S. K. et al. Association of CTLA4 exon-1 polymorphism with the tumor necrosis factor-alpha in the risk of systemic lupus erythematosus among South Indians. Human immunology. 77, 158–164, https://doi.org/10.1016/j.humimm.2015.11.002 (2016).
Postal, M. & Appenzeller, S. The role of Tumor Necrosis Factor-alpha (TNF-alpha) in the pathogenesis of systemic lupus erythematosus. Cytokine. 56, 537–543, https://doi.org/10.1016/j.cyto.2011.08.026 (2011).
Hirankarn, N., Avihingsanon, Y. & Wongpiyabovorn, J. Genetic susceptibility to SLE is associated with TNF-alpha gene polymorphism -863, but not -308 and -238, in Thai population. International journal of immunogenetics. 34, 425–430, https://doi.org/10.1111/j.1744-313X.2007.00715.x (2007).
Sullivan, K. E., Wooten, C., Schmeckpeper, B. J., Goldman, D. & Petri, M. A. A promoter polymorphism of tumor necrosis factor alpha associated with systemic lupus erythematosus in African-Americans. Arthritis and rheumatism. 40, 2207–2211, doi:10.1002/1529-0131(199712)40:12<2207::AID-ART14>3.0.CO;2-Y (1997).
Parks, C. G. et al. Genetic polymorphisms in tumor necrosis factor (TNF)-alpha and TNF-beta in a population-based study of systemic lupus erythematosus: associations and interaction with the interleukin-1alpha-889 C/T polymorphism. Human immunology. 65, 622–631, https://doi.org/10.1016/j.humimm.2004.03.001 (2004).
Das, A. et al. Malaria in India: the center for the study of complex malaria in India. Acta tropica. 121, 267–273, https://doi.org/10.1016/j.actatropica.2011.11.008 (2012).
Banday, M. Z. et al. Tumor necrosis factor-alpha (TNF-alpha)-308G/A promoter polymorphism in colorectal cancer in ethnic Kashmiri population - A case control study in a detailed perspective. Meta gene. 9, 128–136, https://doi.org/10.1016/j.mgene.2016.06.001 (2016).
Rajesh, D., Gurumurthy, R., Kutty, A. V. & Balakrishna, S. Tumor necrosis factor alpha gene promoter -238G/A polymorphism increases the risk of psoriasis vulgaris in Indian patients. Internationa l journal of dermatology. 56, 307–311, doi:10.1111/ijd.13482 (2017).
Yadav, D. K. et al. Association of TNF-alpha −308G > A and TNF-beta +252A > G genes polymorphisms with primary immune thrombocytopenia: a North Indian study. Blood coagulation & fibrinolysis: an international journal in haemostasis and thrombosis. 27, 791–796, https://doi.org/10.1097/MBC.0000000000000492 (2016).
Sikka, R. et al. TNF-alpha (g.-308 G > A) and ADIPOQ (g. + 45 T > G) gene polymorphisms in type 2 diabetes and microvascular complications in the region of Punjab (North-West India). Current eye research 39, 1042–1051, https://doi.org/10.3109/02713683.2014.892998 (2014).
Dhingra, N. et al. Adult and child malaria mortality in India: a nationally representative mortality survey. Lancet. 376, 1768–1774, https://doi.org/10.1016/S0140-6736(10)60831-8 (2010).
Hosking, L. et al. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. European journal of human genetics: EJHG. 12, 395–399, https://doi.org/10.1038/sj.ejhg.52011645201164 (2004).
Fumagalli, M. et al. Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. The Journal of experimental medicine. 206, 1395–1408, doi:jem.2008277910.1084/jem.20082779 (2009).
Peddireddy, V. et al. Association of TNFalpha-308, IFNgamma+874, and IL10-1082 gene polymorphisms and the risk of non-small cell lung cancer in the population of the South Indian state of Telangana. International journal of clinical oncology. 21, 843–852, https://doi.org/10.1007/s10147-016-0972-2 (2016).
Dar, S. A. et al. Tumor necrosis factor (TNF)-alpha -308G/A (rs1800629) polymorphism distribution in North India and its association with pemphigus: Case-control study and meta-analysis. Autoimmunity 49, 179–187, https://doi.org/10.3109/08916934.2015.1134512 (2016).
Karimi, M., Goldie, L. C., Cruickshank, M. N., Moses, E. K. & Abraham, L. J. A critical assessment of the factors affecting reporter gene assays for promoter SNP function: a reassessment of -308 TNF polymorphism function using a novel integrated reporter system. European journal of human genetics: EJHG. 17, 1454–1462, https://doi.org/10.1038/ejhg.2009.80 (2009).
Das, S. N., Baniasadi, V. & Kapuria, V. Association of -308 TNF-alpha promoter polymorphism with type 1 diabetes in North Indians. International journal of immunogenetics. 33, 411–416, doi:10.1111/j.1744-313X.2006.00632.x (2006).
Dutta, D. et al. Tumor necrosis factor alpha -238G/A (rs 361525) gene polymorphism predicts progression to type-2 diabetes in an Eastern Indian population with prediabetes. Diabetes research and clinical practice. 99, e37–41, https://doi.org/10.1016/j.diabres.2012.12.007 (2013).
Greenwood, B. M. Autoimmune disease and parasitic infections in Nigerians. Lancet. 2, 380–382 (1968).
Panda, A. K., Ravindran, B. & Das, B. K. CR1 exon variants are associated with lowered CR1 expression and increased susceptibility to SLE in a Plasmodium falciparum endemic population. Lupus science & medicine 3, e000145, https://doi.org/10.1136/lupus-2016-000145 (2016).
Clatworthy, M. R. et al. Systemic lupus erythematosus-associated defects in the inhibitory receptor FcgammaRIIb reduce susceptibility to malaria. Proceedings of the National Academy of Sciences of the United States of America. 104, 7169–7174, https://doi.org/10.1073/pnas.0608889104 (2007).
Yuan, A. et al. Effect of SOX10 gene polymorphism on early onset schizophrenia in Chinese Han population. Neuroscience letters. 521, 93–97, https://doi.org/10.1016/j.neulet.2012.05.040 (2012).
Panda, A. K. et al. Low producer MBL genotypes are associated with susceptibility to systemic lupus erythematosus in Odisha, India. Human immunology. 74, 114–119, https://doi.org/10.1016/j.humimm.2012.09.003 (2013).
Danchenko, N., Satia, J. A. & Anthony, M. S. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus 15, 308–318 (2006).
Hochberg, M. C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 40, 1725, doi: 10.1002/1529-0131(199709)40:9<1725::AID-ART29>3.0.CO;2-Y (1997).
Pattanaik, S. S., Tripathy, R., Panda, A. K., Sahu, A. N. & Das, B. K. Bacteraemia in adult patients presenting with malaria in India. Acta tropica. 123, 136–138, doi:S0001-706X(12)00166-010.1016/j.actatropica.2012.04.001 (2012).
Xu, Z. & Taylor, J. A. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic acids research. 37, W600–605, https://doi.org/10.1093/nar/gkp290 (2009).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 22, 1790–1797, https://doi.org/10.1101/gr.137323.112 (2012).
Acknowledgements
We would like to thank all patients and controls included in this study. This work was supported by DST-INSPIRE faculty grant (IFA12/LSBM-46) to AKP from the Department of Science and Technology, New Delhi. Centre for Life Sciences is supported by DBT-BUILDER Program (No. BT/PR9028/INF/22/193/2013).
Author information
Authors and Affiliations
Contributions
H.M., B.K.P., M.S., D.D. and A.K.S. performed genotyping and analysis of data. B.R.M. performed in silico analysis, H.M. wrote first draft of the manuscript. B.K.D. and R.T. enrolled patients, clinical categorization and maintain clinical data of patients. A.K.P., R.T. and B.K.D. designed, work supervised and interpret and finalized the paper.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahto, H., Tripathy, R., Meher, B.R. et al. TNF-α promoter polymorphisms (G-238A and G-308A) are associated with susceptibility to Systemic Lupus Erythematosus (SLE) and P. falciparum malaria: a study in malaria endemic area. Sci Rep 9, 11752 (2019). https://doi.org/10.1038/s41598-019-48182-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48182-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.