Abstract
The California poppy (Eschscholzia californica) is renowned for its brilliant golden-orange flowers, though white petal variants have been described. By whole-transcriptome sequencing, we have discovered in multiple white petal varieties a single deletion leading to altered splicing and C-terminal truncation of phytoene synthase (PSY), a key enzyme in carotenoid biosynthesis. Our findings underscore the diverse roles of phytoene synthase in shaping horticultural traits, and resolve a longstanding mystery of the regaled golden poppy.
Similar content being viewed by others
Introduction
The California poppy (Eschscholzia californica), also known as the golden poppy, is native to the West Coast of the United States1,2. The flowers are brilliant golden-orange, instantly recognizable, and widely drawn and photographed. Native Americans valued the golden poppy as a food source. First catalogued from a Russian seafaring expedition to the San Francisco Bay in the early1800s, the golden poppy was designated the state flower of California in 1903. The golden poppy has since been inextricably linked to California pop culture, even eulogized by the novelist John Steinbeck2.
The golden-orange color results from carotenoid pigments3. The carotenoid biosynthetic pathway in plants has been well characterized4,5. The first committed step is the condensation of two geranylgeranyl diphosphate (GGPP) molecules to phytoene (colorless), catalyzed by phytoene synthase (PSY) (Fig. 1a, left). Subsequent enzymatic steps that include desaturation, isomerization, cyclization, hydroxylation and epoxidation sequentially generate carotenoids that appear red (lycopene), orange (α-carotene and β-carotene), and yellow (lutein, zeaxanthin, antheraxanthin, and violaxanthin), and combinations of these pigments create the observed palette. Notably, California poppy petals also contain abundant retro-carotenoids (retro-carotene triol and Eschscholzxanthin), generated from antheraxanthin and violaxanthin by as yet unknown enzymes6,7. Additional proteins have been reported to modulate carotenoid biosynthesis or degradation5,8. Carotenoids serve not only as chromoplast pigments to attract pollinators and horticulturalists, but also as chloroplast accessory pigments and antioxidants crucial for photosynthesis4.
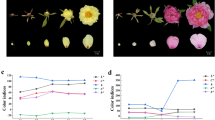
White-petal California poppy varieties show reduced flower PSY transcript. (a) Left, Carotenoid biosynthetic pathway, with pigment colors approximated by colored text. Abbreviations: GGPP, geranylgeranyl diphosphate; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCYE, lycopene ε-cyclase; LCYB, lycopene β-cyclase; CHYB, carotene β-hydroxylase; ZEP, zeaxanthin epoxidase. Right, heatmap depicts flower bud transcript levels of the major carotenoid biosynthetic pathway genes, normalized to housekeeping gene EIF4A2. Note, only phytoene synthase (PSY) transcript levels are significantly altered in white-petal varieties, which show on average 2.5-fold reduction (P = 0.003). (b) Representative California poppy varieties studied (clockwise from top left): single white poppy in a field of golden poppies; Orange; California Golden; Golden West; White; Alba; White Linen; and Ivory Castle.
For those living in or visiting California, it is not uncommon to spot the occasional white-petal California poppy in a field of orange poppies. Indeed, white-petal varieties were described from English garden hybrids as early as the 1880s9, and in scientific literature from the 1930s10. Biochemical and genetic studies ensued, defining the white-petal trait to be recessive and based on near absence of carotenoid pigment11. By crossing different white-petal variants, including 8 originating from natural populations and 7 from commercial sources, Barrell et al.12 reported lack of complementation indicative of a single genetic locus. However, the gene and mutation(s) underlying white-petal variants have yet to be discovered.
Results and Discussion
To investigate the genetic basis of white-petal poppy variants, we carried out transcriptome sequencing (RNAseq) of developing flower buds from four different commercial white poppy varieties displaying varied shades of white: Ivory Castle, White Linen, Alba, and White (Fig. 1b). Three orange-petal poppy varieties (Orange, California Golden, and Golden West) served as controls. RNA was isolated from developing flower buds, where pigment production was presumed ongoing. Since no poppy reference genome was available, RNAseq reads were assembled de novo into transcript contigs, which were then annotated by homology to an orthologous reference transcriptome, for which we selected another eudicot clade flowering plant, the garden tomato (Solanum lycopersicum).
Since white poppy petals are deficient in carotenoid pigments11, we focused on genes of the carotenoid biosynthetic pathway. Comparing expression of carotenoid biosynthesis genes between white and orange poppy varieties, only phytoene synthase (PSY) showed significantly altered expression, with an average 2.5-fold reduced transcript levels in white varieties (P = 0.003, two-sided Student’s t-test) (Fig. 1a, right). While this finding focused attention on PSY, the modest reduction in white varieties was unlikely to account for a near absence of carotenoid pigment.
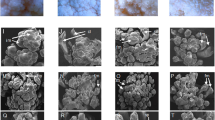
Comparing the aligned PSY transcript reads between white and orange petal poppy varieties, all four white varieties (but none of the three orange varieties) exhibited an apparent 5 bp gap within the PSY transcript (Fig. 2a,b and Supplementary Fig. 1). The gap occurred within the coding region, at the site of an inferred exon-exon junction (by comparison to the tomato reference genome). To define the alteration at the genome level, we designed PCR primers to amplify and sequence across the exon-exon junction from genomic DNA (isolated from poppy seeds). In the white petal varieties, the resultant PCR product was consistently smaller (Fig. 2c). Sequence alignment revealed a 76 bp deletion within the PSY intron, which extended through the 3′ splice acceptor site and 5 bp of the downstream exon (explaining the apparent 5 bp alignment gap from the RNAseq reads) (Figs 2d, 3). By comparing the white-petal PSY genome sequence and assembled transcript contig, loss of the splice acceptor site led to usage of a cryptic splice acceptor site within the intron, resulting in a coding frameshift with early translational termination and predicted C-terminal truncation of the PSY protein (Fig. 2d and Fig. 3). Early translational termination is associated with nonsense-mediated mRNA decay13, consistent with our observed reduced PSY transcript levels. Notably, the C-terminal truncation abolishes a highly-conserved putative enzyme active site (DXXXD motif) in PSY (Fig. 2b,d)14.
White-petal California poppy varieties harbor a frameshifting deletion in PSY. (a) Integrative Genomics Viewer (IGV) coverage plots and alignments for RNAseq reads spanning the PSY coding sequencing, shown each for two representative orange-petal (Orange and California Golden) and white-petal (Ivory Castle and White Linen) varieties. Mismatches (polymorphisms or mutations) relative to the reference (Orange) are indicated by color-coded bars. Note the alignment gap in the two white-petal varieties (black arrow). (b) Close-up view highlighting the 5 bp alignment gap in the white-petal varieties. Note, the alignment gap overlaps with a putative enzyme active site (DXXXD motif, indicated below). (c) PCR across the exon-exon junction (site of alignment gap) results in a shorter PCR product (red arrow), indicative of genomic DNA deletion in the white-petal varieties. Image of full-length gel is available in Supplementary Fig. 4. (d) Illustration summarizing the PSY gene, mRNA and protein products inferred from sequence alignment of the PCR products (PCR primers indicated; see Fig. 3 for annotated cDNA sequences). In all four white-petal varieties, a 76 bp intronic deletion (red hatched box) ablates the 3′ splice acceptor. Usage of a cryptic splice acceptor leads to a coding frameshift with early translational termination. The resulting C-terminal truncation destroys one of two highly-conserved putative enzyme active sites (pink rectangles); brown lines indicate PSY conserved motifs. (e) PCR analysis (and subsequent sequencing) reveals the identical 76 bp intronic deletion in three white-petal poppy plants discovered among ostensibly wild California poppy fields from three geographically distinct locales. Note, a yellow-orange petal poppy plant discovered in Field 1 (also pictured in inset, white arrow) carries both the PSY wildtype and deletion allele, suggesting that it represents an F1 hybrid cross between an orange and white petal variety. Image of full-length gel is available in Supplementary Fig. 4.
Poppy PSY1 cDNA sequences. Shown are the flower bud PSY cDNA sequences (plus one intron) from Orange-petal (top) and White-petal (bottom) varieties, as determined from the RNAseq alignments and exon-spanning PCR. The intron sequences are in lower case text, with splice donor and acceptor sites underlined. The 76 bp sequence deleted in white varieties is indicated in the orange PSY cDNA by purple highlighting. The early termination codon (TGA) in the white PSY cDNA is indicated by red text.
That the PSY deletion is present in all four white petal varieties (minimally 8 alleles if diploid), but in none of the three orange petal varieties (minimally 6 alleles), demonstrates strong segregation with the white petal phenotype (P = 0.0003, two-sided Fisher’s exact test). Together, the genetic and inferred biochemical (predicted loss of active site) data provide strong evidence that the 76 bp deletion underlies the white petal trait. Moreover, that all four white petal poppy varieties (notwithstanding subtle differences in petal hues) harbor precisely the same deletion suggests that they were likely all derived from a single white-petal poppy origin.
In addition to studying commercial varieties, we also sought to examine white-petal specimens among wild poppy plants. To this end, we undertook expeditions to find and collect white petal specimens among California poppy fields across Santa Clara, San Mateo, and Solano counties. Of three specimens collected, all harbored precisely the same PSY mutation (Fig. 2e), suggesting that they likely represent commercial seed contaminants among orange petal varieties that were seeded rather than wild plants. Interestingly, in one field we noted orange and white poppies together with an uncommon yellow-orange petal variant. PCR analysis revealed that the yellow-orange poppy carried both the wildtype and deletion PSY allele, suggesting an F1 hybrid between previously seeded orange and white petal varieties (Fig. 2e).
Carotenoids are flower petal pigments, but they also provide essential roles as accessory pigments and antioxidants in chloroplasts for photosynthesis4. Thus, given the PSY null mutation identified from flower buds, the existence of other PSY encoding genes seemed likely. To investigate that possibility, we carried out RNAseq from green leaf material from orange and white petal poppy varieties. Aligning the reads, only a small fraction of the PSY reads from the white-petal leaf specimen exhibited the deletion (Fig. 4a). A distinct set of single nucleotide polymorphisms (SNPs) present only in the leaf RNA segregated with the non-deletion reads, allowing us to design haplotype-specific PCR primers to amplify across the exon-exon junction. Notably, PCR of genomic DNA using the non-deletion haplotype-specific primers revealed two larger PCR products (Fig. 4b), where sequencing disclosed two different intron sequences (Fig. 4c). This finding indicates the presence of two additional PSY genes (which we have designated PSY1B and PSY1C), expressed in poppy leaves.
Poppy leaves express additional PSY genes. (a) IGV coverage plots and alignments for RNAseq reads spanning a portion of the PSY coding sequencing (bp 885–975), shown for Orange flower, Orange leaf, Ivory Castle flower, and Ivory Castle leaf. Note that the 5 bp alignment gap present in Ivory Castle flower is observed in only a minority of reads from Ivory Castle leaf. Note also in Ivory Castle leaf the presence of 3 SNPs flanking the gap that segregate with the non-deletion reads, permitting design of haplotype-specific (leaf vs. flower expressed) PCR primers (shown below). The 3 SNPs are also present in Orange leaf, but masked by the higher (flower) PSY expression. (b) Haplotype-specific PCR of genomic DNA across the exon-exon junction (site of alignment gap) using leaf-expression specific PCR primers results in two longer PCR products (blue arrows), distinct from the shorter flower-expression specific PCR products, and indicative of additional PSY genes (annotated PSY1B and PSY1C) expressed in poppy leaves. Image of full-length gel is available in Supplementary Fig. 4. (c) Partial genome sequences for PSY1A (top), PSY1B (middle), and PSY1C (bottom), as determined by exon-spanning PCR. Intron sequences are in lower case text.
Based on the relative frequencies of SNPs in the PSY transcripts from petal and leaf tissue (Supplementary Fig. 2 and Supplementary Table 1), we can infer that PSY1A (harboring the mutation in white petal varieties) is the only PSY gene expressed in California poppy petals, while PSY1B and PSY1C (indistinguishable from one another by SNPs) are expressed only in leaves. Nonetheless, PSY1A is also expressed in leaves where indeed it is more abundantly expressed (accounting for 97% of leaf PSY transcripts in orange petal varieties, reduced to 78% in white-petal varieties) compared to PSY1B/1C. California poppy PSY1A exhibits high (99%) homology to PSY1B/1C at the nucleotide sequence level, and 100% identity at the amino acid sequence level, suggesting relatedness by recent gene duplication. However, we note limitations of our analysis, including variable read coverages (particular at the ends of the PSY genes), and the challenges of phasing short reads and assigning SNPs to individual genes. A definitive analysis will require cloning the individual PSY cDNAs and genome loci.
The finding of multiple PSY gene paralogs in plants, first detailed in the tomato15, is now common. Like for the California poppy, some such PSY paralogs are expressed primarily in green (photosynthetic) tissues, while others drive carotenoid accumulation in flowers, fruits, or roots. For example, in the tomato (Solanum lycopersicum), PSY1 is predominantly expressed in the petals and ripening fruit, while PSY2 is predominant in leaves16. In the loquat (Eriobotrya japonica), PSY1 is expressed in the fruit peel, PSY2A in the ripening fruit flesh, and PSY2B in leaves17. And in the carrot (Daucus carota), PSY1 and PSY2 are expressed in the root, while PSY1 is also expressed in leaves18. A comparative analysis of PSY protein sequences among eudicots reveals California poppy PSY to be most closely related to PSY from the recently sequenced opium poppy (Papaver somniferum) genome19, and overall more closely related to the so-called Eudicot PSY1 clade (Fig. 5 and Supplementary Fig. 3)20.
Phylogenetic analysis of PSY protein sequences. Phylogram depicts phylogenic relationship of California poppy PSY and 21 other eudicot PSY proteins. Branch lengths are in units of the number of amino acid substitutions per site. Bootstrap values (percentage of trees in which the associated taxa clustered together in 500 re-samplings) are indicated next to the branches. Clades are based on the Stauder et al. designation20. PSY genes specifically expressed in single carotenoid-rich organs are indicated by red text. Note, PSY gene nomenclature varies considerably by species.
In summary, by transcriptome sequencing of California poppy flower buds, we have identified a frameshifting deletion in phytoene synthase that is common to multiple commercial white petal varieties. All have distinct white hues (likely due to different genetic backgrounds), but nonetheless appear to have been bred from the same common originator. Importantly, the white-petal trait in 15 different natural and commercial California poppy variants was previously shown to map to a single genetic locus12. That study included the Alba and Ivory Castle varieties also analyzed here. Thus, we can infer that PSY1A mutations underlie all previously studied white-petal California poppy variants. Whether other variants share the same 76 bp frameshifting deletion mutation remains to be determined.
PSY variants/mutations have previously been associated with agriculturally important traits, e.g., color variation in tomatoes, peppers, cassavas, and loquats17,21,22,23. We have now also connected PSY mutations to ornamental horticulture. Our discovery resolves a decades old mystery of the molecular underpinnings of white-petal California poppies, and adds to the cultural legacy of the California golden poppy.
Methods
Plant materials
Commercial California poppy varieties were purchased as seeds from Eden Brothers (Ivory Castle, White Linen, Alba, Orange, and Golden West), Vermont Wildflowers (White), and Cornucopia (California Golden). Seeds were germinated in individual pots, and subsequent developing flower buds collected and frozen on dry ice. In some cases, poppy leaf material was also collected. Mature flowers from the same plants were examined and photographed to verify the advertised varieties. We also collected ostensibly wild California poppy flower samples from fields across three San Francisco Bay Area counties. For RNA isolation, plant material (flower buds with calyx caps removed, or leaves) was pulverized in liquid nitrogen using a mortar and pestle, and then RNA prepared using the RNeasy Mini kit (Qiagen). Genomic DNA was isolated from commercial seeds, using the Quick-DNA Plant/Seed Miniprep Kit (Zymo Research).
Transcriptome sequencing
For transcriptome sequencing, RNAseq libraries were generated from 1 µg RNA using Illumina TruSeq RNA Library Prep Kit v2, and barcoded libraries sequenced (101 bp × 2 for flower buds, 50 bp × 1 for leaves) on an Illumina HiSeq 2000 to an average depth of 27 million reads per sample. Reads were then assembled de novo into transcript contigs using Trinity24, implemented within FRAMA25, using the garden tomato (Solanum lycopersicum) transcriptome26 (Assembly SL2.50, accessed from EnsemblePlants) as an orthologous reference to assign gene annotations. Annotated transcripts were quantified as Reads Per Kilobase of transcript per Million mapped reads (RPKMs). Reported transcript levels for carotenoid biosynthetic pathway genes were normalized to the housekeeping gene EIF4A2. Aligned reads were visualized against the Orange (Eden Brothers) variety, using Integrative Genomics Viewer (IGV)27.
PCR analysis
PCR was done using AmpliTaq Gold polymerase and reagents (Applied Biosystems), with 100 ng input DNA and 40 cycles (94 °C 30 s, 54 °C 30 s, 72 °C 60 s). PCR/sequencing primers were PSY-Gap-F 5′-TCAAGCAACGACGGAGAGTA; PSY-Gap-R 5′-CCTTGCCTGCGAATATGTCT; PSY-Flower-F 5c-AAATCAACTCACAAATATTCTTAGAGATGTC; PSY-Flower-R 5′-GCCCTGCCTGTGCTAGC; PSY-Leaf-F 5′-ATCAACTCACAAACATTCTTAGAGATGTT; PSY-Leaf-R 5′-AAGCCCTGCCTGTGCTAGT. PCR products were purified with the QIAquick PCR Purification kit (Qiagen), and then Sanger-sequenced (Quintara Biosciences). Sequence reads were aligned using NCBI BLAST Align two sequences tool.
Phylogenetic analysis
Multiple sequence alignment of PSY proteins was done using Clustal Omega28, using default parameters and the following protein accessions: Arabidopsis thaliana PSY (AAA32836.1); Capsicum annuum (Pepper) PSY1 (ACE78189.1); Cucumis melo (Muskmelon) PSY1 (AEH03200.1), PSY3 (formerly PSY2) (AEH03199.1); Daucus carota (Carrot) PSY1 (ABB52067.1), PSY2 (ABB52068.1), PSY3 (XP_017217851.1); Eriobotrya japonica (Loquat) PSY1 (AIT18246.1), PSY2A (AIT18247.1), PSY2B (AIT18249.1), PSY3 (AIT18250.1); Manihot esculenta (Cassava) PSY1 (ACY42666.1), PSY2 (ACY42670.1); Medicago truncatula (Barrel clover) PSY1 (AES99105.1), PSY2A (KEH33671.1), PSY2B (AET00322.2), PSY3 (AES71870.1); Papaver somniferum (Opium poppy) PSY2 (XP_026387400.1); Solanum lycopersicum (Tomato) PSY1 (P08196.2), PSY2 (ABV68559.1), PSY3 (XP_004228928.1). Phylograms were constructed with MEGA X29, using the Neighbor-Joining method with default parameters.
Accession codes
RNAseq data are available through the NCBI Short Read Archive (accession PRJNA517727). PSY sequences are available through GenBank (accessions MK620867-MK620871).
References
McClintock, E. The California poppy - natural history. Pacific Horticulture 37, 3–5 (1976).
Lack, A. Poppy. 39-41 (Reaktion Books, 2016).
Strain, H. H. Eschscholtzxanthin: a new xanthophyll from the petals of the Californian poppy, Eschscholtzia californica. J Biol Chem 123, 425–437 (1938).
Bartley, G. E. & Scolnik, P. A. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7, 1027–1038 (1995).
Zhu, C. et al. The regulation of carotenoid pigmentation in flowers. Arch Biochem Biophys 504, 132–141 (2010).
Maoka, T. et al. A new retro-carotenoid from the petals of the Californian yellow poppy Eschscholtzia californica. J Nat Prod 63, 1288–1289 (2000).
Zhou, J., Hunter, D. A., Lewis, D. H., McManus, M. T. & Zhang, H. Insights into carotenoid accumulation using VIGS to block different steps of carotenoid biosynthesis in petals of California poppy. Plant Cell Rep 37, 1311–1323 (2018).
Yuan, H., Zhang, J., Nageswaran, D. & Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic Res 2, 15036 (2015).
Gould, R. The California poppy - garden hybrids. Pacific Horticulture 37, 6–8 (1976).
Beatty, A. V. Genetic studies on the California poppy. J Hered 27, 331–336 (1936).
Wakelin, A. M., Lister, C. E. & Conner, A. J. Inheritance and biochemistry of pollen pigmentation in California poppy (Eschscholzia californica Cham.). Int J Plant Sci 164, 867–875 (2003).
Barrell, P. J., Wakelin, A. M., Gatehouse, M. L., Lister, C. E. & Conner, A. J. Inheritance and epistasis of loci influencing carotenoid content in petal and pollen color variants of california Poppy (Eschscholzia californica Cham.). J Hered 101, 750–756 (2010).
Kervestin, S. & Jacobson, A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13, 700–712 (2012).
Shumskaya, M., Bradbury, L. M., Monaco, R. R. & Wurtzel, E. T. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell 24, 3725–3741 (2012).
Bartley, G. E. & Scolnik, P. A. cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J Biol Chem 268, 25718–25721 (1993).
Giorio, G., Stigliani, A. L. & D’Ambrosio, C. Phytoene synthase genes in tomato (Solanumlycopersicum L.) - new data on the structures, the deduced amino acid sequences and the expression patterns. Febs J 275, 527–535 (2008).
Fu, X. et al. Involvement of multiple phytoene synthase genes in tissue- and cultivar-specific accumulation of carotenoids in loquat. J Exp Bot 65, 4679–4689 (2014).
Wang, H., Ou, C. G., Zhuang, F. Y. & Ma, Z. G. The dual role of phytoene synthase genes in carotenogenesis in carrot roots and leaves. Mol Breed 34, 2065–2079 (2014).
Guo, L. et al. The opium poppy genome and morphinan production. Science 362, 343–347 (2018).
Stauder, R. et al. Strigolactone Levels in Dicot Roots Are Determined by an Ancestral Symbiosis-Regulated Clade of the PHYTOENE SYNTHASE Gene Family. Front Plant Sci 9, 255 (2018).
Fray, R. G. & Grierson, D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol 22, 589–602 (1993).
Huh, J. H. et al. A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp.). Theor Appl Genet 102, 524–530 (2001).
Welsch, R. et al. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22, 3348–3356 (2010).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–652 (2011).
Bens, M. et al. FRAMA: from RNA-seq data to annotated mRNA assemblies. BMC Genomics 17, 54 (2016).
TomatoGenomeConsortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192 (2013).
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res (2019).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35, 1547–1549 (2018).
Acknowledgements
We thank the Stanford Genome Sequencing Service Center for providing Illumina sequencing services, and Anna and James Pollack for assistance with spotting and collecting field specimens. This study was supported in part by Stanford Pathology Department funds to J.R.P.
Author information
Authors and Affiliations
Contributions
A.J.P. and J.R.P. conceived and planned the studies; A.J.P. performed experiments; A.J.P. and X.G. analyzed data; A.J.P. and J.R.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pollack, A.J., Gong, X. & Pollack, J.R. A common phytoene synthase mutation underlies white petal varieties of the California poppy. Sci Rep 9, 11615 (2019). https://doi.org/10.1038/s41598-019-48122-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-48122-3
This article is cited by
-
Effect of PI3K/AKT/mTOR signaling pathway-based clustered nursing care combined with papaverine injection on vascular inflammation and vascular crisis after replantation of severed fingers
Molecular and Cellular Biochemistry (2023)
-
Temperature regulation of carotenoid accumulation in the petals of sweet osmanthus via modulating expression of carotenoid biosynthesis and degradation genes
BMC Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.