Abstract
We describe a portable epigenetic switch based on opvAB, a Salmonella enterica operon that undergoes bistable expression under DNA methylation control. A DNA fragment containing the opvAB promoter and the opvAB upstream regulatory region confers bistability to heterologous genes, yielding OFF and ON subpopulations. Bistable expression under opvAB control is reproducible in Escherichia coli, showing that the opvAB switch can be functional in a heterologous host. Subpopulations of different sizes can be produced at will using engineered opvAB variants. Controlled formation of antibiotic-resistant and antibiotic-susceptible subpopulations may allow use of the opvAB switch in the study of bacterial heteroresistance to antibiotics.
Similar content being viewed by others
Introduction
Biosensors able to detect environmental signals are made of a sensor that detects a given input and a reader that responds to the input generating a detectable signal in a quantitative or semi-quantitative fashion1. Classical sensors employ enzymes or whole cells. Enzyme-based biosensors present the advantage of high selectivity but the need for purification can be a drawback due to technical difficulties and high cost. In contrast, whole-cell sensors are often easy to use and inexpensive, especially if microbial strains are used2. A common type of microbial biosensor is an engineered strain that responds to physical or chemical inputs generating electrochemical or optical signals. Sensors of this type often employ a promoter sensitive to a specific input and a reporter gene that produces a detectable signal1,3. The literature contains multiple examples of sensors that detect electrochemical and optical signals, and use of fluorescent proteins has become widespread in the last decade4.
An alternative to genetic circuits able to process information in living cells is the design of epigenetic switches. This approach has received special attention to develop diagnostic tests for human diseases5,6,7, while synthetic biology based on bacterial epigenetics remains largely unexplored. A relevant exception is the recent development of biosensors based on DNA adenine methylation using Escherichia coli as host8.
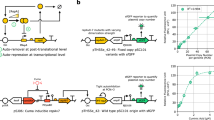
In this study, we describe the construction and application of an epigenetic switch that drives gene expression in a bistable fashion. Bistability generates bacterial subpopulations that differ in a specific phenotypic trait (e. g., antibiotic resistance) and have defined sizes. The switch is based on opvAB, a bacterial operon subjected to epigenetic control by DNA adenine methylation9,10,11. Transcription of opvAB is bistable, with concomitant formation of OpvABOFF and OpvABON cells9. Bistability is controlled by binding of the OxyR transcription factor to a regulatory region upstream of the opvAB promoter (Fig. 1A)10. This region contains four sites for OxyR binding and four GATC motifs. OpvABOFF and OpvABON cell lineages display alternative patterns of OxyR binding, which in turn cause alternative patterns of GATC methylation: in the OFF state, GATC2 and GATC4 are methylated; in the ON state, GATC1 and GACT3 are methylated10. Here, we show that a cassette of 689 nucleotides containing the opvAB promoter and the upstream regulatory region confers bistability to heterologous genes, and describe examples of opvAB-based constructs that produce bacterial subpopulations with distinct phenotypes. One of the examples involves formation of an antibiotic-resistant subpopulation upon cloning of an antibiotic resistance gene downstream of the opvAB promoter. This construct may provide an experimental system to study bacterial heteroresistance (HR) to antibiotics under highly controlled conditions12. HR is a phenotype where a bacterial isolate is characterized by the presence of a main susceptible population and a subpopulation with higher antibiotic resistance. Increasing evidence suggests that heteroresistance can lead to treatment failure12,13,14,15,16,17. Yet, little is known regarding the characteristics of the heteroresistance phenotypes (i.e. the size of the resistant subpopulation or its level of resistance) that are linked to treatment failure. Animal experiments, where infections are started with bacterial cultures that carry an antibiotic resistance gene under control of the opvAB switch, would allow control of the frequency of the resistant subpopulation and determination of how different ratios of resistant:susceptible bacteria influence treatment outcome17. Other potential uses of the opvAB switch in synthetic biology are discussed.
Formation of LacOFF and LacON subpopulations under opvAB control. (A) Diagram of the opvAB promoter and regulatory region, with the GATC sites outlined. (B) Diagram of the wild type opvAB operon and the PopvAB::lacZY construct. (C) Colonies formed on LB + X-gal by a S. enterica strain carrying the lacZY operon under the control of the wild type opvAB control region (SV9700, left), and by S. enterica strains carrying the lacZY operon under the control of mutant opvAB control regions (SV9701, PopvAB GATC1,2::lacZY::gfp, center; SV9702, PopvAB GATC-less::lacZY::gfp, right). (D) Flow cytometry analysis of PopvAB::lacZY expression in strains SV9700, SV9701 and SV9702. The sizes of LacON subpopulations are indicated. (E) Growth of strains SV9700, SV9701 and SV9702 in NCE-lactose. Error bars represent the standard error of the mean from 3 independent replicates. (F) Reversible formation of LacOFF and LacON subpopulations under opvAB control in strain SV9700.
Results
Bistable expression of lacZY under opvAB transcriptional control
The ability of the opvAB epigenetic switch to confer bistable expression to a heterologous locus was tested by engineering a strain that harbored the E. coli lacZY operon downstream of the opvAB promoter and its upstream regulatory region (PopvAB - Fig. 1A,B). To avoid cell-to-cell heterogeneity associated with variations in plasmid copy number, the construct was engineered in the S. enterica chromosome. Construction involved replacement of the opvAB coding region with a promoterless lacZY operon, leaving the opvAB promoter and upstream regulatory region intact. The construct harbored the opvA ribosome binding site (RBS). Plating of the engineered strain on LB containing X-gal yielded Lac+ (blue) and Lac− (white) colonies, thus revealing bistable expression (LacOFF or LacON) of the heterologous lacZY operon (Fig. 1C). Streaking of either Lac+ or Lac− colonies on X-gal agar yielded a mixture of Lac+ and Lac− colonies, thus indicating the occurrence of reversible bistability (“phase variation”) as previously described for the native opvAB locus9.
Calculation of phase variation frequencies indicated a frequency of 1.1 × 10−4 ± 0.3 per cell and generation for the OFF→ON transition, and 3.4 ± 0.1 × 10−2 per cell and generation for the ON→OFF transition. The 300-fold difference between switching rates was two-fold lower than in the native opvAB locus (OFF → ON, 6.1 ± 1.7 × 10−5; ON → OFF 3.7 ± 0.1 × 10−2; 600-fold difference between switching rates)9. The increased size of the LacON subpopulation may result from multiple factors including potential differences in mRNA stability and codon usage constraints.
Variants of the PopvAB::lacZY construct were engineered to further explore the ability of opvAB-driven transcription to confer bistable expression to a heterologous locus. One such variant involved the use of a mutant opvAB regulatory region lacking GATC sites 1 and 2 (GATC1,2), previously shown to increase the size of the OpvABON subpopulation10. As expected, a higher proportion of Lac+ colonies was detected (Fig. 1C). Another variant, used as control, lacked all opvAB GATC sites (GATC-less) and locked lacZY transcription in the ON state (Fig. 1C) as previously described for the native opvAB operon9.
Variants carrying a green fluorescent protein gene (gfp) dowstream of the lacZY operon were also engineered, and assessment of subpopulation sizes by flow cytometry confirmed that the LacON subpopulation formed by the wild type opvAB switch was smaller than that formed by the GATC1,2 variant (Fig. 1D). Furthermore, only cells in the LacON state were detected in the strain that harbored the GATC-less construct, and subpopulation formation was abolished as above (Fig. 1D).
The ability of the opvAB switch to permit selection of one of the subpopulations was examined by testing the ability of strains carrying PopvAB::lacZY::gfp and PopvAB GATC1,2::lacZY::gfp constructs to grow in minimal medium with lactose as sole carbon source. As above, a strain carrying the GATC-less PopvAB::lacZY::gfp construct was included as a control. Assessment of the growth patterns of these strains revealed that the time required for culture saturation was dependent on the size of the LacON subpopulation present at the start of the culture (Fig. 1E). Reversibility of the LacON state was confirmed by growth on NCE-glucose (Fig. 1F).
Bistable expression of the chimaeric opvAB::lacZY operon in a heterologous host, E. coli
The functionality of the opvAB switch in a heterologous host was tested in E. coli. For this purpose, the PopvAB::lacZY::gfp construct and its GATC1,2 and GATC-less variants were introduced into the chromosome of E. coli DR3 (ΔlacZY). Strains carrying the PopvAB::lacZY-gfp and PopvAB GATC1,2::lacZY::gfp constructs (DR22 and DR23, respectively) formed Lac+ and Lac– colonies on X-gal agar, and the number of Lac+ colonies was higher in the strain carrying the PopvAB GATC1,2::lacZY::gfp construct. The strain carrying the GATC-less construct (DR24) formed Lac+ colonies only (Fig. 2A). Flow cytometry assessment of GFP expression upon growth in LB confirmed the occurrence of subpopulations of LacOFF and LacON cells in the strains carrying the PopvAB::lacZY::gfp and PopvAB GATC1,2::lacZY::gfp constructs but not in the strain carrying the GATC-less construct (Fig. 2B). As above, growth pattern assessment revealed that the time required for culture saturation was dependent on the initial size of the LacON subpopulation (Fig. 2C). Altogether, these observations indicated that the opvAB switch is functional in E. coli.
Formation of LacOFF and LacON subpopulations under opvAB control in E. coli. (A) Left: Colonies formed on LB + X-gal by an E. coli strain carrying the lacZY operon under the control of the wild type opvAB control region (strain DR22). Center and right: Colonies formed on LB + X-gal by E. coli strains carrying the lacZY operon under the control of mutant opvAB control regions (DR23 and DR24). (B) Flow cytometry analysis of PopvAB::lacZY expression in strains DR22, DR23 and DR24. The sizes of LacON subpopulations are indicated. (C) Growth of strains DR22, DR23 and DR24 in NCE-lactose. Error bars represent the standard error of the mean from 3 independent replicates.
Bistable expression of antibiotic resistance genes under opvAB control
An additional test of the ability of the opvAB bistable switch to generate bacterial subpopulations was performed by cloning antibiotic resistance genes downstream of the opvAB promoter in the S. enterica chromosome. The antibiotic resistance genes chosen for these experiments were aac3-Ib (henceforth, aac3) and aac(6′)-Ib-cr (henceforth, aac6), which encode aminoglycoside acetyl transferases18, and blaCTX-M-15 (henceforth, ctxM), which encodes an extended-spectrum β-lactamase19. In these constructs, the native ribosome binding sites were replaced with a stronger RBS, named BI20 to adjust the sensitivity of the switch to a level that could permit unambiguous detection of the antibiotic resistance phenotype under study, thus facilitating discrimination between OFF and ON cells. Experiments with strains carrying PopvAB::aac6::gfp and PopvAB::ctxM::gfp fusions (strains SV9703 and SV9706, respectively) yielded bacterial subpopulations resistant to kanamycin and to cefotaxime, respectively (Fig. 3A). Controls using strains that constitutively expressed aac6 and ctxM (SV9705 and SV9707, respectively) showed that the concentrations of antibiotics used permitted growth (Fig. 3A). The wild type strain ATCC 14028 failed to grow under such conditions, confirming that the concentrations of antibiotics used were bactericidal.
(A) Growth of strains SV9703 (PopvAB::aac6::gfp), SV9705 (PopvAB GATC-less::aac6::gfp) SV9706 (PopvAB::ctxM::gfp) and SV9707 (PopvAB GATC-less::ctxM::gfp) in LB and in LB + antibiotic (kanamycin and cefotaxime, respectively). (B) Left: reversibility of formation of Kms and Kmr subpopulations under opvAB control. Right: reversibility of formation of CtxMs and CtxMr subpopulations under opvAB control.
Flow cytometry analysis confirmed that growth in the presence of kanamycin and cefotaxime was a consequence of subpopulation selection (Fig. 3B), excluding the idea that growth might result from selection of mutants present in the inoculum. This conclusion was further strengthened by the observation that growth in LB restored the initial sizes of ON and OFF subpopulations (Fig. 3B).
Use of the OpvAB synthetic switch in generating antibiotic heteroresistance
As a proof of concept, we examined the utility of the OpvAB switch to address antibiotic heteroresistance and the question of what proportions of resistant subpopulations might lead to clinical treatment failure. Specifically, we tested whether the OpvAB switch could generate, in a susceptible main population, defined subpopulations of cells with increased antibiotic resistance. For this purpose, we used a S. enterica strain harboring a PopvAB::BI-aac3::gfp construct (SV9776). Expression of aac3 (Aac3ON) leads to kanamycin resistance (Kmr). The frequency of Kmr cells formed by a pure culture of SV9776 was 1 × 10−2 (Fig. 4A), similar to the frequency of ON cells detected when gfp was cloned behind the opvAB promoter (1.1%: Fig. 1D). To obtain smaller subpopulation sizes without altering other phenotypic traits of the strain, SV9776 was mixed with an isogenic strain that expressed PopvAB::gfp (SV9777) and did not produce any Kmr resistant subpopulation. Mixtures of cells were prepared from overnight cultures in Mueller-Hinton (MH) broth at proportions 1:10, 1:100, 1:1,000, 1:10,000 and 0:1. Population analysis profile (PAP) tests were then performed by plating on MH agar containing increasing concentrations of kanamycin. After overnight incubation, the number of resistant cells and total number of cells were determined to allow calculation of the fraction of resistant cells. The numbers of Kmr colonies detected in the PAP tests were proportional to the amounts of the Aac3ON subpopulations present in each mixture, and ranged from 1 × 10−2 to 1 × 10−6 (Fig. 4A). Epsilometer tests (Etests) further confirmed that the size of the Kmr subpopulation decreased in a manner proportional to dilution (Fig. 4B).
The opvAB switch as a tool for the study of antibiotic heteroresistance. (A) Population analysis profile (PAP) tests of kanamycin resistance in strains SV9776 and SV9777 (carrying PopvAB::BI-aac3::gfp and PopvAB::gfp constructs, respectively). The proportions indicated are those of the mixtures of SV9776:SV9777. Error bars represent standard deviations of three independent mixtures. (B) Epsilometer tests (Etests) performed on the same mixtures of SV9776 and SV9777. The kanamycin resistant subpopulations appear as low-density lawns (mixtures 1:0 and 1:10) or as isolated colonies growing at kanamycin concentrations above the MIC of the main susceptible population ([Km] > 1 mg/L - mixtures 1:10 to 1:10,000). No kanamycin resistant subpopulation exists for the 0:1 mixture.
Discussion
In its native host, the opvAB operon undergoes bistable transcription, which generates OpvABON and OpvABOFF subpopulations9. Bistability is reversible (“phase-variable”) and the switching rate is skewed to OFF in the wild type9,11. In this study, we show that a 689 bp DNA fragment containing the opvAB promoter and the opvAB upstream activating sequence (UAS) confers bistability to genes cloned downstream. For instance, an engineered PopvAB::lacZY operon produces LacOFF and LacON subpopulations (Fig. 1C), and addition of a gfp reporter gene permits discrimination of LacOFF and LacON cells by flow cytometry (Fig. 1D). Utilization of L-lactose sustains growth of LacON cells (Fig. 1E), thereby producing increased fluorescence. However, because the opvAB switch is reversible, in the absence of L-lactose the system slowly returns to its initial state, with a strong predominance of LacOFF cells (Fig. 1F).
The fact that the opvAB cassette is functional in E. coli (Fig. 2) suggests that the switch can be used to generate bistability in other heterologous hosts. However, the need of both Dam methylation and OxyR may be an obvious limitation. Aside from this caveat, the versatility of the switch is reinforced by an additional example of subpopulation formation presented in Fig. 3: PopvAB-driven bistable expression of kanamycin and cefotaxime resistance genes permitted selection of antibiotic-resistant subpopulations in a reversible fashion.
Introduction of mutations in the upstream regulatory region of the native opvAB operon alters the switching rate, yielding OpvABON and OpvABOFF subpopulation sizes that are different from those of the wild type10,11. Hence, variants of the opvAB switch can be engineered to modulate subpopulation sizes at will. For instance, a variant (GATC1,2) that lacks two of the four GATC sites present in the wild type increases the initial size of the ON subpopulation (Figs 1 and 2). Additional UAS variants that yield subpopulations of different sizes have been described10, and their use may allow choice of other switching frequencies. Modification of the ribosome-binding site of genes under PopvAB control can also contribute to adjust the sensitivity of the switch, facilitating detection of the phenotype under study. For instance, use of the BI ribosome binding site20 permitted unambiguous detection of aac3-mediated kanamycin resistance, thereby facilitating discrimination of Kmr cells (Fig. 4).
As a proof of concept, we have used the opvAB switch to produce antibiotic-resistant and antibiotic-susceptible bacterial subpopulations of predetermined sizes. The aim of these experiments was to mimic under laboratory conditions bacterial heteroresistance to antibiotics, a phenomenon where small subpopulations of cells show higher antibiotic resistance than the main population12. Heteroresistance is difficult to detect and study in clinical samples12, and accurate assessment of the frequencies of subpopulation formation and of their antibiotic resistance levels may improve our understanding of heteroresistance as a cause of clinical treatment failure15. Experiments shown in Fig. 4 provide evidence that subpopulation formation under opvAB control allows accurate modulation of the number of resistant cells present in a population. In principle, the method should be applicable to any antibiotic resistance gene. Because we were able to specifically vary the frequency of resistant bacteria in the population, this approach provides a proof of concept to study how different frequencies of resistant subpopulations may affect the outcome of antimicrobial treatment in vivo (e. g., in a murine model). In theory, mixing constitutively resistant and susceptible strains that are otherwise isogenic would also lead to bacterial cultures with pre-defined amounts of resistant bacteria. However, to reach specific frequencies of resistant bacteria our OpvAB-based approach requires mixing bacteria at frequencies 100-fold lower (e. g., to reach frequencies of 1 × 10−6 Kmr resistant bacteria, the PopvAB::BI-aac3 strain was mixed at a frequency of 1 × 10−4). Thus, an advantage of our opvAB switch-based approach is that it can be expected to be less affected by infection bottlenecks that could otherwise eliminate very small subpopulations of bacteria present in the inoculum21. For example, one such bottleneck is observed during cecum colonization by Salmonella in mice 2–4 days after oral infection, and is dependent on the inflammatory response induced by S. enterica invading epithelial cells22,23.
Additional applications of the opvAB switch can be envisaged, including the design of bistable biosensors. For instance, a strain harboring an PopvAB::gfp fusion might be useful to detect bacteriophages in environmental samples using flow cytometry24,25, and to identify DNA methylation inhibitors in screens for novel antimicrobial drugs26,27. Sensors of this kind can be expected to be selective as growth will occur under specific circumstances only. Furthermore, use of fluorescence to monitor growth of ON cells can be expected to be sensitive and rapid, and constitutive expression may contribute to robustness, avoiding the problem of instability of transcription-based gene circuits28. Besides biosensor design, formation of phenotypic subpopulations under epigenetic control might have additional applications in synthetic biology: for instance, division of labour between subpopulations performing distinct segments of a catabolic pathway might optimize biodegradation processes29.
Methods
Strains and strain construction
Strains of Salmonella enterica serovar Typhimurium and Escherichia coli used in this study are listed in Table 1. Strain construction by targeted gene disruption was achieved using plasmids pKD3, pKD4 or pKD13 as templates to generate PCR products for homologous recombination30. Antibiotic resistance cassettes introduced during strain construction were excised by recombination with plasmid pCP2030. Primers used in strain construction are shown in Table 2. For the construction of translational lac fusions on the S. enterica chromosome, FRT sites generated by excision of Kmr cassettes were used to integrate plasmid pCE4031. For construction of fluorescent fusions, a DNA fragment containing a promoterless green fluorescent protein (gfp) gene and a chloramphenicol resistance cassette was PCR-amplified from plasmid pZEP0732, and the resulting PCR product was integrated into the chromosome of each strain. For construction of strains that carry antibiotic resistance genes under PopvAB control, a counterselectable cassette containing sacB and ApR genes was amplified from strain DA52596 using the oligos opvAB-ampsacB-F and ampsacB-gfp-R. The PCR product was integrated into the chromosome of SV6727 and SV6729 respectively, generating the intermediate strains MN441 and MN442, respectively. Antibiotic resistance genes were introduced into these strains by targeted gene disruption30, and transformants in which the ampicillin-sacB cassette had been excised were selected on minimal plates containing sucrose.
Transductional crosses using phage P22 HT 105/1 int20133 were used for transfer of chromosomal markers between S. enterica strains34. To obtain phage-free isolates, transductants were purified by streaking on green plates35. Phage sensitivity was tested by cross-streaking with P22 H5.
Directed construction of point mutations was achieved using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene) using the suicide plasmid pDMS19736 and propagated in E. coli CC118 λ pir. Plasmids derived from pMDS197 (pIZ2224 and pIZ2234) were transformed into E. coli S17-1 λ pir. The resulting strains were used as donors in matings with S. enterica SV9700, selecting tetracycline-resistant transconjugants on minimal plates. One transductant from each mating was propagated as strains SV9701 and SV9702.
Culture media and growth conditions
Bertani’s lysogeny broth (LB)37 was used as standard liquid medium. Solid LB contained agar at 1.5% final concentration. Mueller-Hinton (MH) broth and agar38 was used in antibiotic susceptibility tests. Green plates35 contained methyl blue (Sigma-Aldrich) instead of aniline blue. The indicador of β-galactosidase activity in plate tests was 5-bromo-4-chloro-3indolyl-β-D-galactopyranoside (X-gal; Sigma-Aldrich, 40 mg/L). No-carbon essential (NCE) medium39, supplemented with either glucose (0, 2%) or lactose (0, 2%), was used as minimal medium. When necessary, antibiotics were added to the culture medium at following concentrations: ampicillin (100 mg/L), kanamycin (50 mg/L), chloramphenicol (25 mg/L), and cefotaxamine (40 mg/L).
Flow cytometry
Bacterial cultures were grown in LB at 37 °C until exponential phase (O.D.600 0.3). Cells were then diluted in PBS to a final concentration of approximately 107/ml. Data acquisition and analysis were performed using a Cytomics FC500-MPL cytometer (Beckman Coulter). Data were collected for 100 000 events per sample, and were analyzed with CXP and FlowJo8.7 software. Data are represented by a dot plot (forward scatter [cell size] versus fluorescence intensity.
Growth curves
Plates were incubated at 37 °C with shaking on an automated microplate reader (Synergy HTX Multi-Mode Reader, Biotek), and the absorbance at 600 nm for each well was measured every 30 min. Each sample was assayed by triplicate. Growth of SV9700, SV9701, SV9703, DR22, DR23 and DR24 strains was monitored in NCE-lactose and NCE-glucose. Growth of SV9704, SV9705, SV9706, SV9707 was monitored in LB broth with and without antibiotics.
Calculation of phase variation rates
Phase variation rates were estimated as described by Eisenstein40. Briefly, a strain harboring a lacZY fusion was plated on LB + X-gal and colonies displaying an ON or OFF phenotype after 16 h growth at 37 °C were selected, resuspended in PBS and re-spread on fresh LB + X-gal plates. Phase variation frequencies were calculated using the formula (M/N)/g where M is the number of cells that underwent phase variation, N the total number of cells, and g the total number of generations that gave rise to the colony.
Epsilometer (E) tests of antibiotic resistance
Etest strips were purchased from bioMérieux. Mixtures of overnight cultures of bacteria grown in MH broth were diluted 1:25 in phosphate buffered saline (PBS) to reach cell densities of 0.5 MacFarland or about 1.5 × 108 CFU/mL. Bacteria were plated onto MH agar plates using sterile cotton swabs dipped in the cell suspensions, and a Etest strip was applied on top. Plates were incubated 18 h at 37 °C before reading the results and taking pictures.
Population analysis profile (PAP) tests
PAP tests were performed on MH agar plates supplemented with increasing amounts of kanamycin (Sigma Aldrich) as described previously15. Five µl of overnight cultures in MH broth (containing approx. 3 × 109 cells/ml) and serial dilutions (down to 10−6) were spread on MH plates containing no antibiotics (for total CFU determination) or different concentrations of kanamycin. The plates were incubated overnight and the colonies were counted. Colony numbers were plotted in a graph to determine if the PAP fulfilled the criteria for heteroresistance (at least 8-fold difference in antibiotic concentration between the highest non-inhibitory concentration and the highest inhibitory concentration).
To prepare mixtures of resistant and susceptible cells, three isolated colonies of SV9776 (PopvAB::BI-aac3::gfp, kanamycin resistant in the ON state) and SV9777 (PopvAB::gfp, always kanamycin susceptible) were grown overnight in 2 mL MH broth at 37 °C under shaking. Pure cultures of each overnight or three independent sets of SV9776:SV9777 mixtures at proportions ranging from 1:10 to 1:10,000 were used for PAP tests.
Change history
25 November 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Bradley, R. W., Buck, M. & Wang, B. Tools and principles for microbial gene circuit engineering. J Mol Biol 428, 862–888 (2016).
Park, M., Tsai, S. L. & Chen, W. Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors (Basel) 13, (5777–5795 (2013).
Engstrom, M. D. & Pfleger, B. F. Transcription control engineering and applications in synthetic biology. Synth Syst Biotechnol 2, 176–191 (2017).
Kremers, G. J., Gilbert, S. G., Cranfill, P. J., Davidson, M. W. & Piston, D. W. Fluorescent proteins at a glance. J Cell Sci 124, 157–160 (2011).
Huertas, C. S. et al. Label-free DNA-methylation detection by direct ds-DNA fragment screening using poly-purine hairpins. Biosens Bioelectron 120, 47–54 (2018).
Yamatsugu, K., Kawashima, S. A. & Kanai, M. Leading approaches in synthetic epigenetics for novel therapeutic strategies. Curr Opin Chem Biol 46, 10–17 (2018).
Damayanti, N. P., Buno, K., Voytik Harbin, S. L. & Irudayaraj, J. M. K. Epigenetic process monitoring in live cultures with peptide biosensors. ACS Sens 4, 562–565 (2019).
Maier, J. A. H., Mohrle, R. & Jeltsch, A. Design of synthetic epigenetic circuits featuring memory effects and reversible switching based on DNA methylation. Nat Commun 8, 15336 (2017).
Cota, I., Blanc-Potard, A. B. & Casadesus, J. STM2209-STM2208 (opvAB): a phase variation locus of Salmonella enterica involved in control of O-antigen chain length. PLoS One 7, e36863 (2012).
Cota, I. et al. OxyR-dependent formation of DNA methylation patterns in OpvABOFF and OpvABON cell lineages of Salmonella enterica. Nucleic Acids Res 44, 3595–3609 (2016).
Cota, I. et al. Epigenetic control of Salmonella enterica O-antigen chain length: a tradeoff between virulence and bacteriophage resistance. PLoS Genet 11, e1005667 (2015).
El-Halfawy, O. M. & Valvano, M. A. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28, 191–207 (2016).
Band, V. I. et al. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1, 16053 (2016).
Band, V. I. et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9, e02448–17 (2018).
Nicoloff, H., Hjort, K., Levin, B. R. & Andersson, D. I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4, 504–514 (2019).
Hjort, K., Nicoloff, H. & Andersson, D. I. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol 102, 274–289 (2019).
Andersson, D. I., Nicoloff, H. & Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol 17, 479–496 (2019).
Dubois, V. et al. Molecular characterization of a novel class 1 integron containing bla(GES-1) and a fused product of aac3-Ib/aac6′-Ib’ gene cassettes in Pseudomonas aeruginosa. Antimicrob Agents Chemother 46, 638–645 (2002).
Duan, R. S. et al. Escherichia coli producing CTX-M beta-lactamases in food animals in Hong Kong. Microb Drug Resist 12, 145–148 (2006).
Eckdahl, A. J., Neal, R., Campbell, A. M. & Eckdahl, T. T. rClone: A synthetic biology tool that enables the research of bacterial translation. J Young Inv 32, 12–19 (2017).
Grant, A. J. et al. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol 6, e74 (2008).
Maier, L. et al. Granulocytes impose a tight bottleneck upon the gut luminal pathogen population during Salmonella typhimurium colitis. PLoS Pathog 10, e1004557 (2014).
Lim, C. H. et al. Independent bottlenecks characterize colonization of systemic compartments and gut lymphoid tissue by salmonella. PLoS Pathog 10, e1004270 (2014).
Muniesa, M. et al. Bluephage: A rapid method for the detection of somatic coliphages used as indicators of fecal pollution in water. Water Res 128, 10–19 (2018).
Vinay, M. et al. Phage-based fluorescent biosensor prototypes to specifically detect enteric bacteria such as E. coli and Salmonella enterica Typhimurium. PLoS One 10, e0131466 (2015).
Ceccaldi, A. et al. Identification of novel inhibitors of DNA methylation by screening of a chemical library. ACS Chem Biol 8, 543–548 (2013).
Mashhoon, N., Pruss, C., Carroll, M., Johnson, P. H. & Reich, N. O. Selective inhibitors of bacterial DNA adenine methyltransferases. J Biomol Screen 11, 497–510 (2006).
Purcell, O. & Lu, T. K. Synthetic analog and digital circuits for cellular computation and memory. Curr Opin Biotechnol 29, 146–155 (2014).
Nikel, P. I., Silva-Rocha, R., Benedetti, I. & de Lorenzo, V. The private life of environmental bacteria: pollutant biodegradation at the single cell level. Environ Microbiol 16, 628–642 (2014).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 90, 6640–6645 (2000).
Ellermeier, C. D., Janakiram, A. & Slauch, J. M. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290, 153–161 (2002).
Hautefort, I., Proenca, M. J. & Hinton, J. C. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol 69, 7480–7491 (2003).
Schmieger, H. Phage P22 mutants with increased or decreased transducing abilities. Mol Gen Genet 119, 75–88 (1972).
Garzón, A., Cano, D. A. & Casadesus, J. Role of Erf recombinase in P22-mediated plasmid transduction. Genetics 140, 427–434 (1995).
Chan, R. K., Botstein, D., Watanabe, T. & Ogata, Y. Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology 50, 883–898 (1972).
Edwards, R. A., Keller, L. H. & Schifferli, D. M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157 (1998).
Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic. Escherichia coli. J Bacteriol 62, 293–300 (1951).
Mueller, J. H. & Hinton, J. A protein-free medium for primary isolation of the gonococcus and meningococcus. Exp Biol Med 48, 330–333 (1941).
Vogel, H. J. & Bonner, D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218, 97–106 (1956).
Eisenstein, B. I. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214, 337–339 (1981).
Acknowledgements
This study was supported by grant BIO2016-75235-P from the Ministerio de Ciencia, Innovación y Universidades of Spain (to JC) and the Swedish Research Council grant 2017-01527 (to DIA). We are grateful to Elena Puerta-Fernández for advice and discussions, and to Modesto Carballo, Laura Navarro, and Cristina Reyes of the Servicio de Biología, CITIUS, Universidad de Sevilla, for help with experiments performed at the facility.
Author information
Authors and Affiliations
Contributions
D.R.O., H.N., M.A.S. and I.C. carried out the experiments; D.R.O., H.N., M.A.S., I.C., D.I.A. and J.C. designed the experiments and interpreted results; D.R.O., H.N., D.I.A. and J.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Olivenza, D.R., Nicoloff, H., Sánchez-Romero, M.A. et al. A portable epigenetic switch for bistable gene expression in bacteria. Sci Rep 9, 11261 (2019). https://doi.org/10.1038/s41598-019-47650-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47650-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.