Abstract
Successful primer design for polymerase chain reaction (PCR) hinges on the ability to identify primers that efficiently amplify template sequences. Here, we generated a novel Taq PCR data set that reports the amplification status for pairs of primers and templates from a reference set of 47 immunoglobulin heavy chain variable sequences and 20 primers. Using logistic regression, we developed TMM, a model for predicting whether a primer amplifies a template given their nucleotide sequences. The model suggests that the free energy of annealing, ΔG, is the key driver of amplification (p = 7.35e-12) and that 3′ mismatches should be considered in dependence on ΔG and the mismatch closest to the 3′ terminus (p = 1.67e-05). We validated TMM by comparing its estimates with those from the thermodynamic model of DECIPHER (DE) and a model based solely on the free energy of annealing (FE). TMM outperformed the other approaches in terms of the area under the receiver operating characteristic curve (TMM: 0.953, FE: 0.941, DE: 0.896). TMM can improve primer design and is freely available via openPrimeR (http://openPrimeR.mpi-inf.mpg.de).
Similar content being viewed by others
Introduction
Polymerase chain reaction (PCR) forms the foundation for a multitude of a variety of molecular methods (e.g. determining drug resistance1,2 and viral loads3). Primers – short nucleotide oligomers complementary to template DNA – are critical for the effective amplification of templates through PCR. For example, the optimization of primers targeting immunoglobulin variable gene sequences is critical for the identification of novel antibodies such as broadly neutralizing antibodies targeting HIV-14. Models that estimate PCR efficiencies can guide primer design for quantitative PCR (qPCR)5,6,7,8, while models estimating the likelihood of amplification can guide primer design for conventional PCR9. These models need to consider the two consecutive molecular interactions that determine whether a primer allows for the amplification of a PCR template. In the first reaction, the primer anneals to the template to form the primer-template heteroduplex. In the second reaction, polymerase attaches to the partial heteroduplex and elongates the oligonucleotide to a complementary full-length sequence10.
Efficient primer annealing is largely determined by the complementarity of primer and template11, a characteristic that is captured by the free energy of annealing. Therefore, non-complementary bases in the nucleotide sequences of primers and templates (mismatches) should be avoided. Mismatches within the 3′ hexamer of the primer-template duplex (i.e. the terminal six nucleotides) are especially detrimental as they can disrupt polymerase binding5,6,12,13,14,15. The impact of 3′ mismatches increases with growing proximity to the 3′ terminus13,15. Moreover, the extent at which 3′ terminal mismatches decrease PCR efficiency critically depends on the type of mismatch (e.g. an A/G mismatch is substantially more detrimental than an A/C mismatch)12,13,16,17,18,19,20. To stabilize the 3′region, primers are often designed to exhibit a GC clamp21,22,23 consisting of one to three Gs or Cs at the 3′ end of the primer.
Primer binding events can be identified using thermodynamic or statistical models24. To our best knowledge, the thermodynamic model provided by DECIPHER8 (DE) is the only model that is currently available. DECIPHER incorporates empiric evidence about the impact of position- and nucleotide-specific mismatches within the last seven positions of the 3′ region. These data were gathered by measuring the elongation efficiency of Taq polymerase in PCRs performed with 171 primers exhibiting different binding properties. The model considers three reactions: the interaction between primer and template, unimolecular folding of the primer, and unimolecular folding of the template. Based on the underlying kinetic differential equations for these reactions, the concentrations of the considered molecular states are mechanistically computed for inferring the efficiency of PCR.
Here, we present a novel Taq PCR data set providing the amplification status for 47 immunoglobulin heavy-chain variable (IGHV) genes. Triplicate measurements were performed with primers from two sets. Set 1 consists of 16 forward primers that have been recently designed using openPrimeR25, while Set 2 is a well-established set of 4 forward primers26. PCR was performed for each combination of the 20 primers and 47 templates giving rise to a total of 940 triplicate measurements. In contrast to other studies investigating PCR amplification, which are largely based on qPCR, this data set provides the amplification status according to gel electrophoresis. Using statistical methods, we analyzed the data set with three goals in mind. First, to investigate which physicochemical properties of primer-template pairs (PTPs) exert the greatest influence on the PCR amplification status. Second, to develop a new logistic regression model for predicting the amplification of a template. Third, to compare available models for determining amplification events.
Results
Having selected 908 PTPs from the PCR data set, we classified the amplification status of each PTP either as Amplified or Unamplified depending on the result of gel electrophoresis (Fig. 1). To investigate which properties of PTPs are associated with the amplification status, we computed their physicochemical properties using openPrimeR, most notably, the free energy of annealing, ΔG [kcal/mol], and three features related to 3′ mismatches: z ∈ {0, 1}6, \({X}_{N}\in {{\mathbb{N}}}_{0}\), and iX ∈ {0, 1, …, 6} (Fig. 2). We used these features to train a logistic regression model for predicting the amplification status and validated the model by comparing its performance with that from DECIPHER and an approach relying only on ΔG.
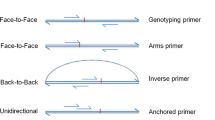
Examples for encoding mismatches within the 3′ hexamer region. Primers are indicated as arrows and templates are indicated as horizontal bars. Arrowheads indicate the 3′ hexamer region. Mismatches within the 3′ hexamer are encoded via z ∈ {0, 1}6, \({X}_{N}\in {{\mathbb{N}}}_{0}\), and iX ∈ {0, 1, …, 6}. While z uses a binary encoding to indicate the presence of mismatches within the 3′ hexamer, XN gives the total number of 3′ hexamer mismatches, and iX indicates the position of the 3′ hexamer mismatch closest to the 3′ terminus. (a) Absence of 3′ terminal mismatches between primer and template. (b) Mismatches in the 3′ hexamer at positions 4 and 6.
Properties of the data set
Table 1 shows the distribution of the physicochemical properties of PTPs in the data set. The primers from Set 1 and Set 2 are characterized by contrasting rates of amplification. While 165 of 188 PTPs (87.8%) in Set 2 were labeled as Amplified, only 217 of 720 (30.1%) observations in Set 1 set were labeled as Amplified. Accordingly, PTPs from Set 1 exhibited a greater number of mismatches and higher free energies. The PTPs from Set 1 had an average of 2.3 mismatches in the 3′ hexamer, while the PTPs from Set 2 had an average of 0.5 mismatches in this region. Moreover, while samples from Set 2 had a ΔG inter-quartile range (IQR) of [−8.6 kcal/mol, −5.2 kcal/mol], the samples from Set 1 were associated with a higher range of [−4.9 kcal/mol, −2.0 kcal/mol].
Table 2 shows the relationship between the number of primer-template mismatches, free energy of annealing, and the rate of amplification. In our data set, primers with at most 3 mismatches had a 100% amplification rate. It is noteworthy that even primers binding with as many as 6 mismatches obtained a high amplification rate of 83.3%. Note that, for any given number of mismatches, the primers from Set 2 consistently exhibit a greater rate of amplification than the primers from Set 1. Comparing amplified and unamplified PTPs (Fig. 3), we found that the ΔG IQR of observations labeled as Unamplified was higher and more concentrated ([−2.17 kcal/mol, −1.69 kcal/mol]) than for those labeled as Amplified ([−12.70 kcal/mol, −5.21 kcal/mol]). Amplified samples generally exhibited fewer mismatches in the 3′ hexamer (XN IQR of [0, 1] vs [2, 4]) and particularly fewer mismatches close to the 3′ terminus (iX IQR of [0, 3] vs [5, 6]) than unamplified samples. Applying two-sided Wilcoxon rank-sum tests revealed that there is a significant difference between Amplified (N = 382) and Unamplified (N = 526) observations concerning both ΔG (p-value 1.68e-107) and iX (p-value 1.51e-91).
Impact of the free energy of annealing (ΔG) and 3′ terminal mismatches on the amplification of templates. The x-axis indicates, for every PTP, the mismatch position closest to the primer 3′ terminus such that position 1 in the plot corresponds to iX = 6 and position 6 corresponds to iX = 1. PTPs with zero mismatches are denoted by None. Every point represents a primer-template pair. Pairs that are labeled as Amplified are shown in blue, while those that are labeled as Unamplified are shown in red. Observations from Set 1 are indicated by circles and those from Set 2 by triangles. The dashed lines indicate cutoffs that are suitable for separating observations according to their amplification status. The vertical dashed line indicates the end of the 3′ hexamer, while the horizontal dashed line indicates a free energy of −5 kcal/mol.
Logistic regression models
We used logistic regression in order to identify the features that are predictive of successful PCR amplification events. Since considered primers shared similar physicochemical properties (Table 3), we only considered properties relating to PTPs when defining the two logistic regression models LR1 and LR2 (Table 4). LR1 was defined using the features z, XN, and ΔG. For LR2, a term modeling the 3′ mismatch closest to the 3′ terminus, iX, and a term ΔGix modeling the interaction of ΔG and ix were additionally included. Since LR1 was not corrected for the association between ΔG and iX, only z6 (p = 8.25e-08) and ΔG (p < 2e-16) were found to be significantly predictive of the amplification status. Based on LR2, on the other hand, only ΔG (p = 1.78e-11) and ΔGix (p = 5.12e-05) were found to be significantly predictive of the amplification status. This finding indicates that mismatches within the 3′ hexamer are not independent predictors of the amplification status but dependent on ΔG.
Evaluated models and classifiers
In order to form a generalizable logistic regression model for predicting the likelihood of amplification, features were eliminated by performing backward stepwise selection on a model trained using the features considered in LR2. The selection procedure reduced the Akaike Information Criterion (AIC) of the initial logistic regression model from 112.34 to 102.38. Besides the intercept, the following three features were selected: ΔG, iX, and the interaction term ΔGiX. In the following, this logistic regression model is called the thermodynamic mismatch model (TMM).
In order to assess the predictive performance of available approaches for predicting the likelihood of PCR amplification, we considered three models: The model DE from DECIPHER8, a model solely based on the free energy (FE), and TMM. Besides evaluating the quantitative output of these approaches, we also evaluated the performance of classifiers corresponding to these models by calculating a cutoff based on the estimates of each model in order to classify PTPs either as Amplified or Unamplified. Two types of cutoffs were selected for each model, one optimized for overall accuracy (by maximizing Youden′s index) and another optimized for specificity (Table 5). Classifiers optimized for overall performance and classifiers optimized for high specificity are denoted by subscription of Y or s, respectively. For example, TMMs denotes the high-specificity TMM classifier and TMMY denotes the TMM classifier that was optimized for overall performance.
Comparison of model and classifier performance
Quantitative model responses were compared with the categorical amplification status from gel electrophoresis according to the area under the receiver operating characteristic curve (AUC). TMM achieved the highest AUC (0.953) but was closely followed by FE (0.941), and DE (0.896). For all models, predictive performance was higher for observations from Set 2 than for those from Set 1 (Table 6). The classifier performance was evaluated with respect to sensitivity, specificity, and the F1 score (Fig. 4). Among high-performance classifiers, TMMY had a larger F1 score than DEY and FEY (90% vs 88% and 88%). Among high-specificity classifiers, TMMs and DEs outperformed FEs with respect to sensitivity (76% and 78% vs 64%).
Performance of three models for identifying primer amplification events. TMM indicates our newly developed logistic regression model, DE refers to the approach from DECIPHER, and FE is solely based on the free energy of annealing. Models subscripted with s use cutoffs optimized for high specificity, while models subscripted with Y use cutoffs optimized for overall performance.
Interpretation of the TMM model
For interpreting and deploying TMM, a final model was trained on the full data set. The model can be specified in the following way (Table 7). Let p = Pr(yi = Amplified) denote the probability that a template is amplified. Given ΔG and iX, the model estimates \(\hat{p}\) = Pr(yi = Amplified) according to its coefficients β0 = −5.62, β1 = −1.55, β2 = 0.33 and β3 = 0.18 in the following way:
The intercept of the model is β0 = −5.62, which indicates that the odds of template amplification are low if the other terms are negligible (i.e. for ΔG → 0 and iX → 0). The second term, (−1.55 + 0.18 iX)⋅ ΔG, is controlled by the free energy of annealing. For typical negative values of ΔG, the odds of amplification increase with decreasing ΔG because −1.55 + 0.18 iX is always negative since 0 ≤ iX ≤ 6. The presence of 3′ terminal mismatches (iX ≠ 0), however, reduces the odds of amplification. The third term, 0.33 iX, increases the odds if a 3′ mismatch is present (iX ≠ 0). This term can be interpreted as a correction factor, which models that there is an overrepresentation of PTPs with high ΔG (e.g. −5 kcal/mol) and high iX.
The model can be visualized as a cube (Fig. 5) whose three dimensions correspond to ΔG, iX, and the estimated likelihood of amplification, \(\hat{p}\), for the PTPs in the IGHV data set. For low and high free energies (e.g. at −20 and −5 kcal/mol), ΔG dominates \(\hat{p}\), while iX influences \(\hat{p}\) mostly at intermediate values of ΔG (e.g. at −10 kcal/mol).
Visualization of the TMM model. Individual dots show the prediction function of the model. Red dots indicate low probabilities of amplification while blue dots indicate high probabilities. The rectangles show the model estimate for the observations contained in the data set. Here, red points indicate primer-template pairs that were labeled as Unamplified, while blue points indicate observations labeled as Amplified.
Discussion
In this work, we presented a novel PCR data set providing the amplification status for all combinations of 47 IGHV templates and 20 primers. Using these data, we investigated the interplay of the free energy of annealing and the presence of 3′ terminal mismatches and found that both factors should be considered in dependence of each other. Based on this insight, we developed TMM, a logistic regression model for predicting amplification events.
In our analysis of the IGHV data, we could mostly confirm the established factors governing the efficiency of PCR. More specifically, we could show that templates whose amplification could not be detected via gel electrophoresis are a result of primer-template conformations exhibiting high free energies, an increase in the number of mismatches within the 3′ hexamer, and a tendency for displaying mismatches close to the 3′ terminus. For the present data, however, we found that terminal mismatches by themselves are not significantly predictive of the amplification status when correcting for their association with the free energy of annealing. This finding suggests that a mismatch at the 3′ terminus does not preclude detection via gel electrophoresis as long as primer and template are otherwise highly complementary.
The newly developed TMM model for predicting amplification events has several advantages over the other models. First, since the model is based only on ΔG and iX, it is easily interpretable and it is unlikely that the model suffers from overfitting. Second, the model estimates the probability of amplification, which is a more intuitive measure than the efficiency of amplification from DE. Third, TMM achieved the largest AUC and its high-specificity classifier achieved the highest sensitivity among all classifiers. Since the present data set contains only primers exhibiting specific properties such as the absence of self-dimers and the presence of a GC clamp (Table 3), TMM neither considers primer- nor template-specific properties. Thus, it is likely that TMM overestimates the likelihood of amplification for primers exhibiting less favorable properties or when templates exhibit secondary structures27,28,29. Indeed, a previously described logistic regression model proposed by Yuryev et al.9 considered a larger number of features than TMM. Their model, however, was developed for primer genotyping assays, which renders it inappropriate for applications where several primer-template mismatches need to be considered.
Overall, all three methods achieved high predictive performances on the IGHV data set. Although the predictive performance of FEY was surprisingly high, the considerably lower performance of FEs indicates that the free energy of annealing by itself lacks robustness. In contrast to DE, which estimates the efficiency of polymerase elongation according to the impact of position- and base-specific effects in the 3′ region, TMM considers only the position of 3′ mismatches. The following two observations could explain why the consideration of base-specific effects did not provide an advantage over TMM, although their influence is extensively described in the literature. First, none of the primers contained in the IGHV data set displayed terminal nucleotides other than G or C (Table 3). Second, since base-specific differences in amplification efficiencies have only been reported for qPCR8, these difference may simply not be observable with data from gel electrophoresis. Additionally, the present data (Table 2 and Fig. 3) suggest that even simple stringent approaches can be used to ensure high rates of amplification, for example, requiring free energies less than −10 kcal/mol or allowing at most three mismatches.
In order to select a suitable prediction model, its field of application should be carefully deliberated. For example, for multiplex primer design, false positive predictions should be avoided at all costs because they may preclude the amplification of templates that are not redundantly covered. False negative predictions, on the other hand, are much more tolerable. Our analysis suggests that high-specificity classifiers such as TMMs or DEs are most appropriate in this scenario. In multiplex scenarios where it is not necessary to amplify all templates, smaller primer sets can be designed by choosing a model with greater sensitivity.
Although models that estimate the likelihood of amplification should be an integral part of rational primer design approaches, there are few available models for this task. The lack of publicly available PCR data is not only a limiting factor for model development but also for improving our understanding of the molecular characteristics that govern PCR amplification. Only when enough data are available will it be possible to devise more comprehensive models that consider all relevant properties concerning primers, templates, and their interaction. Here, we presented a novel PCR data set on which basis we developed TMM, a model for predicting the PCR amplification status, which is freely available via openPrimeR (http://openprimer.mpi-inf.mpg.de/).
Materials and Methods
Template design and PCR measurements
We cloned 47 heavy chain fragments from naive B cells into pCR4-TOPO-vector backbones. Each fragment comprises a different functional IGHV gene with the complete leader (L) region, the complete V region and a short part of the constant region. The individual V genes served as representative templates for two different IGHV-specific primer sets. Set 1 is a set of 16 forward primers that was recently designed using openPrimeR25, while Set 2 consists of 4 forward primers that were described previously26. We performed three independent PCR reactions for each of the 20 primers on all 47 templates with the same IgM constant region-specific reverse primer (GGTTGGGGCGGATGCACTCC)30. All primers used in the experiments are listed in Table 3. PCRs were performed in 25 µL reactions with 2U/rxn Platinum Taq (Thermofisher), 0.2 µM forward and reverse primer, 0.2 mM dNTPs, 1.5 mM MgCl2, and 6% Kb extender under the following cycling conditions: 2 min initial denaturation at 94 °C followed by 25 cycles of 30 s at 94 °C, 30 s at 57 °C (Set 2) or 55 °C (Set 1), and 55 s at 72 °C. The expected 600–700 bp fragments were visualized on a 2% agarose gel supplemented with SYBR Safe (Thermofisher) and documented with the BioRAD Gel DocTM XR + Imaging system.
Data set construction
Template sequences were retrieved by Sanger sequencing and annotated with IgBlast31. Every considered PTP \(i\in {\mathbb{N}}\) was assigned a label yi ∈ {Amplified, Unamplified} based on the evaluation of gel electrophoresis by five persons. Each of the five reviewers visually inspected the gels and independently classified the amplification status. If a band was visible in a gel, the corresponding measurement was labeled as Amplified and otherwise as Unamplified (Fig. 1). The following procedure was used to identify yi,j, the label of PTP i according to reviewer j ∈ {1, …, 5} from a set of triplicate measurements. If at least two of three measurements were labeled as Amplified, yi,j was set to Amplified. Otherwise, yi,j was set to Unamplified. Let ni,A = |{yi,j|yi,j = Amplified}| and ni,U = |{yi,j|yi,j = Unamplified}| indicate the number of times that PTP i was labeled as Amplified or Unamplified, respectively. By setting
we labeled PTP i as Amplified only if the majority of reviewers had labeled the PTP as Amplified.
We used openPrimeR to enrich the PCR data with physicochemical properties relating to primers and PTPs. The most likely binding mode for every PTP was identified by selecting the binding conformation minimizing the number of mismatches. Since the exact annealing site of primers is uncertain for PTPs subject to many mismatches, we excluded PTPs with more than 12 mismatches. This reduced the size of the data set from 940 to 908 observations. Based on the determined binding conformation, we derived further properties such as the position of primer-template mismatches. The free energy of annealing ΔG was computed with OligoArrayAux32 using temperatures of 55 °C and 57 °C for PTPs from Set 1 and Set 2, respectively. Additionally, the following primer-specific properties were computed: primer length, extent of GC clamp, GC ratio, melting temperature, number of repeats/runs, free energy of secondary structures, and self-dimerization.
For model development purposes, we split the data set into three distinct parts (Table 8). To obtain an independent data set for the selection of classifier cutoffs, 25% of the observations were randomly sampled for inclusion in the validation set. We randomly selected 50% of the remaining observations for inclusion in the training data set, which was used for forming a supervised learning model, and the remainder for inclusion in the test data set, which was used for evaluating model performance.
Feature encoding
In order to investigate the impact of 3′ terminal mismatches, we implemented several encodings, which are illustrated in Fig. 2. The mismatch feature vector z ∈ {0, 1}6 relies on a binary encoding to indicate whether a mismatch was identified at the j-th position in the 3′ hexamer via
Here, j ∈ {1, 2, … 6} identifies the 3′ hexamer position such that j = 1 indicates the first position in the 3′ hexamer and j = 6 indicates the 3′ terminal position. To explicitly model the augmenting effect of co-occurring mismatches in the 3′ hexamer8, the total number of 3′ hexamer mismatches was encoded as \({X}_{N}=\sum _{j}{z}_{j}\).
Since positions closer to the 3′ terminus deteriorate PCR efficiency to a greater degree5,6,12,13,14,15, we encoded the 3′ hexamer mismatch closest to the 3′ terminus by setting
For example, a primer without 3′ mismatches has iX = 0, while a primer exhibiting mismatches at positions 4 and 6 in the 3′ hexamer has iX = 6.
Logistic regression models
We used multivariate logistic regression models in order to investigate the influence of individual features on the template amplification status. Logistic regression is a commonly used approach for problems with categorical outcomes. In this case, we would like to estimate the amplification status yi ∈ {Amplified, Unamplified }. Let p = Pr(yi = Amplified) denote the probability that a template is amplified and let \(\hat{p}\) indicate the corresponding estimated likelihood. Further, let β0 indicate the model intercept and let βi with \(i\in {\mathbb{N}}\) indicate the weight associated with the i-th feature xi. Then the logistic regression model can be formulated as
Due to the small number of evaluated primers, only terms relating to PTPs were considered as features for the logistic regression models. The logistic regression models LR1 and LR2 were used for studying feature importance. While LR1 was defined using the mismatch feature vector z ∈ {0, 1}6, the number of mismatches in the 3′ hexamer (XN), and the free energy of annealing ΔG, LR2 additionally included the terms ix and ΔGiX in order to correct for the association between ΔG and iX.
For the definition of a logistic regression model estimating the probability of amplification, we formulated TMM by performing feature selection using backward stepwise selection. This process was guided by the AIC33, which is defined as
where k is the number of model parameters and \(\hat{L}\) indicates the maximum value of the likelihood function. Starting from a model trained on the LR2 features in the validation set, variables were iteratively eliminated in order to minimize the AIC, thereby ensuring that the final model obtains the best possible fit at the lowest possible complexity.
Further models and classifiers
In addition to TMM, we considered two additional approaches for predicting template amplification status: FE and DE. FE was selected as baseline model because it relies solely on the free energy of annealing ΔG. The model DE is the thermodynamic model of DECIPHER8, which considers the impact of mismatches on the efficiency of polymerase elongation. Since all models provide quantitative outputs, we transformed them to classifiers in the following manner. For FE, we applied the classification rule
where ΔG(x) is the free energy of annealing of sample x and ΔGc is a cutoff on the free energy of annealing. For DE, we performed classification by applying a cutoff ηc on the PCR efficiency η(x) computed by DECIPHER:
Finally, for TMM, we applied a cutoff \({\hat{p}}_{c}\) on the estimated likelihood of amplification \(\hat{p}\):
We selected two cutoffs for each approach: one cutoff ensuring an empiric specificity of at least 99% (denoted by s) and another cutoff maximizing Youden’s index Y = sensitivity + specificity − 1. For FE and DE, which did not require model training, we selected optimal cutoffs by maximizing the two criteria on a data set containing training and validation observations. For TMM, cutoffs were chosen by performing 10 runs of 5-fold cross validation on the validation data set. Finally, all model and classifier performances were determined on the independent test set.
Data Availability
The IGHV data set is available via openPrimeR and figshare (https://doi.org/10.6084/m9.figshare.6736175 for the raw PCR data, https://doi.org/10.6084/m9.figshare.6736232 for the feature matrix). The code pertaining to the analyses is available at http://www.github.com/matdoering/openPrimeR-User/tree/master/src/primerAmplification. The IGHV data set was annotated using the following code: http://www.github.com/matdoering/openPrimeR/tree/master/data-raw/RefCoverage.R.
References
Petropoulos, C. J. et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44, 920–8 (2000).
Hertogs, K. et al. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42, 269–76 (1998).
Foulongne, V., Montes, B., Didelot-Rousseau, M.-N. & Segondy, M. Comparison of the LCx human immunodeficiency virus (HIV) RNA quantitative, RealTime HIV, and COBAS AmpliPrep-COBAS TaqMan assays for quantitation of HIV type 1 RNA in plasma. J. Clin. Microbiol. 44, 2963–6 (2006).
Scheid, J. F. et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011).
Klein, D. et al. Influence of preassay and sequence variations on viral load determination by a multiplex real-time reverse transcriptase-polymerase chain reaction for feline immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 26, 8–20 (2001).
Whiley, D. M. & Sloots, T. P. Sequence variation in primer targets affects the accuracy of viral quantitative PCR. J. Clin. Virol. 34, 104–107 (2005).
Gibson, N. J. The use of real-time PCR methods in DNA sequence variation analysis. Clin. Chim. Acta 363, 32–47 (2006).
Wright, E. S. et al. Exploiting extension bias in polymerase chain reaction to improve primer specificity in ensembles of nearly identical DNA templates. Environ. Microbiol. 16, 1354–1365 (2014).
Yuryev, A. et al. Predicting the success of primer extension genotyping assays using statistical modeling. Nucleic Acids Res. 30, 131e–131 (2002).
Pan, W. et al. DNA polymerase preference determines PCR priming efficiency. BMC Biotechnol. 14, 10 (2014).
Sipos, R. et al. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol. Ecol. 60, 341–350 (2007).
Stadhouders, R. et al. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J. Mol. Diagnostics 12, 109–17 (2010).
Kwok, S. et al. Effects of primer-template mismatches on the polymerase chain reaction: Human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18, 999–1005 (1990).
Bru, D., Martin-Laurent, F. & Philippot, L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl. Environ. Microbiol. 74, 1660–1663 (2008).
Ghedira, R. et al. Assessment of Primer/Template Mismatch Effects on Real-Time PCR Amplification of Target Taxa for GMO Quantification. J. Agric. Food Chem. 57, 9370–9377 (2009).
Ayyadevara, S., Thaden, J. J. & Shmookler Reis, R. J. Discrimination of Primer 3′-Nucleotide Mismatch by Taq DNA Polymerase during Polymerase Chain Reaction. Anal. Biochem. 284, 11–18 (2000).
Day, J. P., Barany, F., Bergstrom, D. & Hammer, R. P. Nucleotide analogs facilitate base conversion with 3′ mismatch primers. Nucleic Acids Res. 27, 1810–1818 (1999).
Huang, M.-M., Arnheim, N. & Goodman, M. F. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 20, 4567–4573 (1992).
Li, B., Kadura, I., Fu, D.-J. & Watson, D. E. Genotyping with TaqMAMA. Genomics 83, 311–320 (2004).
Wu, J.-H., Hong, P.-Y. & Liu, W.-T. Quantitative effects of position and type of single mismatch on single base primer extension. J. Microbiol. Methods 77, 267–275 (2009).
Analytical Methods Committee. PCR – the polymerase chain reaction. Anal. Methods 6, 333–336 (2014).
Lorenz, T. C. Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies. J. Vis. Exp. e3998, https://doi.org/10.3791/3998 (2012).
Thornton, B. & Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 39, 145–154 (2011).
Yuryev, A. PCR Primer Design, https://doi.org/10.1007/978-1-59745-528-2 (Humana Press Inc., 2007).
Döring, M. & Pfeifer, N. openPrimeR, https://doi.org/10.18129/B9.bioc.openPrimeR (2017).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008).
Lvovsky, L. et al. Interdependence between DNA template secondary structure and priming efficiencies of short primers. Nucleic Acids Res. 26, 5525–5532 (1998).
Snyder, T. M., Tse, B. N. & Liu, D. R. Effects of template sequence and secondary structure on DNA-templated reactivity. J. Am. Chem. Soc. 130, 1392–1401 (2008).
Gardner, S. N. et al. Multiplex primer prediction software for divergent targets. Nucleic Acids Res. 37, 6291–6304 (2009).
Ippolito, G. C. et al. Antibody Repertoires in Humanized NOD-scid-IL2Rγnull Mice and Human B Cells Reveals Human-Like Diversification and Tolerance Checkpoints in the Mouse. PLoS One 7, e35497 (2012).
Ye, J., Ma, N., Madden, T. L. & Ostell, J. M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013).
Markham, N. R. & Zuker, M. In Bioinformatics 3–31, https://doi.org/10.1007/978-1-60327-429-6_1 (Humana Press, 2008).
Akaike, H. In Selected Papers of Hirotugu Akaike (eds Parzen, E., Tanabe, K. & Kitagawa, G.) 199–213, https://doi.org/10.1007/978-1-4612-1694-0_15 (Springer, New York, NY, 1998).
Acknowledgements
We would like to acknowledge Nora Speicher for helpful discussions. We acknowledge support by Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Tübingen.
Author information
Authors and Affiliations
Contributions
M.D. developed openPrimeR, performed the analysis of the IGHV data set, and drafted the manuscript. C.K. planned the IGHV template generation and the PCR experiments. C.K. and N.L. generated IGHV templates. N.L. performed PCR reactions. C.K., M.D., and N.L. evaluated PCR reactions. F.K. initiated the project and guided the work. N.P. contributed to the development of openPrimeR and supervised the analysis of the data. All authors contributed to the writing of the manuscript and reviewed the final version.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Döring, M., Kreer, C., Lehnen, N. et al. Modeling the Amplification of Immunoglobulins through Machine Learning on Sequence-Specific Features. Sci Rep 9, 10748 (2019). https://doi.org/10.1038/s41598-019-47173-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47173-w
This article is cited by
-
Effective high-throughput isolation of fully human antibodies targeting infectious pathogens
Nature Protocols (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.