Abstract
Statins have been reported to increase the plasma concentration of arachidonic acid (AA), an omega-6 long chain polyunsaturated fatty acid (LCPUFA) in several clinical studies indicating that statins affect the endogenous synthesis of LCUFAs. In the present study, we investigated the roles of the intrinsic mevalonate cascade and Rho-dependent pathway in LCPUFA synthesis, especially focusing on fatty acid desaturases (Fads) 2, using the human hepatocellular carcinoma cell line HepG2. Cell number and the activity of caspase-3 and 7 (caspase-3/7) was measured using a commercial kit. Gene expression was analyzed by quantitative real-time PCR. Protein expression was detected by Western blot analysis. Atorvastatin decreased cell viability and increased caspase-3/7 activity in a dose-dependent manner. At lower concentrations, atorvastatin stimulated both mRNA and protein expression of Fads2, and increased mRNA expression of FADS1 and ELVOL5. Both mevalonate and geranylgeranyl-pyrophosphate (GGPP), but not cholesterol, fully reversed atorvastatin-induced upregulation of Fads2, and mevalonate-effected reversal was inhibited by treatment with the Rho-associated protein kinase inhibitor Y-27632. These data clearly demonstrated that in human HepG2 cells, statins affect the endogenous synthesis of LCPUFAs by regulation of not only Fads2, but also Fads1 and Elovl5, through the GGPP-dependent Rho kinase pathway.
Similar content being viewed by others
Introduction
Long chain polyunsaturated fatty acids (LCPUFAs) have been demonstrated to play critical roles in the regulation of many biological functions such as brain development, cognition, emotion, reproduction, inflammation, and homeostasis. Arachidonic acid (AA, 20:4 n-6), a 20-carbon 4-double-bond omega-6 LCPUFA, is an “essential fatty acid” not synthesized by the human body. AA is one of the crucial components of membrane phospholipids and the major source of eicosanoids, both of which are lipid modulators of vascular function. Eicosanoids mainly act locally by signaling via specific receptors, and modulate many functions such as vasomotor tone, hemostasis, inflammation, and cell proliferation. In the pathological setting of atherosclerosis, eicosanoids play significant roles since they contribute to endothelial dysfunction and cause inflammation, arterial smooth muscle cell hyperplasia and thrombosis1.
The landmark epidemiological study in Greenlandic Eskimos demonstrated the key role of fish oil (omega-3 LCPUFAs) in the prevention of atherosclerotic diseases2. Following this study, the health benefits of omega-3 LCPUFAs, consisting mainly of eicosapentaenoic acid (EPA, 22:5 n-3) and docosahexaenoic acid (DHA, 22:6 n-3), as part of a fatty acid-rich diet have been researched extensively. Large-scale epidemiological studies, clinical outcome trials, and meta-analyses have demonstrated significant relative cardiovascular risk reduction by intake of omega-3 LCPUFAs3,4,5,6,7. Studies have also shown that omega-3 and omega-6 LCPUFAs have opposing effects on metabolic functions in the body, and the balance between EPA or DHA and AA is likely to be important for regulating the production of mediators and subsequently vascular function in the human body. Indeed, serum EPA to AA ratio (EPA/AA) has been found to be a good biomarker for the risk of cardiovascular disease not only in the general population8, but also in a post-hoc analysis of the results of a clinical trial9.
The important roles of statins for both primary and secondary prevention of cardiovascular disease have been established10, and low-density-lipoprotein cholesterol (LDL-C)-lowering therapy with statins has been used as the first-line treatment. Despite significant LDL-C lowering with statins, substantial residual cardiovascular risk remains11, and several risk factors such as low level of high-density-lipoprotein cholesterol and high level of triglycerides have attracted attention. A particularly interesting finding is that increase in plasma AA concentration and decrease in plasma omega-3 fatty acid concentration and/or plasma omega-3/AA ratio have been observed in patients treated with statins12,13,14,15,16,17, indicating that statins may regulate the endogenous biosynthesis of LCPUFAs. Actually statins have been reported to increase synthesis of AA from linoleic acid (LA, 18:2 n-6) in vivo18,19. Although these effects of statins on endogenous biosynthesis of LCPUFAs may be involved in the residual risk after initiation of statin treatment, its precise molecular mechanism remains unknown.
LCPUFAs are endogenously biosynthesized from omega-6 and omega-3 LCPUFA precursors by position-specific desaturation and carbon-chain elongation reactions20. Endogenous synthesis of LCPUFAs and the degree of unsaturation of the biological membranes depend largely on the actions of fatty acid desaturases Fads1 (∆5-desaturase), Fads2 (∆6-/∆8-/∆4-desaturase) and putatively Fads3, as well as elongation of very long-chain fatty acids proteins (Elovls)21. Recent genome-wide association studies demonstrated that the genes mediating endogenous synthesis of LCPUFAs contribute to wide variability in the efficiency of LCPUFA synthesis, and that synthesis is likely controlled at the levels of Fads2 and Fads1 as well as by the elongases depending on genotype and metabolic state21.
Since Fads2 is known to play an important role in endogenous biosynthesis of AA, EPA and DHA21, we studied the effect of atorvastatin on gene and protein expression of this gene, and its regulatory mechanism via the mevalonate cascade. We used human hepatocellular carcinoma cell line HepG2 in this study, because the liver is the major organ for endogenous biosynthesis of LCPUFAs22, and HepG2 cells have been used extensively in studies of human liver lipid and lipoprotein metabolisms23.
Results
Effect of atorvastatin on HepG2 cell viability
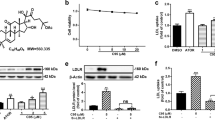
At 48 h, ATR decreased cell viability significantly to 68.0 ± 5.7% at 25 μM, 35.3 ± 2.6% at 50 μM and 12.8 ± 1.5% at 100 μM, apparently in a dose-dependent manner (Fig. 1A). On the contrary, caspase 3/7 activity was significantly stimulated by atorvastatin at 25 μM, and peaked at 50 μM (Fig. 1A).
Effects of atorvastatin on cell viability and caspase3/7 activity in HepG2 cells. (A) Dose-dependent effect of atorvastatin. (B) Effect of mevalonate under treatment with atorvastatin (30 μM). ATR: atorvastatin, MVA: mevalonolactone. Data are presented as mean ± SD of 5 experiments. *p < 0.05, **p < 0.01; one-way ANOVA followed by Bonferroni multiple comparison test.
Both ATR-induced decease in cell viability and increase in caspase 3/7 activity were reversed by the addition of MVA in a dose-dependent manner as shown in Fig. 1B, although MVA did not affect cell viability and caspase 3/7 activity in the absence of ATR (data not shown).
Effect of atorvastatin and mevalonate on FADS1, FADS2, and ELOVL5 expression in HepG2 cells
We examined the effects of ATR on mRNA expression of FADS1, FADS2 and ELOVL5 in HepG2 cells (Fig. 2A). FADS2 mRNA expression increased markedly by the addition of ATR at lower concentrations (871 ± 11.3% at 12.5 μM and 945 ± 15.0% at 25 μM, relative to control), and was followed by a decline at higher concentrations of ATR (517 ± 6.3% at 50 μM, and 91.1 ± 2.8% at 100 μM, relative to control). The decline in FADS2 mRNA expression at higher concentrations of ATR appeared to be associated with decreased cell viability (Fig. 1A). Similarly, ATR-induced increases in FADS1 mRNA expression (292 ± 29.3% at 12.5 μM, 334 ± 50.9% at 25 μM, 229 ± 18.6% at 50 μM, and 86.8 ± 20.7% at 100 μM) and ELOVL5 mRNA expression (208 ± 9.2% at 12.5 μM, 219 ± 10.0% at 25 μM, 241 ± 21.2% at 50 μM, and 154 ± 22.3% at 100 μM) were observed (Fig. 2A). These decreased expression of FADS1, FADS2, and ELOVL5 at higher concentrations of ATR might be linked with its cell toxicity.
Effects of atorvastatin and mevalonate on FADS1, FADS2, and ELOVL5 mRNA and protein expression in HepG2 cells. (A) Dose-dependent effect of atorvastatin on FADS2, FADS1 and ELOVL5 mRNA expression. (B) Effect of mevalonate on FADS2 mRNA and FADS2 protein expression in HepG2 cells treated with ATR (25 μM). The grouping of gel/blots cropped from different gels. ATR: atorvastatin, MVA: mevalonolactone, Data are presented as mean ± SD of 6 experiments. *p < 0.05, **p < 0.01; one-way ANOVA followed by Bonferroni multiple comparison test.
The effects of MVA on ATR-stimulated expression of FADS2 mRNA and protein expression were examined. While ATR at 25 μM stimulated FADS2 mRNA and protein expression, MVA at higher concentrations (1000 and 10000 μM) reversed the upregulation of both mRNA and protein expression in a dose-dependent manner (Fig. 2B). The fulll length gels of protein expression in Western blot analysis were shown in supplementary information.
Effect of isoprenoids and Rho kinase inhibitor Y-27632 in HepG2 cells treated with ATR
Since ATR affected cell viability, caspase3/7 activity and FADS2 mRNA expression, we examined the roles of metabolites in the mevalonate cascade. Although cholesterol did not affect the ATR-induced decrease in cell viability or increase in caspase3/7 activity or FADS2 mRNA expression, 1 mM of MVA and 10 μM of GGPP both reversed all the ATR-induced changes in cell viability, caspase3/7 activity and FADS2 mRNA expression, and 10 μM of FPP reversed some of these changes (Fig. 3A).
Effects of isoprenoids and Rho kinase inhibitor Y-27632 in HepG2 cells treated with ATR. (A) Effect of isoprenoids on cell viability, caspase3/7 activity, and FADS2 mRNA expression in HepG2 cells treated with ATR (25 μM). Data are expressed as mean ± SD of 6 experiments. ATR: atorvastatin, MVA: mevalonolactone, GGPP: geranylgeranyl pyrophosphate, FPP: farnesyl pyrophosphate, Chol: cholesterol. (B) Effect of the Rho kinase inhibitor Y-27632 on FADS2 mRNA and FADS2 protein expression in HepG2 cells treated with ATR (25 μM) and MVA (1 mM). The grouping of gel/blots cropped from different gels. Data are expressed as mean ± SD of 3 experiments. *p < 0.05, **p < 0.01; one-way ANOVA followed by Bonferroni multiple comparison test.
The above results showed that replenishment of MVA and GGPP in culture medium reversed the ATR-induced changes, suggesting that GGPP may be depleted by ATR. Since GGPP is responsible for posttranslational activation of the small GTPase Rho A, it is possible that ATR-induced depletion of GGPP leads to inhibition of Rho A and its effector Rho-associated protein kinase (ROCK), one of the major downstream targets of Rho A. Therefore, we examined the effect of Y-27632, a Rho kinase inhibitor. We found that Y-27632 dose-dependently inhibited the MVA-effected reversal of ATR-induced upregulation of FADS2 mRNA and protein expression in HepG2 cells (Fig. 3B). The fulll length gels of protein expression in Western blot analysis were shown in supplementary information.
Effect of LCPUFAs in HepG2 cells treated by ATR
Since upregulated Fads1, Fads2, and Elovl5 expression may accelerate the metabolism of both omega-3 and omega-6 LCPUFAs, we studied the effect of LCPUFAs such as AA (omega-6), EPA and DHA (both omega-3) on the ATR-induced upregulation of FADS2 mRNA expression in HepG2 cells. EPA and DHA both significantly suppressed FADS2 mRNA expression, but AA did not (Fig. 4).
Effects of DHA, EPA, and AA on FADS2 mRNA expression in HepG2 cells treated with ATR. ATR: atorvastatin, DHA: docosahexaenoic acid, EPA: eicosapentaenoic acid, AA: arachidonic acid. Data are expressed as mean ± SD of 6 experiments. *p < 0.05, **p < 0.01; one-way ANOVA followed by Bonferroni multiple comparison test.
Materials and Methods
HepG2 cell culture
Human hepatocellular carcinoma cell line HepG2 was obtained from the JCRB Cell Bank (Osaka, Japan), and cultured in basal medium consisting of Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 100 IU/mL penicillin-streptomycin (Gibco, Thermo Fisher Scientific, Inc.) at 37 °C in 5% CO2 atmosphere. For the experiments, the basal medium was changed to the experimental medium (DMEM containing 0.5% FBS). Cells were exposed to atorvastatin (ATR; Wako Pure Chemical Industries, Ltd., Osaka, Japan) for 48 h in combination with various agents: mevalonolactone (MVA), geranylgeranyl-pyrophosphate (GGPP), farnesyl pyrophosphate (FPP), cholesterol, DHA, EPA, and AA (all from Sigma-Aldrich; Merck KGaA). To examine the involvement of Rho kinase signaling, the cells were also incubated with Y27632 (Wako Pure Chemical Industries, Ltd.), the selective Rho-associated protein kinase inhibitor, for 24 h after exposure to ATR and MVA.
Cell number and assay of caspase activity
Cell number was assessed at 48 h following the addition of various reagents, by measuring mitochondrial activity that reduces 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt (WST-8) to formazan, using the Cell Counting Kit-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to manufacturer’s protocol.
The activity of caspase-3 and 7 (caspase-3/7) was assessed at 48 h following the addition of various reagents, using the Caspase-Glo 3/7 assay kit (Promega, Corp., Madison, WI) according to manufacturer’s protocol. The assay uses a DEVD-linked luminogenic substrate that is cleaved upon exposure to active caspase 3/7. Cleavage of the substrate results in a luminescent signal generated by activated luciferase.
Quantitative real-time PCR
Total cellular RNA was extracted from HepG2 cells using the RNeasy kit (Qiagen S.A.S., Courtaboeuf, France). The RNA concentration of each sample was determined spectrophotometrically by measuring absorbance at 260 nm. Reverse transcription was performed using the PrimeScript® RT reagent kit (Takara Bio, Inc., Otsu, Japan). The temperature protocol used was as follows: reverse transcription at 37 °C for 15 min, inactivation of reverse transcriptase by heat treatment at 85 °C for 5 s, and storage at 4 °C. Quantification of mRNA expression was performed using SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) on a Mx3005P qPCR system (Agilent Technologies Japan, Ltd., Tokyo, Japan). Thermocycling conditions were as follows: enzyme activation at 95 °C for 30 s, initial denaturation at 95 °C for 5 s, followed by 40 cycles of annealing/extension at 60 °C for 30 s. This was followed by a melt curve analysis according to the manufacturer’s protocol to ensure specific amplification. The following primers were used: human FADS1 sense, 5′-CAG CTT TGA GCC CAC CAA GAA-3′ and antisense, 5′-AGC AAG ATG TGC AGC AGG TAC AG-3′; human FADS2 sense, 5′-TCA TGA CCA TGA TCG TCC ATA AGA A-3′ and antisense, 5′-GCT CCC AGG ATG CCG TAG AA-3′; human ELVOL5 sense, 5′-TGA GGC AGT GGT CAA ACA GGT A-3′ and antisense, 5′-AGA TAT GTC ATG AGT GGT TCC AAGA-3′; human GAPDH sense, 5′-GCA CCG TCA AGG CTG AGA AC-3′ and antisense, 5′-TGG TGA AGA CGC CAG TGGA-3′; and β-actin sense, 5′-TGGCACCCAGCACAATGAA-3′ and antisense 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. In the present study, GAPDH expression was used as an internal standard as reported by others24, since we compared the gene expression of GAPDH and β-actin as internal standards, and both expression was closely correlated in the preliminary experiment. Relative mRNA expression was calculated using the comparative Cq method and was normalized to GAPDH expression25.
Western blot analysis
Whole-cell extracts were prepared in 1× RIPA buffer (Cell Signaling Technology, Inc., Danvers, MA, USA) supplemented with 1 mM PMSF (Cell Signaling Technology, Inc.). Lysates were sonicated briefly and cleared by full-speed centrifugation for 10 min at 4 °C. Protein quantification was performed using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL, USA). Equal amounts of protein (20 to 60 μg) were denatured and resolved by SDS-PAGE on a 10% acrylamide gel, and transferred to PVDF membranes. The membranes were incubated with the following primary antibodies: rabbit antiserum against human FADS2 (1:500; Abcam) and mouse antiserum against human GAPDH (1:5000; Wako Pure Chemical Industries Ltd., Osaka, Japan) as negative control, followed by the following HRP-conjugated secondary antibodies: donkey antiserum against rabbit IgG (1:2000; (GE Healthcare BioSciences, Little Chalfont, UK) and sheep antiserum against mouse IgG (1:2000; GE Healthcare). Bound antibodies were visualized using ECL Prime Western Blotting Detection Reagent (GE Healthcare) and imaged with the ChemiDoc XRS + imaging system (BioRad, Munich, Germany). The fulll length gels of protein expression in Western blot analysis were shown in supplementary information.
Statistical analysis
All data are expressed as mean ± standard deviation (SD). SPSS software (version 11.5, Chicago, IL, USA) was used for statistical processing. Treatment effects were evaluated using one-way ANOVA followed by Bonferroni multiple comparison test, and p values less than 0.05 were considered significant.
Discussion
The main objective of the current study was to verify whether the gene and protein expression of Fads2 is affected by treatment with ATR, a HMG-CoA reductase-specific inhibitor26, and the molecular mechanism involved. This study using HepG2 cells demonstrated that ATR at lower concentrations (12.5 and 25 μM) markedly upregulated the gene and protein expression of Fads2, although the upregulation was attenuated at higher concentrations of ATR (50 and 100 μM), coinciding with markedly decreased cell viability at those concentrations, consistent with previous report27. Atorvastatin at lower concentration also increased mRNA expression of FADS1 and ELVOL5. Moreover, addition of mevalonate or its intermediate metabolite GGPP to cell culture reversed the ATR-induced upregulation of Fads2 and decrease of cell viability, suggesting that the ATR-induced changes may be caused by depletion of mevalonate and GGPP in the cells. Since GGPP is responsible for the posttranslational activation of Rho A, ATR-induced depletion of GGPP probably leads to inhibition of Rho A and its effector ROCK. Indeed, we observed that treatment with a ROCK inhibitor (Y-27632) reversed the MVA-effected suppression of FADS2 gene expression in HepG2 cells treated with ATR. In addition, ATR-induced upregulation of FADS2 gene was suppressed by treatment with EPA and DHA, but not AA.
The effect of statins on endogenous synthesis of LCPUFAs was first reported by Hrboticky et al.18. Using human monocytic Mono Mac 6 (MM6) cells and human hepatoma Hep G2 cells, they found that lovastatin increased AA levels to stimulate thromboxane synthesis, indicating that statin regulates the endogenous biosynthesis of LCPUFAs. Similar change in endogenous synthesis of LCPUFAs induced by simvastatin was also reported19. The in vivo effect of statin on endogenous synthesis of PUFAs was demonstrated in patients treated with simvastatin for 2 months, and simvastatin treatment was associated with an increase in AA content in erythrocyte membrane28. In Japanese patients with hyperlipidemia, plasma AA concentration increased significantly and EPA to AA ratio decreased significantly following statin treatment29. Similar results have been confirmed by other investigators12,13,14,15,16,30. Although these data strongly indicate upregulation of endogenous biosynthesis of LCPUFAs by statins, the molecular mechanism remains unknown.
Since endogenous synthesis of LCPUFAs depends largely on the actions of Fads and Elovls proteins21, the regulation of gene and protein expression of these enzymes may be critical for endogenous synthesis of LCPUFAs. For example, a meta-analysis of 51 human studies on the LCPUFA contents in plasma, erythrocyte or adipose tissue has shown that the proportion of DHA (22:6 n-3) is 37% lower in men than in women31, indicating the important roles of sex hormone in endogenous synthesis of LCPUFAs. An in vitro study demonstrated that progesterone upregulated omega-3 LCPUFA biosynthesis by increasing the mRNA expression of genes involved in this pathway in human liver cells32. Therefore, it is conceivable that statins may affect the expression and functions of Fads and Elovls. In the present study, we clearly demonstrated upregulation of gene and protein expression of FADS2 by atorvastatin in HepG2 cells. Previous study has also shown that simvastatin increases FADS1 and FADS2 gene expression in human lymphoblasts33.
Molecular mechanism of statin-induced upregulation of Fads2 in HepG2 cells remains unclear. Statin-induced suppression of cell proliferation and viability, and statin-induced apoptosis have been reported to be dependent on mevalonic acid and GGPP in various cell types26,34,35 including HepG2 cells27. In the present study, we also demonstrated that co-incubation of HepG2 cells with MVA and its metabolite GGPP, but not cholesterol, fully reversed atorvastatin-induced decrease in cell viability and upregulation of FADS2 gene expression. In addition, the ROCK inhibitor Y-27632 abolished the MVA-effected reversal of atorvastatin-induced upregulation of FADS2 gene in HepG2 cells, which is consistent with our recent report that atorvastatin increases FADS1, FADS2 and ELVOL5 gene expression via the GGPP-dependent Rho kinase pathway in 3T3-L1 cells36. We used 3T3-L1 adipocytes in our previous study34 because these cells are known to have a functional Fads pathway. However, endogenous biosynthesis of LCPUFAs is known to vary in various cell systems18,19. We therefore examined HepG2 cells in this study and found that atorvastatin also increased not only FADS2, but also FADS1 and ELOVL5 gene expression in a GGPP-dependent manner in these cells (data not shown).
Our present study clearly demonstrates that statin upregulates protein and gene expression of FADS2 probably through the GGPP-dependent ROCK pathway in HepG2 cells (Fig. 5), in a similar manner to a recent report of fibulin-2 expression in human coronary artery smooth muscle cells, in which simvastatin increased mRNA level of the ECM protein fibulin-2 through Rho A and Rho kinase-mediated pathway37. Rho A/ROCK signaling pathways play central roles in regulating cell adhesion, migration, motility, contraction, apoptosis and proliferation38,39. Rho A/ROCK signaling pathway is capable of controlling gene and protein expressions40,41, although the mechanism of how RhoA/ROCK affects protein and gene expression of FADS2 is not known from the present study.
It is well known that the activity of Fads is primarily regulated at the transcriptional level by transcription factors such as SREBP 1c42, PPAR α43,44, and Elk45. From this point of view, there were the quite interesting studies, in those the statin-induced inhibition of the Rho-signaling pathway was reported to activate PPAR α activity46, and the treatment of statin increased the PPARα mRNA expression in HepG2 cells47. Although the mRNA expression of neither SREBP 1c, PPARα, nor Elk was not affected by the treatment of ATR in our study (data not shown), the involvement of PPARα remained to be possible since we did study the PPAR α activity in this study.
Another interesting finding in the present study is that EPA and DHA, both omega-3 LCPUFA, significantly suppressed the atorvastatin-induced FADS2 gene upregulation, whereas AA, an omega-6 LCPUFA, showed a tendency of suppression, but was not significant. It is comprehensible that the downstream metabolites of Fads2, such as EPA and DHA, suppress the atorvastatin-induced upregulation of Fads2 in a negative feedback manner, but it is unclear why there is a difference in effect among the downstream metabolites of LCPUFAs (EPA and DHA versus AA).
Recently, the dietary effects of omega-3 and omega-6 LCPUFAs [studied using food with different linoleate/ α-linolenate (omega-6/omega-3) ratios] on gene expression of FADS1, FADS2, ELVOL2 and ELVOL5, as well as LCPUFA composition of liver membrane phospholipids (PLs) were studied in wild type mice (WT) and heterozygous Fads2-null-mice (HET)48. In that study, three types of food with omega-6/omega-3 ratio of 1;1 (control), 7:1 (moderate), and 44:1 (high) were used. A diet with high omega-6/omega-3 ratio dramatically affected liver PLs; increasing AA and decreasing EPA and DHA in a dose-dependent manner. Moreover, the decrease of EPA was markedly greater than the decrease of DHA in both WT and HET mice. Specifically, AA concentration in liver PLs of mice fed a diet high in omega-6/omega-3 ratio (44:1) was three times higher compared to control diet, and EPA and DHA in liver PLs were reduced to 2.5% and 40.7%, respectively, compared to control diet in WT mice. On the contrary, a diet high in omega-6/omega-3 ratio significantly increased the gene expression of FADS1, FADS2, ELVOL2 and ELVOL5 in a dose-dependent manner in the liver of both WT and HET mice. These data indicate that deficiency of omega-3 in the diet causes a reduction of EPA and DHA in liver PLs to increase gene expression of FADS1, FADS2, ELVOL2 and ELVOL5 in the liver. The greater reduction of EPA than DHA may indicate that DHA has physiologically important roles to regulate homeostasis among the omega-3 metabolites. These findings suggest that omega-3 metabolites such as EPA and DHA, may be strongly involved in upregulation of the transcription of Fads and Elovl. Their findings may account for our result that EPA and DHA, but not AA, suppressed the atorvastatin-induced upregulation of these genes.
From the clinical point of view, the observations of increase in plasma AA concentration and decreases in plasma omega-3 LCPUFA concentration and/or omega-3/AA ratio in patients treated with statins are particularly interesting12,13,14,15,16,29,30. Although statins upregulate FADS1, FADS2 and ELVOL5 gene expression to increase endogenous synthesis of LCPUFAs (Fig. 5), it remains unclear from the present study why statin treatment predominantly increases omega-6 (AA), but reduces omega-3 (EPA and DHA) LCPUFAs in patients. There are several possibilities. The substrates for synthesis of omega-6 PUFAs, such as linoleic acid (LA, 18:2n-6), are present in much larger quantities than those for synthesis of omega-3 LCPUFAs, such as α-linolenic acid (ALA, 18:3n-3), in both in vivo and in vitro systems. As a result, conversion to AA may be dominant. This possibility is supported by a report that α-linoleic acid supplementation reduces lovastatin-induced elevation in AA and increases in cellular lipoprotein EPA and DHA levels in Hep G2 cells49. The other possibility is that syntheses of omega-6 and omega-3 PUFAs differ dependent on cell type. In one report, LA, ALA and stearic acid were metabolized differently in THP-1 and HepG2 cells, and simvastatin increased their conversion to a less extent in HepG2 than in THP-1, although the precise mechanism was unknown19.
The AA-dominant endogenous synthesis of LCPUFAs resulting in decreased plasma omega-3 concentration and/or omega-3/AA ratio during statin treatment may be clinically very important, because serum EPA/AA ratio has been reported to be a good biomarker of the risk of cardiovascular disease not only in the general population8, but also in a post-hoc analysis of the results of a clinical trial9. Therefore, it seems reasonable to recommend omega-3 LCPUFAs supplementation for patients on statin treatment, in order to maintain the plasma omega-3 concentration and omega-3/AA ratio, and to suppress the atorvastatin-induced upregulation of Fads and Elovl as demonstrated in the present study. Actually, a clinical study demonstrated the effectiveness of EPA in reducing cardiovascular events in Japanese patients treated with statin4.
The present study has some limitations. We used HepG2 cells as an in vitro model to investigate the endogenous synthesis of LCPUFAs, since it is well known that LCPUFAs in plasma are largely produced by the liver, and hepatic regulation of endogenous synthesis of LCPUFA may be critical. Since HepG2 cell is a cell line derived from hepatoblastoma, and it might be possible that the cancer cell line has the alterations in the metabolic pathways, our data should be evaluated with the consideration from these points. Therefore, it might be good to use the immortalized hepatocyte cell line in future, and it is also necessary to demonstrate statin-induced upregulation of FADS1, FADS2, and ELVOL5 gene expression via the GGPP-dependent Rho kinase pathway in an in vivo model. In addition, another possibility that statin upregulates hepatic lipoprotein clearance to increase delivery of the fatty acids has been reported18. Although we focused on the lower concentrations of ATR due to the reduction of cell viability at its higher concentrations, the study of FADS2 expression and its molecular mechanism at higher concentrations of ATR should have been remained.
In conclusion, statin may affect the endogenous synthesis of LCPUFAs by regulating FADS1, FADS2 and ELVOL5 gene expression via the GGPP-dependent Rho kinase pathway in HepG2 cells.
Date Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Weksler, B. B. Prostanoids and NSAIDs in cardiovascular biology and disease. Curr Atheroscler Rep. 17, 41 (2015).
Bang, H. O., Dyerberg, J. & Nielsen, A. B. Plasma lipid and lipoprotein pattern in Greenlandic West-coast Eskimos. Lancet. 1, 1143–1145 (1971).
Tavazzi, L. et al. Effect of n-3 polyunsaturated fatty acids in patients with chronic heart failure (the GISSI-HF trial): a randomised, double-blind, placebo-controlled trial. Lancet. 372, 1223–1230 (2008).
Yokoyama, M. et al. Effects of eicosapentaenoic acid on major coronary events in hypercholesterolaemic patients (JELIS): a randomised open-label, blinded endpoint analysis. Lancet. 369, 1090–1098 (2007).
Dietary supplementation with n-3 polyunsaturated fatty acids and vitamin E after myocardial infarction: results of the GISSI-Prevenzione trial. Gruppo Italiano per lo Studio della Sopravvivenza nell’ Infarto miocardico. Lancet. 354, 447–455 (1999).
Morris, M. C. et al. Fish consumption and cardiovascular disease in the physicians’ health study: a prospective study. American journal of epidemiology. 142, 166–175 (1995).
Burr, M. L. et al. Effects of changes in fat, fish, and fibre intakes on death and myocardial reinfarction: diet and reinfarction trial (DART). Lancet. 2, 757–761 (1989).
Ninomiya, T. et al. Association between ratio of serum eicosapentaenoic acid to arachidonic acid and risk of cardiovascular disease: the Hisayama Study. Atherosclerosis. 231, 261–267 (2013).
Itakura, H. et al. Relationships between plasma fatty acid composition and coronary artery disease. J Atheroscler Thromb. 18, 99–107 (2011).
Fulcher, J. et al. Efficacy and safety of LDL-lowering therapy among men and women: meta-analysis of individual data from 174,000 participants in 27 randomised trials. Lancet. 385, 1397–1405 (2015).
Sampson, U. K., Fazio, S. & Linton, M. F. Residual cardiovascular risk despite optimal LDL cholesterol reduction with statins: the evidence, etiology, and therapeutic challenges. Curr Atheroscler Rep. 14, 1–10 (2012).
Nozue, T. et al. Comparison of effects of serum n-3 to n-6 polyunsaturated fatty acid ratios on coronary atherosclerosis in patients treated with pitavastatin or pravastatin undergoing percutaneous coronary intervention. Am J Cardiol 111, 1570–1575 (2013).
Kurisu, S. et al. Effects of lipid-lowering therapy with strong statin on serum polyunsaturated fatty acid levels in patients with coronary artery disease. Heart Vessels. 28, 34–38 (2013).
Jula, A. et al. Effects of diet and simvastatin on fatty acid composition in hypercholesterolemic men: a randomized controlled trial. Arterioscler Thromb Vasc Biol. 25, 1952–1959 (2005).
de Lorgeril, M. et al. Lipid-lowering drugs and essential omega-6 and omega-3 fatty acids in patients with coronary heart disease. Nutr Metab Cardiovasc Dis 15, 36–41 (2005).
Harris, J. I. et al. Statin treatment alters serum n-3 and n-6 fatty acids in hypercholesterolemic patients. Prostaglandins Leukot Essent Fatty Acids. 71, 263–269 (2004).
Nakamura, N. et al. Joint effects of HMG-CoA reductase inhibitors and eicosapentaenoic acids on serum lipid profile and plasma fatty acid concentrations in patients with hyperlipidemia. Int J Clin Lab Res. 29, 22–25 (1999).
Hrboticky, N. et al. Lovastatin increases arachidonic acid levels and stimulates thromboxane synthesis in human liver and monocytic cell lines. J Clin Invest. 93, 195–203 (1994).
Rise, P. et al. Differential modulation by simvastatin of the metabolic pathways in the n-9, n-6 and n-3 fatty acid series, in human monocytic and hepatocytic cell lines. Biochem Pharmacol. 69, 1095–1100 (2005).
Abedi, E. & Sahari, M. A. Long-chain polyunsaturated fatty acid sources and evaluation of their nutritional and functional properties. Food Sci Nutr. 2, 443–463 (2014).
Zhang, J. Y., Kothapalli, K. S. & Brenna, J. T. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr Opin Clin Nutr Metab Care. 19, 103–110 (2016).
Jump, D. B. Fatty acid regulation of hepatic lipid metabolism. Curr Opin Clin Nutr Metab Care. 14, 115–120 (2011).
Javitt, N. B. Hep G2 cells as a resource for metabolic studies: lipoprotein, cholesterol, and bile acids. FASEB J. 4, 161–168 (1990).
Singh, A. B. et al. SREBP2 Activation Induces Hepatic Long-chain Acyl-CoA Synthetase 1 (ACSL1) Expression in Vivo and in Vitro through a Sterol Regulatory Element (SRE) Motif of the ACSL1 C-promoter. J Biol Chem. 291, 5373–5384 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408 (2001).
Siddals, K. W. et al. Abrogation of insulin-like growth factor-I (IGF-I) and insulin action by mevalonic acid depletion: synergy between protein prenylation and receptor glycosylation pathways. J Biol Chem. 279, 38353–38359 (2004).
Maeda, A. et al. Down-regulation of RhoA is involved in the cytotoxic action of lipophilic statins in HepG2 cells. Atherosclerosis. 208, 112–118 (2010).
Dwight, J. F., Mendes Ribeiro, A. C. & Hendry, B. M. Effects of HMG-CoA reductase inhibition on erythrocyte membrane cholesterol and acyl chain composition. Clin Chim Acta. 256, 53–63 (1996).
Nakamura, N. et al. Effect of HMG-CoA reductase inhibitors on plasma polyunsaturated fatty acid concentrations in patients with hyperlipidemia. Int J Clin Lab Res. 28, 192–195 (1998).
Nozue, T. & Michishita, I. Statin treatment alters serum n-3 to n-6 polyunsaturated fatty acids ratio in patients with dyslipidemia. Lipids Health Dis. 14, 67 (2015).
Lohner, S. et al. Gender differences in the long-chain polyunsaturated fatty acid status: systematic review of 51 publications. Ann Nutr Metab. 62, 98–112 (2013).
Sibbons, C. M. et al. Effect of sex hormones on n-3 polyunsaturated fatty acid biosynthesis in HepG2 cells and in human primary hepatocytes. Prostaglandins Leukot Essent Fatty Acids. 90, 47–54 (2014).
Reardon, H. T. et al. Insertion-deletions in a FADS2 intron 1 conserved regulatory locus control expression of fatty acid desaturases 1 and 2 and modulate response to simvastatin. Prostaglandins Leukot Essent Fatty Acids. 87, 25–33 (2012).
Tatsuno, I. et al. Geranylgeranylpyrophosphate, a metabolite of mevalonate, regulates the cell cycle progression and DNA synthesis in human lymphocytes. Biochem Biophys Res Commun. 241, 376–382 (1997).
Hirai, A. et al. Geranylgeranylated rho small GTPase(s) are essential for the degradation of p27Kip1 and facilitate the progression from G1 to S phase in growth-stimulated rat FRTL-5 cells. J Biol Chem. 272, 13–16 (1997).
Ishihara, N. et al. Atorvastatin increases Fads1, Fads2 and Elovl5 gene expression via the geranylgeranyl pyrophosphate-dependent Rho kinase pathway in 3T3-L1 cells. Mol Med Rep. 16, 4756–4762 (2017).
Serra, N. et al. Simvastatin Increases Fibulin-2 Expression in Human Coronary Artery Smooth Muscle Cells via RhoA/Rho-Kinase Signaling Pathway Inhibition. PLoS One. 10, e0133875 (2015).
Etienne-Manneville, S. & Hall, A. Rho GTPases in cell biology. Nature 420, 629–635 (2002).
Takai, Y. et al. Rho as a regulator of the cytoskeleton. Trends Biochem Sci. 20, 227–231 (1995).
Cai, A., Li, L. & Zhou, Y. Pathophysiological effects of RhoA and Rho-associated kinase on cardiovascular system. J Hypertens. 34, 3–10 (2016).
O’Cochlain, D. F. et al. Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myopathy and hypotension traits of myotonic dystrophy. Hum Mol Genet. 13, 2505–2518 (2004).
Nara, T. Y. et al. The E-box like sterol regulatory element mediates the suppression of human Delta-6 desaturase gene by highly unsaturated fatty acids. Biochem Biophys Res Commun. 296, 111–117 (2002).
Li, Y., Nara, T. Y. & Nakamura, M. T. Peroxisome proliferator-activated receptor alpha is required for feedback regulation of highly unsaturated fatty acid synthesis. J Lipid Res. 46, 2432–2440 (2005).
Matsuzaka, T. et al. Dual regulation of mouse Delta(5)- and Delta(6)-desaturase gene expression by SREBP-1 and PPARalpha. J Lipid Res. 43, 107–114 (2002).
Lattka, E. et al. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 51, 182–191 (2010).
Martin, G. et al. Statin-induced inhibition of the Rho-signaling pathway activates PPARalpha and induces HDL apoA-I. J Clin Invest. 107, 1423–1432 (2001).
Seo, M. et al. Statins Activate Human PPARalpha Promoter and Increase PPARalpha mRNA Expression and Activation in HepG2 Cells. PPAR Res. 2008, 316306 (2008).
Su, H. et al. Compensatory induction of Fads1 gene expression in heterozygous Fads2-null mice and by diet with a high n-6/n-3 PUFA ratio. J Lipid Res. 57, 1995–2004 (2016).
Hrboticky, N., Zimmer, B. & Weber, P. C. α-Linoleic acid reduces the lovastatin-induced rise in arachidonic acid and elevates cellular and lipoprotein eicosapentaenoic acid and docosahexaenoic acid levels in Hep G2 cells. J Nutr Biochem. 7, 452–471 (1996).
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
Author information
Authors and Affiliations
Contributions
S.T., N.I., S.S., T.Y., T.T. and I.T. conceived and designed the study. Y.W., D.N., M.O., A.S. and I.T. analyzed and interpreted the data. N.I., S.S., S.T., Y.W., D.N., A.S., T.T. and I.T. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
Ichiro Tatsuno received lecture fees from Takeda Pharmaceutical Co., Ltd., and received research grants from Takeda Pharmaceutical Co., Ltd. and Mochida Pharmaceutical Pharma Co., Ltd. The remaining authors declared they do not have anything to disclose regarding conflict of interest with respect to the subject matter or materials discussed in the manuscript.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tanaka, S., Ishihara, N., Suzuki, S. et al. Fatty acid desaturase 2 is up-regulated by the treatment with statin through geranylgeranyl pyrophosphate-dependent Rho kinase pathway in HepG2 cells. Sci Rep 9, 10009 (2019). https://doi.org/10.1038/s41598-019-46461-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-46461-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.