Abstract
We recently identified inhibitors targeting Mycobacterium marinum MelF (Rv1936) by in silico analysis, which exhibited bacteriostatic/bactericidal activity against M. marinum and M. tuberculosis in vitro. Herein, we evaluated the effect of best four inhibitors (# 5175552, # 6513745, # 5255829, # 9125618) obtained from the ChemBridge compound libraries, on intracellular replication and persistence of bacteria within IFN-γ activated murine RAW264.7 and human THP-1 macrophages infected with M. marinum. Inhibitors # 5175552 and # 6513745 significantly reduced (p < 0.05) the intracellular replication of bacilli during day 7 post-infection (p.i.) within RAW264.7 and THP-1 macrophages infected at multiplicity of infection (MOI) of ~1.0. These observations were substantiated by electron microscopy, which revealed the protective effect of # 5175552 in clearing the bacilli inside murine macrophages. Strikingly, # 6513745 displayed synergism with isoniazid against M. marinum in murine macrophages, whereas # 5175552 significantly suppressed (p < 0.05) the persistent bacilli during day 10–14 p.i. in infected RAW264.7 and THP-1 macrophages (MOI of ~ 0.1). Moreover, # 5175552 and # 6513745 were non-cytotoxic to host macrophages at both 1X and 5X MIC. Further validation of these inhibitors against M. tuberculosis-infected macrophages and animal models has potential for development as novel anti-tubercular agents.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is a leading public health problem. In 2017, approximately 10 million people developed TB throughout the world1. There were ~1.3 million deaths among HIV-negative individuals and ~0.3 million deaths among HIV-positive co-infected TB individuals. India accounted for the highest TB burden (27%), including MDR (multi-drug resistant)-TB followed by China and Indonesia. The U.S Food and Drug Administration (FDA) has recently recommended the use of two drugs, i.e. bedaquiline and delamanid for individuals diagnosed with MDR-TB, extensively-drug resistant TB (XDR-TB) and totally-drug resistant TB. However, high doses of antibiotics, long duration of treatment, failure to kill persistent bacteria effectively by the currently used anti-tubercular drugs (ATDs) and the emergence of drug-resistant forms of Mycobacterium tuberculosis necessitates to immediately design new drugs, which shall be effective against all forms of TB2. Various strategies are employed to design ATDs, including the whole cell screening (WCS), target based screening (TBS) and the computer aided drug designing2,3. In WCS, cell-based high throughput screening (HTS) of compound libraries is used to identify inhibitors with whole-cell activity against M. tuberculosis in vitro4. On the contrary, in the genome-derived TBS strategy, compounds that inhibit the biochemical function of a target are identified by HTS of compound libraries or by structure-based drug designing but it does not assure the druggability of target5,6. Though WCS has been more successful in giving possible hits as TBS does not take into consideration the poor penetration and efflux problems, it lacks precise target knowledge4. Virtual ligand screening (VLS) is also considered as an effective tool for in silico screening of large compound libraries and small molecule chemical libraries for obtaining the potential leads against biological targets7,8.
The designing of new ATDs is considerably difficult primarily due to slow growth of M. tuberculosis. M. marinum, a relatively fast-grower has recently been shown to be a useful model (which requires BSL-2 facility) to evaluate the activity of ATDs within infected macrophages9. Zebrafish-M. marinum infection model has also been employed for evaluating ATDs in vivo as a HTS system10. Interestingly, mycobacterial mel2 locus (Rv1936-1941) is M. marinum and M. tuberculosis specific and absent in other pathogenic and nonpathogenic mycobacterial species, which can tolerate reactive oxygen species (ROS) and reactive nitrogen species (RNS) stress response8,11,12. Notably, M. marinum melF of mel2 locus displayed a high similarity to luxA of Vibrio harveyi, which has a significant role in resistance to ROS in bioluminescent bacteria11. M. marinum melF mutants were shown to be defective for bacterial growth in activated murine macrophages and revealed a reduced bacterial growth at late stages in the C57BL/6 mouse footpad model of infection12. Furthermore, the association of mel2 locus in resistance to ROS has been documented during the persistence stage of M. tuberculosis infection in mice13. Using in silico VLS and TBS approach, we identified potent inhibitors targeting M. marinum MelF (~84 kDa, a dimeric protein) that significantly suppressed the flavin oxidoreductase activity of MelF and exhibited bacteriostatic/bactericidal effect against M. marinum and M. tuberculosis in an in vitro system7, however, the effect of these inhibitors against mycobacterium-infected macrophages was not examined. Indeed, macrophages are the key effectors cells to control mycobacterial infections and that also have a niche for their replication14,15, which produce antibacterial ROS and RNS molecules through NADPH oxidase and inducible nitric oxide synthase8,16. Mostly, mycobacteria are resistant to ROS, but RNS display bacteriostatic/bactericidal effect within activated macrophages. We hypothesized that targeting and inhibiting mycobacterial MelF (with anti-ROS/RNS activity) would facilitate efficient clearance of bacteria from the invading macrophages. Therefore, this study was designed to evaluate the effect of in silico designed inhibitors targeting MelF on entry, intracellular replication and persistence of bacteria within IFN-γ activated macrophages infected with M. marinum.
Results
In preliminary studies, the production of ROS levels in all the experiments were evaluated in IFN-γ activated RAW264.7 and phorbol 12-myristate 13-acetate (PMA)-treated THP-1 macrophages by measuring H2O2 production12, which was found to be in the range of 10–12 µM (data not shown).
Evaluation of cytotoxicity of the inhibitors in RAW264.7 and THP-1 macrophages
The in silico designed inhibitors of Chembridge compound libraries are detailed in a recent report7. Among these, the best four inhibitors (# 5175552, # 5255829, # 6513745 and # 9125618) were evaluated for cytotoxicity in RAW264.7 and THP-1 macrophages in this study. We chose 1X and 5X MIC (minimum inhibitory concentration) values of inhibitors ranging from 1.56–62.5 µg/mL to examine cytotoxicity by MTT (3-(4,5-dimethyl thiazol-2yl)-2, 5-diphenyl tetrazolium bromide) assay (Fig. 1). It was observed that >90% of both THP-1 and RAW264.7 macrophages survived at a concentration of 62.5 µg/mL, while >95% of these cells survived at lesser concentrations (7.8–31.25 µg/mL, Fig. 1). Interestingly, each inhibitor was found to be non-cytotoxic (as viability of cells was >95%) even up to 5 X MIC.
Determination of cytotoxicity of inhibitors by MTT assay: (A) Survival of human THP-1 macrophages and (B) murine RAW264.7 macrophages in presence of varying concentrations of inhibitors. Rifampicin was taken as a positive control. Data represent mean ± SD from two independent experiments done in triplicates.
Cell association/entry of M. marinum within activated murine RAW264.7 macrophages
To mimic the actual immune response elicited during an infection, macrophages were activated with IFN-γ before infection to create stringent growth conditions for M. marinum inside the host macrophages12. We evaluated the entry of WT (wild-type) M. marinum, M. marinum ΔMelF (MelF mutant) and WT + inhibitors at 1X MIC in activated murine RAW264.7 macrophages. It was observed that 9.75% of the WT inoculum could associate/enter within RAW264.7 cells at an MOI of ~ 0.2 (Table 1). On the other hand, significantly lesser (p < 0.05) inoculum of ΔMelF, WT + # 5175552 and WT + # 6513745 could associate/enter within murine macrophages as compared to WT alone. It is pertinent to mention that we did not incorporate the aminoglycoside antibiotic such as amikacin/gentamicin in such assays to kill extracellular bacteria for determining the true entry/invasion17,18,19, since presence of amikacin/gentamicin would otherwise give incorrect results. Therefore, we used the term cell association (entry) for such assays.
Similarly, we did not incorporate amikacin/gentamicin in cell culture medium to study the effect of inhibitors on intracellular replication/persistence of M. marinum bacilli within macrophages.
Intracellular replication of M. marinum within IFN-γ activated macrophages
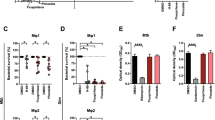
The inhibitors permeated the mycobacterial cells within activated RAW264.7 and THP-1 macrophages at both 1X and 5X MIC (Fig. 2 and Suppl. Table 1). By day 4 p.i., we observed 25.2-fold increase in CFU for WT within RAW264.7 cells, which declined thereafter (Fig. 2A,B). We compared the CFU data of ΔMelF and of WT + inhibitors, with the WT alone. There was 2.5- and 7.4-fold decrease in CFU for ΔMelF on day 4 and 7 p.i., respectively in RAW264.7 macrophages (Fig. 2A,B). However, 3.5-fold decrease in CFU was observed for inhibitor # 5175552 (1X MIC) and 1.9 log decrease in CFU for # 6513745 (1X MIC) within RAW264.7 macrophages on day 4 p.i., compared to WT alone. In a similar manner, 2.2 and 1.9 log differences in CFU were observed for # 5175552 and # 6513745, respectively at 1X MIC on day 7 p.i. However, at 5X MIC, # 5175552 and # 6513745 displayed 3 and 3.5 log differences in CFU, respectively, as compared to WT alone on day 4 p.i.; while 2.6 and 2.8 log decrease in CFU were observed for # 5175552 and # 6513745, respectively within RAW264.7 macrophages on day 7 p.i. thus indicating dose-dependent inhibition of M. marinum bacilli by these inhibitors.
Intracellular replication of M. marinum-infected macrophages at MOI of ~1.0 during 7 days experiment: Intracellular replication of WT M. marinum, ΔMelF, Rifampicin treated WT (Rifampicin) and inhibitor-treated WT (# 5175552, # 5255829, # 6513745, # 9125618) within IFN-γ activated RAW264.7 and THP-1 macrophages at 1X MIC (A,C) and 5X MIC (B,D) on day 0, 2, 4 and 7 p.i. A significant difference (p < 0.05) in intracellular replication was observed between the WT and inhibitor-treated WT (# 5175552, # 5255829 and # 6513745) on day 4 and 7 p.i. Data represent mean ± SD of two independent experiments done in triplicates.
We observed 3.6- and 8.3-fold decrease in CFU for ΔMelF on day 4 and 7 p.i., respectively, compared to WT in THP-1 macrophages (Fig. 2C,D). However, at 1X MIC, CFU reduced to 1.8 and 2 log for inhibitors # 5175552 and # 6513745, respectively, as compared to WT alone on day 7 p.i., while at 5X MIC, 1.5 and 1.8 log decrease in CFU, respectively were observed within THP-1 macrophages on day 7 p.i.
Synergism of # 6513745 with isoniazid (INH) against M. marinum within activated RAW264.7 macrophages
Notably, the inhibitor # 6513745 displayed a synergistic effect with INH against M. marinum within activated RAW264.7 macrophages on day 7 p.i. as INH + # 6513745 revealed a significantly diminished (p < 0.05) bacillary load than INH alone (Fig. 3). Moreover, no cytotoxicity was observed on host macrophages (data not shown).
Synergism of # 6513745 + INH on intracellular replication of M. marinum within activated RAW264.7 macrophages: Growth inhibition of WT M. marinum within murine macrophages at an MOI of ~1.0 in presence of INH (6.25 µg/mL) alone, # 6513745 alone (6.25 µg/mL) and # 6513745 (6.25 µg/mL) + INH (6.25 µg/mL) on day 2 and 7 p.i. Data represent mean ± SD of two independent experiments done in triplicates.
Intracellular survival/killing of persistent M. marinum by the inhibitors within activated macrophages
During the 14 days persistence experiment, WT bacilli increased 15.7- and 13.2-fold by day 7 p.i. within activated RAW264.7 and THP-1 macrophages, respectively and later the bacillary load declined and became steady during day 10–14 p.i. (Fig. 4A–D). On the other hand, ΔMelF revealed 6.3-, 4- and 10.9-fold decrease in CFU within RAW264.7 macrophages on day 7, 10 and 14 p.i., respectively as compared to WT alone (Fig. 4A,B and Suppl. Table 2). Similarly, 4.9-, 6.3- and 9.1-fold decrease in CFU was observed for ΔMelF within THP-1 macrophages on day 7, 10 and 14 p.i., respectively (Fig. 4C,D). Interestingly, a dose-dependent inhibitory effect of # 5175552 was observed in both activated RAW264.7 and THP-1 macrophages since greater (2.85 and 3.4 log) reductions in persistent bacilli were observed with # 5175552 at 5X MIC on day 14 p.i. as compared to 1X MIC (Fig. 4B,D).
Persistence of M. marinum-infected macrophages with MOI of ~ 0.1 during 14 days experiment: Intracellular survival/persistence of WT M. marinum, ΔMelF, Rifampicin-treated WT (Rifampicin) and inhibitor-treated WT (# 5175552, # 5255829, # 6513745 and # 9125618) within IFN-γ activated RAW264.7 and THP-1 macrophages at 1X MIC (A,C) and 5X MIC (B,D) on day 7, 10 and 14 p.i. A significant difference (p < 0.05) in intracellular survival/persistence was observed between the WT and inhibitor-treated WT (# 5175552, # 6513745 and # 9125618) on day 7, 10 and 14 p.i. Data represent mean ± SD of two independent experiments done in triplicates.
Strikingly, the inhibitory effect of # 6513745 was less pronounced as compared to # 5175552 in persistence experiments within both RAW264.7 and THP-1 macrophages on day 10 and 14 p.i. (Fig. 4A–D), thus implying that this inhibitor was less effective against the persistent bacilli as compared to its effect on intracellular replication. Overall, #5175552 seems to be the best inhibitor (followed by # 6513745), since it efficiently inhibited intracellular replication and cleared persistent M. marinum bacilli at both 1X and 5X MIC.
Electron microscopy
Transmission electron microscopy (TEM) clearly revealed the presence of M. marinum bacilli inside both vacuoles and cytoplasm of macrophages infected with WT, ΔMelF, RIF-treated WT and # 5175552-treated WT (Fig. 5). Strikingly, distinct formation of bacterial septa was observed on day 4 and 7 p.i. (shown by arrowheads in Fig. 5E,F) in WT M. marinum-infected RAW264.7 macrophages, which indicated bacterial replication inside the host cell. On day 7 p.i., degraded regions of the host cell infected with WT were clearly visible along with simultaneous lysis of the host cell membrane, which led to cytoplasmic streaming and extrusion of bacilli into the extracellular space (Fig. 5F). In contrast, features such as large number of intracellular bacteria, septum formation, host cell lysis and cytoplasmic streaming were not observed in the uninfected macrophages (UI, Fig. 5A–C) and the macrophages infected with ΔMelF (Fig. 5G–I), RIF-treated WT (Fig. 5J–L) and # 5175552-treated WT (Fig. 5M–O). Also, M. marinum bacilli were found to be tightly apposed to the vacuolar membranes in WT, RIF- and #5175552-treated WT on day 7 p.i. (Fig. 5F,L,O).
TEM analysis: TEM images were acquired on day 2, 4 and 7 p.i. after infection of murine IFN-γ treated RAW264.7 macrophages with WT M. marinum at an MOI of ~1.0. Majority of bacilli were found in vacuoles and cytoplasm, distinct from lysosomes in WT-, ΔMelF-, Rifampicin (RIF)-treated WT- and # 5175552-treated WT-infected macrophages (D–O). Bacilli were also found to be tightly apposed to the vacuolar membranes on day 7 p.i. in WT-, RIF-treated WT- and #5175552-treated WT-infected macrophages (F,L,O). (A–C) Uninfected RAW264.7 macrophages (UI) on day 2, 4 and 7 p.i. Macrophages were healthy with intact cell membrane throughout day 2–7 (A–C). UI macrophages showed distinct plasma membrane, vacuole, nucleus and lysosome. (D–F) WT M. marinum infected RAW264.7 cells. WT infected macrophages showed damaged plasma membrane on day 4 and 7 p.i. (E,F). On day 7 p.i., ruptured plasma membrane, nuclear condensation and cytoplasmic streaming were observed with extrusion of intracellular bacteria and degraded cellular materials (F). Notably, WT M. marinum bacillary replication was evident from the appearance of bacterial septa on day 4 and 7 p.i. (E,F). (G–I) M. marinum ΔMelF infected RAW264.7 cells. ΔMelF infected macrophages showed protection to host cells as evident from lesser cellular damage. However, on day 7 p.i., the plasma membrane of host cell showed a punctate appearance. As compared to WT, lesser number of intracellular bacilli were observed on day 2, 4 and 7 p.i. (G,H,I). Also, the host cell membrane integrity remained intact with no visible damage even on day 4 and 7 p.i. (H,I). (J–L) RIF-treated WT infected RAW264.7 cells. Lesser number of M. marinum bacilli was observed in RIF-treated WT infected host cells than with WT alone on day 2, 4 and 7 p.i. (J-L). Bacterial debris were found to be present both within and outside the vacuoles on day 2, 4 and 7 p.i. In addition, cell membrane of host cells remained intact and the macrophages appeared relatively healthier, as compared to that with WT alone (J–L). (M–O) # 5175552-treated WT infected RAW264.7 cells. Lesser intracellular bacilli were observed in # 5175552-treated WT infected host cells and the macrophages appeared relatively healthier as compared to that with WT, ΔMelF and RIF-treated WT on day 2, 4 and 7 p.i. Interestingly, more extracellular bacilli than intracellular bacilli were distinctly visible on day 4 p.i. (N). Furthermore, bacterial debris were observed in both vacuoles and cytoplasm of infected macrophage on day 7 p.i. (O). Respective scale bar has been indicated in each micrograph.
In the ΔMelF-infected macrophages, lesser number of bacilli were observed on day 2, 4 and 7 p.i. (Fig. 5G–I), as compared to that with WT, which corroborates with the CFU data. The membrane integrity remained intact, without any visible damage to the host cell till day 7 p.i. (Fig. 5G–I). Similarly on day 2, 4 and 7 p.i., RIF- and # 5175552-treated WT infected macrophages showed intact nucleus and host cell membrane integrity with presence of bacillary debris and lesser number of intracellular bacilli (Fig. 5J–O), as compared to that with WT. Of note is the fact that within # 5175552-treated WT infected macrophages, more number of bacilli was observed outside the host cell rather than inside the cell on day 4 p.i., (Fig. 5N). In addition, bacterial debris was observed in both vacuoles and cytoplasm on day 7 p.i. (Fig. 5O). These observations clearly indicate the protective effect of RIF- and #5175552-treated WT in clearing the M. marinum bacilli within RAW264.7 macrophages. To add, # 5175552 exhibited better clearance of M. marinum bacilli than RIF-treated WT.
Discussion
ROS and RNS are the most effective antimycobacterial molecules generated by the host macrophages during infection. Infection of Phox−/− and NOS−/− mice as well as bone marrow-derived macrophages infected with M. tuberculosis clearly suggest that the mel2 locus imparts pathogenesis through its ability to resist host ROS stress8,13. M. tuberculosis is also constantly exposed to endogenous ROS including the production of superoxide radicals during normal aerobic respiration20. We previously demonstrated that the M. marinum MelF mutant was defective for growth in activated J774A.1 cells, which was annulled by inhibitors of ROS scavengers or nitric oxide synthase inhibitors12. In this study, we examined the efficacy of in silico designed inhibitors targeting mycobacterial MelF on intracellular replication and persistence of bacteria within IFN-γ activated RAW264.7 and THP-1 macrophages-infected with M. marinum7.
Comparable to M. marinum strain M-infected RAW264.7 and THP-1 macrophage model of this study, intracellular replication of M. marinum BAA-535 strain within J774A.1 macrophages was previously used to assess the inhibitory activity of compounds targeting isocitrate lyase (ICL, Rv0467)9. IMBI-3, a competitive inhibitor was found to be the most effective compound with an inhibition constant (Ki) of 1.85 μM9. Interestingly, # 5175552 used in this study is also a competitive inhibitor targeting flavin oxidoreductase of MelF with Ki of 68 μM, while # 6513745 is a non-competitive inhibitor with Ki of 18 μM7. Furthermore, Liu et al.9 demonstrated the susceptibility of M. marinum BAA-535 strain to14 of first-line and second-line ATDs (both intracellular and extracellular), thus establishing the possibility of using M. marinum BAA-535 to evaluate anti-TB activity within murine macrophages. Similar to non-replicating M. tuberculosis bacilli which are resistant to INH, M. marinum BAA-535 bacilli persisting inside the J774A.1 macrophages were also found to be resistant to INH, but not to RIF; thus proposing it as a surrogate model to assess the activity of inhibitors on persistent M. tuberculosis9. In this study, we also evaluated the efficacy of MelF inhibitors against the persistent M. marinum strain M within RAW264.7 and THP-1 activated macrophages infected at low MOI of ~ 0.1 during the duration of 14 days p.i.
Similarly, a BSL-2 compatible model for testing the efficacy of drugs has been developed using THP-1 cells-passaged gfp expressing M. tuberculosis mc26206 (ΔpanCD, ΔleuCD), which formed densely packed M. tuberculosis/macrophage aggregate structures. M. tuberculosis mc26206, a derivative of H37Rv strain contains all but four genes, i.e. panCD and leuCD, which are not involved in virulence and drug resistance21,22. The MIC of standard ATDs against this auxotrophic strain was determined and its requirement of l-leucine and pantothenate for replication did not affect the drug susceptibility as compared to the parent H37Rv strain. However, the macrophage-passaged M. tuberculosis might revert back to the virulent BSL-3 compatible mycobacterium, which would then involve a high risk for the handling personnel. Moreover, this auxotroph does not replicate during macrophage infection thus a higher MOI is needed to mimic the intracellular replication observed with the WT M. tuberculosis21. Although M. smegmatis (a fast grower that lacks most M. tuberculosis virulence genes) and M. bovis BCG also surrogate M. tuberculosis and can be handled in BSL-2 conditions; these mycobacterial species have limitations. For example, M. bovis does not naturally infect humans and M. smegmatis has different antibiotic resistance profiles and is killed by the activated macrophages21,23. Therefore, M. marinum-infected macrophages used in this study seem to be an alternative useful model to assess the activity of inhibitors.
Our data on entry and intracellular replication of WT M. marinum and ∆MelF within activated RAW26.7 macrophages (Table 1 and Fig. 2) are in agreement with the previous reports12,19,24, however, intracellular replication was previously examined up to 48 h p.i. only. Bacterial multiplication of WT M. marinum within vacuoles and cytoplasm of RAW264.7 cells, septum formation as well as host cell lysis and cytoplasmic streaming on day 4 and 7 p.i. observed by TEM analysis (Fig. 5) are in accordance with the earlier observations made with M. tuberculosis-infected J774A.1 macrophages and the cultured human lung microvascular endothelial cells17,18. Interestingly, M. marinum bacilli were found to be tightly apposed to the vacuolar membranes of RAW264.7 macrophages infected with WT, RIF- and # 5175552-treated WT on day 7 p.i., which also corroborates with the earlier report25, wherein the virulent mycobacteria survived and probably replicated within a unique tight vacuole in the infected macrophages of the lungs. We demonstrated that the inhibitors # 5175552 and # 6513745 were quite effective in inhibiting the intracellular replication during 7-day and clearing the bacillary persistence during 14-day experiments in IFN-γ activated RAW264.7 macrophages (Figs 2, 4), which was dose-dependent as higher inhibition/clearance of bacillary load was observed at 5X MIC than at 1X MIC. Although these inhibitors were effective in activated THP-1 macrophages but the dose-dependent effect was not observed during 7-day intracellular bacillary replication, whereas the clearance of persisting bacilli during 14-day experiment was dose-dependent. In a similar manner, tamoxifen (a synthetic anti-estrogen) significantly reduced both drug-sensitive and drug-resistant intracellular M. tuberculosis bacilli within RAW264.7 macrophages in a dose-dependent manner26. In addition, thiosulfinate-derivative rich extract of Allium sativum showed better inhibitory activity than its isolated constituents against M. tuberculosis probably due to synergism amongst the constituents of garlic extract, which also revealed a dose-dependent effect within RAW264.7 macrophages27 with no cytotoxicity in host macrophages as determined by MTT assay, whereas our study demonstrated no cytotoxicity with inhibitors on RAW264.7 and THP-1 macrophages even at 5X MIC (Fig. 1). Consistent with the CFU data, the efficient clearance of intracellular M. marinum bacilli within RAW264.7 macrophages was also observed with inhibitor # 5175552 on day 7 p.i. as revealed by TEM analysis (Fig. 5).
Interestingly, the combination of # 6513745 with INH was significantly more effective (p < 0.05) in clearing the bacterial load within activated RAW264.7 macrophages on day 7 p.i. as compared to INH alone (Fig. 3), which could be due to the facilitated penetration of # 6513745 inside the M. marinum bacilli in presence of INH, that acts primarily on mycobacterial cell wall28. These findings are in agreement with our observations with # 6513745 + INH against M. marinum in vitro7. Such synergy could also be due to activation of INH through ROS generator, i.e. # 6513745 (by inhibiting MelF possessing anti-ROS/RNS machinery), thus leading to high susceptibility of mycobacteria within RAW264.7 macrophages. Moreover, compounds such as clofazimine and plumbagin capable of generating ROS through production of superoxide radicals, were previously shown to reduce the MIC of INH for M. tuberculosis8,29. In a similar manner, inhibitor IMBI-3 targeting ICL exhibited a synergistic effect in combination with INH against persistent M. marinum within J774A.1 macrophages9. Also, the efflux pump inhibitors such as verapamil, timcodar, thioridazine, etc. have been shown to synergize with the existing ATDs30,31. For example, Pieroni et al.31 synthesized a number of putative efflux inhibitors based on the structure of thioridazine. Some of those synthesized compounds revealed higher efflux inhibition, lesser cytotoxicity and synergistic bactericidal effect with the existing ATDs against M. tuberculosis in vitro as well as ex vivo (inside the infected human monocyte-derived macrophages)31. Furthermore, timcodar (earlier known as VX-853) displayed synergy with RIF, moxifloxacin and bedaquiline against M. tuberculosis infection within human THP-1 macrophages30.
During the long-term persistence experiments at low MOI of ~0.1 in cultured RAW264.7 and THP-1 macrophages, # 5175552 was found to be more efficacious than # 6513745 in clearing the non-multiplying persistent bacteria during day 10–14 p.i. (Fig. 4). The different effects of inhibitor # 6513745 could be due to different growth conditions, i.e. non-replicating versus replicating within macrophages. In a similar manner, the inhibitory activity of FDA approved bedaquiline (TMC207, diarylyquinoline) against M. tuberculosis in bacteriological liquid medium (7H9 broth) initially revealed a bacteriostatic phase for 7 days and later a dose-dependent bactericidal activity was observed32. However, the same drug initially showed a negligible bacteriostatic phase in murine peritoneal macrophages and J774 macrophages, while the bactericidal effect was noticed on day 5–7 p.i., which was probably due to low bacterial ATP levels in presence of TMC207, thus demonstrating different effects of drug in different growth conditions (in vitro and ex vivo). Moreover, pyrazinamide acts primarily on the non-multiplying persistent bacteria but is inactive against the actively replicating M. tuberculosis bacteria33.
TB treatment becomes considerably difficult due to tolerance displayed by the persistent M. tuberculosis bacilli towards the conventional ATDs and there is an urgent need to devise new drugs that would specifically kill the persisting bacteria located deep inside the tissues. During the persistent stage also, the bacilli are continually exposed to ROS and RNS generated as part of the host response and redox-defense mechanisms are considered vital for the survival of the mycobacterium34. Similar to mel2 locus that is involved in persistence of the tubercle bacilli13, the sulfur metabolism and de novo cysteine biosynthesis pathways are involved in maintaining the redox homeostasis for the persistent tubercle bacilli34, which are thus suggested as potential drug targets to treat latent TB. Protein kinase G, a thioredoxin-fold containing eukaryotic like serine/threonine protein kinase of M. tuberculosis (MtPknG, Rv0410c) also mediates persistence under stressful conditions like hypoxia and assists in drug tolerance35. In an attempt to design inhibitors against MtPknG, Singh et al.36 selected six ligands by employing a sequential pharmacophore-based VLS protocol, wherein three of them exhibited significant MtPknG inhibition, while we employed the structure-based VLS approach for shortlisting MelF inhibitors7. Furthermore, a significant inhibition of the M. bovis BCG bacilli was observed in THP-1 macrophages by the compound NRB04248 with no cytotoxicity to host macrophages36, as observed with inhibitors # 5175552 and # 6513745 on M. marinum-infected THP-1 macrophages in this study. However, the effect of NRB04248 on M. bovis BCG-infected macrophages was examined till 48 h p.i. only36, whereas the long-term intracellular replication and persistence experiments were not performed. A recent trend in ATD development has also been to target the host molecules that are considered crucial for intracellular survival of M. tuberculosis, e.g. tyrosine kinase Src inhibitors significantly reduced the survival of H37Rv as well as MDR and XDR strains of M. tuberculosis within human THP-1 macrophages37, which may develop into host-directed ATDs.
In brief, this study demonstrates that in silico designed inhibitor # 5175552 (N~1~,N~1~-diethyl-N~4~-{6-methoxy-2-[2-(4-nitrophenyl)vinyl]-4-quinolinyl}-1,4-pentanediamine) targeting MelF significantly suppressed the intracellular replication and persistence of M. marinum within activated murine RAW264.7 and human THP-1 macrophages with no cytotoxicity to host macrophages. Similarly, # 6513745 ([3-(4-isobutoxyphenyl)-1-phenyl-1H-pyrazol-4-yl] methanol) suppressed the intracellular replication of M. marinum within activated macrophages and displayed synergism with INH. Validation of these findings in M. tuberculosis-infected macrophages and animal models in BSL-3 facility certainly needs further investigations to find the suitability of these candidates for development as anti-tubercular agents. Nonetheless, this study would undoubtedly set up new directions in designing more ROS generators, which may also improve the effectiveness of existing ATDs.
Methods
Bacterial strains and growth conditions
Wild-type (WT) M. marinum strain M and M. marinum melF mutant (ΔMelF) were received from Dr. Cirillo’s laboratory, Texas A & M University, College Station, TX, USA, which were grown at 33 °C for ~7 days in 7H9 Middlebrook medium supplemented with 0.5% glycerol, 10% albumin dextrose complex (ADC) and 0.05% Tween-80 (M-ADC-TW) and M-ADC-TW supplemented with 10 µg/mL of kanamycin, respectively as detailed in previous papers7,11. M. marinum colony-forming units (CFU) were enumerated on 7H10 (M-ADC) agar7,11, while CFU enumeration of ΔMelF was carried out on 7H10 (M-ADC) agar + kanamycin (10 µg/mL). This work was carried out in Dr. Pandey’s laboratory, after taking approval from the Departmental Committee of THSTI, Faridabad to work on M. marinum in biosafety-hood.
In silico designed inhibitors
We employed a bi-pronged protocol for identifying the potential M. marinum MelF inhibitors based on the consensus between the docking results of DOCK 3.6 and Vina 1.0.2 docking methods as detailed in a recent paper7. Briefly, ~1 million compounds from the ChemBridge compound libraries were filtered using FAFDrugs2 to eliminate compounds with poor ADMETox indices by applying hard-filters. The sdf files of these shortlisted compounds (~ 250,000) were subjected to PDB format using Open Babel 2.2.338. The compounds were subjected to VLS with Vina 1.0.2 and DOCK 3.6. Top 1000 consensus-ranked molecules were shortlisted as putative MelF inhibitors. Among these, 178 molecules were shortlisted for experimental testing based on their docking score, scaffold diversity and the manual assessment of their fit in the targeted site. These 178 inhibitors were procured from Chembridge Corp., San Diego, USA and tested for diminished flavin oxidoreductase activity against the purified M. marinum MelF, which were further shortlisted on the basis of MIC and MBC (minimum bactericidal concentration) values determined against M. marinum and M. tuberculosis in vitro7. Best four inhibitors, i.e. # 5175552, # 5255829, # 6513745 and # 9125618 were evaluated against the M. marinum-infected macrophages in this study. These inhibitors were received as dry powder in vials (each containing 5 mg). A stock solution of 5 mg/mL for each inhibitor was prepared by dissolving in 100% DMSO. The respective stock solutions were further diluted in cell culture medium to attain the desired concentrations for macrophage experiments. The final concentrations of DMSO for these inhibitors were in the range of 0.01–0.5%, which is non-cytotoxic for the cell culture.
Maintenance of RAW264.7 and THP-1 cell lines
Murine RAW264.7 macrophages were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% foetal bovine serum (FBS) and incubated at 37 °C in 5% CO2 with humidified atmosphere. Similarly, human THP-1 monocytic cells were maintained in RPMI-1640 medium supplemented with 10% FBS and incubated at 37 °C in 5% CO2 with humidified atmomsphere.THP-1 monocytic cells were treated with 10 ng/mL PMA to convert them into THP-1 macrophages. During cell association/entry assays, the culture medium was supplemented with 10% FBS. However, the culture medium supplemented with 1% FBS was used for the intracellular replication and persistence experiments in order to slow the growth of the cell culture thus permitting experiments to proceed for 7 and 14 days, respectively17,19. Viability of RAW264.7 and THP-1 macrophages was >95% and >80% during 7-day and 14-day experiments, respectively as assessed by trypan blue exclusion. The MIC of RIF/INH and the inhibitors were employed as reported in a previous paper (0.39 µg/mL for RIF, 6.25 µg/mL for INH, 1.56 µg/mL for # 5175552, 12.5 µg/mL for # 5255829, 6.25 µg/mL for # 6513745, 6.25 µg/mL for # 9125618)7. During the infection studies, cultured macrophages were maintained at 33 °C to provide optimal growth conditions for WT M. marinum and ΔMelF12,39.
Cytotoxicity assay
Cytotoxicity of the inhibitors was studied in murine RAW264.7 and human THP-1 macrophages by MTT (3-(4,5-dimethyl thiazol-2yl)-2,5-diphenyl tetrazolium bromide) assay as described in previous papers with little modifications7,40. Briefly, RAW264.7 and THP-1 cells were plated at a concentration of 1 × 105/well and 2 × 105/well, respectively in 100 µL of cell culture medium +10% heat-inactivated FBS in 96-well plates. After overnight incubation at 37 °C in 5% CO2 incubator with humidified atmosphere, medium was removed and different concentrations of inhibitors were added in 100 µL of culture medium containing 10% heat-inactivated FBS. Also, various concentrations of rifampicin were included in such assays, which served as a positive control. After incubation for 20 h, cells were washed with warm cell culture medium (without FBS) and 100 μL of MTT (0.5 mg/mL) was added. The plates were further incubated for 4 h followed by the addition of 50 µL of DMSO per well, which were mixed thoroughly by a micropipette and left for 45 s. Presence of viable cells was visualized by the development of a purple color due to formation of formazan crystals. The assays were carried out in triplicates and repeated twice to ensure the reproducibility of results. The OD values were read at 570 nm using DMSO as a blank. A standard graph was plotted between the concentration of inhibitors and the relative cell viability, which was calculated as follows:
Entry and intracellular replication of M. marinum in cultured RAW264.7 and THP-1 cells
RAW264.7 and THP-1 macrophages were infected with WT M. marinum, ΔMelF and WT + inhibitors as described previously12,41 with some modifications. Briefly, murine macrophages were seeded in 24-well plates at a concentration of 5 × 105 cells/well and the cells were activated overnight with 100 U/mL of murine IFN-γ (Sigma-Aldrich) prior to infection. A single-cell suspension of dispersed M. marinum growth (OD600 = 0.5–0.6) was used to infect the macrophages. For entry assays, an MOI of ~ 0.2 was employed and 1X MIC of inhibitors was added per well followed by infection with M. marinum for 30 min at 33 °C in 5% CO2 with humidified atmosphere. Following incubation, extracellular bacteria were removed by washing with warm medium (without FBS) and then 1 ml of 0.1% Triton X-100 was added/well. After rigorous pipetting, serial dilutions of lysates were plated for CFU enumeration. The assays were carried out in triplicates and repeated twice to ensure the reproducibility of results.
For intracellular replication/persistence experiments, 106 human THP-1 monocytic cells were pre-treated with 10 ng/mL PMA overnight to result in adherent macrophages in 24-well plates and activated overnight with 100 U/mL of human IFN-γ (Sigma-Aldrich). Both activated RAW264.7 and THP-1 macrophages were infected with M. marinum at an MOI of ~1.0 and ~ 0.1, for intracellular replication and persistence experiments, respectively in 24-well plates for 30 min as described above, followed by washing with warm medium (without FBS) and addition of fresh medium + 1% FBS in presence of 1X and 5X MIC of inhibitors. In a similar manner, RAW264.7 and THP-1 macrophages were also infected with WT and ΔMelF, which served as positive and negative controls, respectively. Since ΔMelF displayed a significantly lesser cell association/entry within the RAW264.7 cells as compared to WT (Table 1), we infected the macrophages with ΔMelF at an MOI of ~1.8 and ~ 0.18 for intracellular replication and persistence experiments, respectively to get approximately the same numbers of bacteria on day 0 p.i. The synergistic effect of # 6513745 (at 1X MIC) in combination with INH at 1X MIC was also evaluated against the intracellular replication of M. marinum for 7 days within RAW264.7 macrophages. We included INH alone (6.25 µg/mL) and # 6513745 (6.25 µg/mL) alone in such experiments at their respective MIC. The 24-well plates were incubated at 33 °C in 5% CO2 with humidified atmosphere for intracellular replication and persistence experiments for 7 and 14 days, respectively. On every alternate day, the media was removed and replenished with fresh warm media containing the respective inhibitors (at 1X and 5X MIC). On different days p.i., the infected monolayers were washed with warm medium (without FBS) and lysed with 1 mL of 0.1% Triton X-100 for 5 min to release the intracellular mycobacteria. Lysis of macrophage monolayer was ascertained by viewing the 24-well plate under a phase-contrast microscope and the serial dilutions of lysates were plated for CFU enumeration. The assays were carried out in triplicates and repeated twice to ensure the reproducibility of results.
Transmission electron microscopic (TEM) analysis
Murine IFN-γ activated RAW264.7 cells were infected with WT M. marinum in T-75 flasks at an MOI of ~1.0 in the absence and presence of # 5175552 (1X MIC). Similarly, RAW264.7 cells were infected with ΔMelF at an MOI of ~1.8 to get the same number of bacteria on day 0 p.i. Infected RAW264.7 cells were harvested on day 2, 4 and 7 p.i., washed thrice with PBS and fixed in freshly prepared 4% paraformaldehyde. The monolayers were incubated at RT for 30 min, washed thrice with PBS and post-fixed with 1% osmium tetraoxide in 0.1 M phosphate buffer at 4 °C for 1 h19,42. Samples were dehydrated with graded acetone, cleared with toluene and infiltrated with toluene and araldite mixture at 25 °C and then kept overnight at 50 °C in pure araldite. Later, the samples were embedded in 1.5 mL eppendorf tubes with pure araldite mixture at 60 °C for ultra-thin section cutting with the help of ultra-microtome (Ultra-microtome Leica EM UC6). Sections were placed on copper grid (3.05 mm diameter and 200 mesh), stained with uranyl acetate and observed at 120 kV under TEM (Model # JEOL2100F with accelerating voltage 80 kV–200 kV and Magnification 50 to 1,500,000X).
Statistical analyses
Statistical analyses were carried out for cytotoxicity, entry, intracellular replication and persistence experiments. Two independent experiments were performed in triplicates assuming that the observations resulted from normal population and homogeneous variance. The average of these independent experiments along with the standard deviation (SD) was calculated for each assay. Student’s t-test was performed to check if there was any significant difference between the mean values of two groups. To show the significance and reproducibility of experiments, R software was employed for all analyses and p-value calculations43. A p-value of <0.05 indicated a significant difference between two independent groups of observations.
Ethics approval and consent to participate
The Departmental committee of THSTI, Faridabad has given ethical approval to work on M. marinum M strain in biosafety-hood.
Data Availability
All the data is available in the manuscript.
References
World Health Organization. Global Tuberculosis Report 2018. https://www.who.int/tb/publications/global_report/en/.
Dharra, R. & Mehta, P. K. Strategies for designing novel anti-tubercular drugs with special reference to mycobacterial MelF (Rv1936) as a target. Mycobact. Dis. 8, 1–3 (2018).
Chetty, S., Ramesh, M., Singh-Pillay, A. & Soliman, M. E. S. Recent advancements in the development of anti-tuberculosis drugs. Bioorganic and Medicinal Chemistry Letters 27, 370–386 (2017).
Mdluli, K., Kaneko, T. & Upton, A. The tuberculosis drug discovery and development pipeline and emerging drug targets. Cold Spring Harb Perspect Med 5, a021154 (2015).
Krieger, I. V. et al. Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase. Chem. Biol. 19, 1556–1567 (2012).
Chim, N. et al. The TB structural genomics consortium: a decade of progress. Tuberculosis 91, 155–172 (2011).
Dharra, R. et al. Rational design of drug-like compounds targeting Mycobacterium marinum MelF protein. PLoS One 12, e:0183060 (2017).
Mehta, P. K., Dharra, R. & Kulharia, M. Could mycobacterial MelF protein (Rv1936) be used as a potential drug target? Future Microbiol. 13, 1211–1214 (2018).
Liu, Y. et al. Identification of a novel inhibitor of isocitrate lyase as a potent antitubercular agent against both active and non-replicating Mycobacterium tuberculosis. Tuberculosis 97, 38–46 (2016).
Carvalho, R. et al. A high-throughput screen for tuberculosis progression. PLoS One 6, e16779 (2011).
Subbian, S., Mehta, P. K., Cirillo, S. L. G., Bermudez, L. E. & Cirillo, J. D. A Mycobacterium marinum mel2 mutant is defective for growth in macrophages that produce reactive oxygen and reactive nitrogen species. Infect. Immun. 75, 127–134 (2007).
Subbian, S., Mehta, P. K., Cirillo, S. L. & Cirillo, J. D. The Mycobacterium marinum mel2 locus displays similarity to bacterial bioluminescence systems and plays a role in defense against reactive oxygen and nitrogen species. BMC Microbiol 7, 4 (2007).
Cirillo, S. L. G. et al. Protection of Mycobacterium tuberculosis from reactive oxygen species conferred by the mel2 locus impacts persistence and dissemination. Infect. Immun. 77, 2557–2567 (2009).
Behar, S. M. et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 4, 279–287 (2011).
Ramakrishnan, L. Revisiting the role of the granuloma in tuberculosis. Nature Reviews Immunol. 12, 352–366 (2012).
Ehrt, S. & Schnappinger, D. Mycobacterial survival strategies in the phagosome: Defence against host stresses. Cell.Microbiol. 11, 1170–1178 (2009).
Mehta, P. K., King, C. H., White, E. H., Murtagh, J. J. & Quinn, F. D. Comparison of in vitro models for the study of Mycobacterium tuberculosis invasion and intracellular replication. Infect. Immun. 64, 2673–2679 (1996).
Mehta, P. K. et al. Identification of Mycobacterium marinum macrophage infection mutants. Microb. Pathog. 40, 139–151 (2006).
Mehta, P. K., Karls, R. K., White, E. H., Ades, E. W. & Quinn, F. D. Entry and intracellular replication of Mycobacterium tuberculosis in cultured human microvascular endothelial cells. Microb. Pathog. 41, 119–124 (2006).
Tyagi, P., Dharmaraja, A. T., Bhaskar, A., Chakrapani, H. & Singh, A. Mycobacterium tuberculosis has diminished capacity to counteract redox stress induced by elevated levels of endogenous superoxide. Free Radic. Biol. Med. 84, 344–354 (2015).
Schaaf, K. et al. A macrophage infection model to predict drug efficacy against Mycobacterium tuberculosis. Assay Drug Dev. Technol. 14, 345–54 (2016).
Sampson, S. L. et al. Protection elicited by a double leucine and pantothenate auxotroph of Mycobacterium tuberculosis in guinea pigs. Infect. Immun. 72, 3031–37 (2004).
Jordao, L., Bleck, C. K. E., Mayorga, L., Griffiths, G. & Anes, E. On the killing of mycobacteria by macrophages. Cell. Microbiol. 10, 529–548 (2008).
El-Etr, S. H., Subbian, S., Cirillo, S. L. G. & Cirillo, J. D. Identification of two Mycobacterium marinum loci that affect interactions with macrophages. Infect. Immun. 72, 6902–6913 (2004).
Moreira, A. L. et al. Sequestration of Mycobacterium tuberculosis in tight vacuoles in vivo in lung macrophages of mice infected by the respiratory route. Infect. Immun. 65, 305–308 (1997).
Jang, W. S. et al. Anti-mycobacterial activity of tamoxifen against drug-resistant and intra-macrophage Mycobacterium tuberculosis. J. Microbiol. Biotechnol. 25, 946–950 (2015).
Nair, S. S., Gaikwad, S. S., Kulkarni, S. P. & Mukne, A. P. Allium sativum constituents exhibit anti-tubercular activity in vitro and in RAW 264.7 mouse macrophage cells infected with Mycobacterium tuberculosis H37Rv. Pharmacogn. Mag. 13, S209–S215 (2017).
Favrot, L. & Ronning, D. R. Targeting the mycobacterial envelope for tuberculosis drug development. Expert Rev. Anti. Infect. Ther. 10, 1023–36 (2012).
Bulatovic, V. M. et al. Oxidative Stress increases susceptibility of Mycobacterium tuberculosis to isoniazid. Antimicrob. Agents Chemother. 46, 2765–2771 (2002).
Grossman, T. H. et al. The efflux pump inhibitor timcodar improves the potency of antimycobacterial agents. Antimicrob. Agents Chemother. 59, 1534–1541 (2015).
Pieroni, M. et al. Rational design and synthesis of thioridazine analogues as enhancers of the antituberculosis therapy. J. Med. Chem. 58, 5842–5853 (2015).
Dhillon, J., Andries, K., Phillips, P. P. J. & Mitchison, D. A. Bactericidal activity of the diarylquinoline TMC207 against Mycobacterium tuberculosis outside and within cells. Tuberculosis 90, 301–305 (2010).
Diacon, A. H. et al. Bactericidal activity of pyrazinamide and clofazimine alone and in combinations with pretomanid and bedaquiline. Am. J. Respir. Crit. Care Med. 191, 943–953 (2015).
Schnell, R., Sriram, D. & Schneider, G. Pyridoxal-phosphate dependent mycobacterial cysteine synthases: Structure, mechanism and potential as drug targets. Biochim. Biophys. acta1854, 1175–1183 (2015).
Khan, M. Z. et al. Protein kinase G confers survival advantage to Mycobacterium tuberculosis during latency-like conditions. J. Biol. Chem. 292, 16093–16108 (2017).
Singh, N., Tiwari, S., Srivastava, K. K. & Siddiqi, M. I. Identification of novel inhibitors of Mycobacterium tuberculosis PknG using pharmacophore based virtual screening, docking, molecular dynamics simulation, and their biological evaluation. J. Chem. Inf. Model 55, 1120–1129 (2015).
Chandra, P., Rajmani, R. S., Verma, G., Bhavesh, N. S. & Kumar, D. Targeting drug-sensitive and -resistant strains of Mycobacterium tuberculosis by inhibition of Src family kinases lowers disease burden and pathology. mSphere 1, e00043–15 (2016).
O’Boyle, N. M. et al. Open Babel: An open chemical toolbox. J. Cheminf. 3, 33 (2011).
Tobin, D. M. & Ramakrishnan, L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell.Microbiol. 10, 1027–1039 (2008).
Senthilraja, P. & Kathiresanb, K. In vitro cytotoxicity MTT assay in Vero, HepG2 and MCF -7 cell lines study of Marine Yeast. J. Appl. Pharm. Sci. 5, 080–084 (2015).
Chen, Y., Yang, F., Wu, S., Lin, T. & Wang, J. Mycobacterium marinum mmar_2318 and mmar_2319 are responsible for lipooligosaccharide biosynthesis and virulence toward Dictyostelium. Front. Microbiol. 6, 1458 (2016).
Walker, L. A. et al. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4, e1000040 (2008).
R Core Team. R: a language and environment for statistical computing, www.R-project.org/ (R Foundation for Statistical Computing, Vienna, Austria, 2018).
Acknowledgements
R.D. acknowledges D.B.T. for providing Junior/Senior Research Fellowship, and MDU, Rohtak for providing University Research Scholarship. T.P. acknowledges JNU-DST-PURSE Phase-II and JNU-UPoE-II (ID: 161) grant support. This work was financially supported by the Department of Biotechnology (D.B.T.), Govt. of India, New Delhi (BT/PR5641/MED/29/557/2012) to P.K.M. and M.K.
Author information
Authors and Affiliations
Contributions
R.D., P.K.M., V.S.R. and T.P. carried out majority of the experiments, participated in data analysis and preparation of the manuscript. Z.T. carried out some experiments. T.P., A.K.P., J.D.C., A.S. and M.K. participated in data analysis. P.K.M. conceived the study, designed the experiments and prepared the final draft of the manuscript. All the authors read the manuscript, participated in editing the manuscript and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dharra, R., Radhakrishnan, V.S., Prasad, T. et al. Evaluation of in silico designed inhibitors targeting MelF (Rv1936) against Mycobacterium marinum within macrophages. Sci Rep 9, 10084 (2019). https://doi.org/10.1038/s41598-019-46295-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-46295-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.