Abstract
Sedimentary bacteria play a role in polymetallic nodule formation and growth. There are, however, limited reports on bacterial diversity in nodule-rich areas of the Central Indian Ocean Basin (CIOB). In this study, bacterial abundance in thirteen sediment cores collected from the CIOB was enumerated, followed by phylogenetic characterisation and, screening of select heterotrophic bacteria for extracellular enzyme activities. Total bacterial counts (TBC) were in the order of 107 cells g−1; there was a significant difference (p > 0.05) among the cores but not within the sub-sections of the cores. The retrievable heterotrophic counts ranged from non-detectable to 5.33 × 105 g−1; the heterotrophic bacteria clustered within the phyla Firmicutes, Proteobacteria and Actinobacteria. Bacillus was the most abundant genus. The extracellular enzyme activities were in the order: amylase > lipase > protease > phosphatase > Dnase > urease. Major findings are compared with previous studies from the CIOB and other areas.
Similar content being viewed by others
Introduction
Polymetallic nodules (PMNs) are found on the seabed and believed to replace the land-based deposits as sources of metals in future1. PMNs are majorly composed of concentric layers of iron and manganese oxides around a core, and they are abundantly found at 4000–6000 m water-depth in major oceans2. PMNs are rich in metals such as manganese (Mn), nickel (Ni), copper (Cu), cobalt (Co) etc. Regions in the Pacific and Indian Oceans are under consideration for industrial-level mining of PMNs1. India is presently conducting environmental impact assessment (EIA) studies in the CIOB (0°–25°S and 70°–90°E)3 to evaluate the possible impact of mining of PMNs on the deep-sea environment (Fig. 1)1. PMNs are formed abiotically, involving hydrogenetic and diagenetic processes, on the seabed. It is hypothesised that microbial activities, especially of sedimentary bacteria, may contribute towards precipitation of metal hydroxides and play a role in formation and growth of the nodules4. These bacteria possibly adsorb metals, including Mn, Fe, Co, Cu and Ni from the deep-sea water and enrich the nodules5,6. Sedimentary bacteria are already known to play crucial roles in nutrient cycling in the deep-sea environment7, with the help of extracellular enzymes8.
Map showing sampling locations in the Central Indian Ocean Basin. The map was prepared using the tool Ocean Data View40 (version 4.7.10), following the attribution guidelines (Schlitzer, R., Ocean Data View, odv.awi.de, 2019).
Chandramohan et al.9, in one of the earliest studies from the Indian Ocean (02°N, 58°E), characterised bacteria present in nodules and associated sediments. They reported the presence of both Mn(II)-oxidising and MnO2-reducing psychrotrophic bacteria in the nodules. Bacterial taxa Arthrobacter, Bacillus, “coryneforms”, Enterobacteriaceae, Micrococcus, Staphylococcus and Vibrio were identified from the nodules and associated sediments. Later, Loka Bharathi & Nair10 reported Acinetobacter, “coryneforms”, Enterobacter, Micrococcus, Moraxella, Pseudomonas and Staphylococcus from the sediments of the CIOB. Naik et al.11 isolated and identified 17 bacterial genera from the sediment samples of the southern and northern regions of the CIOB (Supplementary Table S1).
There are few reports on gene sequence-based identification and taxonomy of culturable bacteria from the CIOB, though there are reports on metagenomic studies [e.g. Das et al.12, Sujith et al.13 and Wang et al.14]. Das et al.12 performed 454 pyrosequencing of bacterial communities of red clays of the CIOB (16°S, 75.5°E) and reported 14 phyla, 25 classes, 38 orders, 52 families and 62 genera. Sujith et al.13 studied bacteria from sea-mount samples from the CIOB (03°S, 83°E) and reported the dominance of Proteobacteria and bacterial genera Halomonas and Thiomicrospira. Recently, Wang et al.14 characterised the microbial communities in a gravity core sediment sample from rare earth element (REY)-rich mud of the Indian Ocean (22°S, 81°E) using Illumina high-throughput sequencing of 16S rRNA genes and revealed the dominance of Proteobacteria, followed by Firmicutes and Actinobacteria (Supplementary Table S1).
During a cruise (SSD-013) to collect baseline geochemical, biochemical and microbiological data from the First Generation Mining site and the Preservation Reference Zone (PRZ) site, we aimed to enumerate down-core variation of bacterial abundance in the deep sub-seafloor sediments of the CIOB. This was followed by 16S rRNA gene-based phylogenetic characterisation and screening of heterotrophic bacterial cultures for extracellular enzyme activities, including amylase, protease, lipase, DNAse, urease, alkaline phosphatase and nitrate reduction.
Results
Abundance of sedimentary bacteria
The water depth at the sampling locations ranged from 4856–5297 m. Down-core variation of total bacterial counts (TBC) in the 13 sediment box-cores is presented in Fig. 2. The TBC were in the order of 107 cells g−1 of dry sediment and higher at 0–4 cm bsf. Highest TBC of 8.12 × 107 cells g−1 of dry sediment were observed in the BC-10, subsection 6–8 cm bsf. Conversely, lowest TBC of 1.10 × 107 cells g−1 of dry sediment were observed in the BC-13, subsection 25–30 cm bsf. Two way ANOVA analysis showed that there is a significant difference (p < 0.05) in the TBC between the sediment cores. However, no significant difference in the TBC was observed within the core sub-sections.
The retrievable counts of heterotrophic bacteria (colony forming units, CFUs) performed in duplicates and averaged later are presented in Fig. 3. The CFUs ranged from non-detectable to 5.33 × 105 g−1 of dry sediment. Down-core variation in the CFUs was higher at 4–6 cm bsf. The highest CFUs were recovered from the BC-02, sub-section 4–6 cm bsf. Higher CFUs were recovered from the upper 10 cm, compared to the deeper sub-sections of the cores (Fig. 3). The heterotrophic bacteria were not detectable in the cores BC-09 and BC-10, though the TBC from the cores were in the order of 107 g−1 of dry sediment.
Phylogenetic characterisation of heterotrophic bacteria
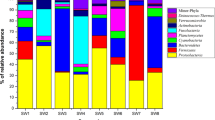
In total, 62 heterotrophic bacteria were isolated. Only 43 bacterial cultures were included in the phylogenetic analysis due to poor viability of the 19 cultures. Phylogenetic relationships of the bacteria, inferred based on 16S rRNA gene sequence-data analysis, are presented in Fig. 4. The system output details retrieved from MEGA 715 about the tree construction are in Supplementary Document. In the NJ tree (Fig. 4), 43 newly generated gene-sequences clustered within three bacterial phyla, Actinobacteria, Firmicutes and Proteobacteria (subphyla α-Proteobacteria and γ-Proteobacteria). The phylogenetic analysis recovered 15 bacterial clades, including Bacillus clades I-V, Oceanobacillus, Staphylococcus (Firmicutes), Alteromonas, Glycocaulis, Paracoccus, Pseudomonas (Proteobacteria), Brachybacterium, Brevibacterium, Micrococcus and Streptomyces (Actinobacteria) (Fig. 4). Location-wise and subsection-wise groupings of the identified clades are presented in Supplementary Tables S2 and S3, respectively.
Screening for extracellular enzyme activities
The enzyme activities of the 43 bacterial cultures followed the order: amylase > lipase > protease > phosphatase > Dnase > urease. Nine cultures were positive for nitrate reduction. Nine Bacillus cultures isolated from 4–6 cm subsection bsf were positive for most of the enzyme activities (Table 1). Streptomyces sp. VJ21 was positive for all the extracellular enzyme activities and nitrate reduction. Staphylococcus sp. VJ01 was positive for only nitrate reduction. Alteromonas sp. VJ25 was negative for all the enzyme activities and nitrate reduction.
Discussion
This is the first study reporting 16S rRNA gene-based phylogenetic characterisation of heterotrophic bacteria from sub-seafloor sediments of PMN-rich areas in the CIOB. The forty-three heterotrophic bacteria clustered within three phyla and 11 known genera (Table 1). Bacillus species were majorly isolated, which formed 5 distinct clades. Bacillus is widely-reported from the CIOB and other parts of the Indian Ocean9,10,11,12,13,16,17, especially in PMNs and associated sediments. Further, four bacteria grouped within Oceanobacillus. Oceanobacillus is a new record from the sediments of the CIOB. It appears that culturable bacilli are abundant in the PMN-rich sediments of the CIOB. Similar patterns of dominance by Firmicutes, including bacilli, were reported by Sujith et al.13, Wang et al.14, and Khandeparker et al.16 from the Indian Ocean. Bacilli are known to survive and grow under physicochemical extremities and with high metabolic versatility. This possibly can explain the abundance of culturable bacilli in the CIOB sediments.
To the best of our knowledge, Brachybacterium, a member of the phylum Actinobacteria, has not been reported from the CIOB (Supplementary Table S1), even though they are known from deep-sea sediments of Southern Ocean18 and Pacific Ocean deep-sea Mn nodule sediment19. This genus is reported for the first time from the CIOB sediment samples. Interestingly, Brachybacterium sp. isolated from the nodules in the Pacific Ocean19 reportedly showed high Mn(II) resistance and Mn(II)-oxidizing/removing abilities. Brevibacterium, a member of “coryneforms” bacteria reported in this study, might have been also isolated from the CIOB sediments and classified under “coryneforms” by Loka Bharathi & Nair10. Chandramohan et al.9 and Sujith et al.17 reported “coryneforms” and Brevibacterium members from the nodules and basal fragments collected from the non-CIOB regions of the Indian Ocean, respectively (Supplementary Table S1).
Glycocaulis, to the best of our knowledge, has not been previously reported from the CIOB (Supplementary Table S1). The genus was originally described from hot water plume of a deep-sea hydrothermal vent from Canada and it reportedly lacks phospholipids20. Role of bacterial phospholipids in deep-sea ecology is subject to future studies. Micrococcus is previously reported from the sediments of the CIOB by Loka Bharathi & Nair10 and Naik et al.11. Chandramohan et al.9 reported this genus from nodules and associated sediments from a non-CIOB region in the Indian Ocean. Micrococcus species are known to have the ability to adapt to elevated pressure21, while some species of Micrococcus may produce antibiotics22.
Pseudomonas species have been reported from the CIOB sediments (Supplementary Table S1) and also from a few non-CIOB regions. Pseudomonas species are reportedly involved in Mn-oxidation5,6, and they possibly adsorb metals such as Co, Cu, and Ni. Interestingly, they may form biofilms which help in the growth of nodules23. Paracoccus has not been previously reported from the CIOB sediments, though the genus is known from deep sediments of the Arctic Ocean24.
Staphylococcus has been reported from the CIOB and the non-CIOB regions (Supplementary Table S1). Recently, Wang et al.25 reported the whole genome sequence of a Mn(II) tolerant Staphylococcus sp. isolated from deep-sea sediment in the Clarion-Clipperton Zone in the Pacific ocean, which exhibited efficient Mn(II) oxidation. Further studies are required from the CIOB about molecular mechanisms and pathways involved in Mn(II) oxidation by bacteria such as Staphylococcus spp. Bacterial genus Streptomyces is reported for the first time from the CIOB sediments. Streptomyces is known for the production of secondary metabolites. Recently, Rodríguez et al.26 reported Streptomyces cyaneofuscatus isolated from a Gorgonian coral collected at 1500 m depth in the Avilés submarine Canyon which produced antibiotic Anthracimycin B. Role of bacterial secondary metabolites, including antibiotics, in deep-sea ecology of the CIOB, needs further investigations.
In our study, we observed that the TBC were two orders higher than the CFUs. The TBC were in the order 107 g−1 of dry sediment. Similar counts were reported by Naik et al.11 from the CIOB, while Raghukumar et al.27 reported TBC two orders higher than our findings. There was no significant difference in the TBC within the core sub-sections, suggesting a homogeneous distribution pattern of total bacterial abundance. Loka Bharathi and Nair10 and Raghukumar et al.27 reported similar homogeneous distribution patterns. Geological studies from the CIOB indicate that sediment characteristics (e.g. porous, mottled and loose)28 and disturbance of sediment layers by biological activities such as bioturbation11 could be responsible for this kind of homogeneous distribution pattern. ISBA guidelines (2018) recommend the use of molecular tools to quantify bacterial abundance in the proposed PMN-mining areas. Traditional methods, therefore, may be replaced with modern molecular tools in microbial abundance studies, in near future E.g. Cho et al.29.
The cultivability of heterotrophic bacteria in our study was about 1% of the TBC. Similar results were reported by Nair et al.30 and Raghukumar et al.27 from the CIOB and by Chandramohan et al.9 from a non-CIOB region. Naik et al.11, however, reported CFU counts one order lesser than the present study. The retrievable counts of heterotrophic bacteria have been used to quantify variations in bacterial abundance under the influence of disturbance in environmental conditions. For example, Loka Bharathi and Nair10 observed an increase in retrievable counts from 102 to 104 g−1 of dry sediment in response to benthic disturbance in the CIOB. Culture-based CFU studies also result in microbial cultures which could be employed in ecological and biotechnological studies.
The 43 heterotrophic bacterial cultures screened for seven extracellular enzymatic activities showed promising results. Hydrolytic degradation of polymers such as starch, protein and lipids by bacterial cultures is a possible indicator of mineralisation of polymers and regeneration of nutrients in the deep-sea27. In this study, 25 bacterial cultures exhibited amylase activity. Chandramohan et al.9, Raghu kumar et al.27 and Loka bharathi and Nair10 have previously reported amylase activity from the bacterial cultures from the Indian Ocean. Loka Bharathi and Nair10 suggested that amylase activity by bacterial cultures from the sediments indicates the availability of free sugar in the CIOB sediments. Lipase activity was observed in 20 bacterial cultures (Table 1), suggesting in situ lipolytic activity of the bacteria9,10,27. Seventeen cultures were positive for protease activity, and possibly these bacteria are involved in break-down of complex proteins present in the sediments into simpler peptides and amino acids. Fourteen cultures were positive for phosphatase activity. Recently, Biche et al.31 reported phosphatase activities of the sediment samples as an indication of available phosphate in the CIOB sediments. The bacterial enzymatic activities of phosphatases and nitrate reductases in the present study indicate that the sedimentary bacterial communities possibly participate in nutrient recycling. These are preliminary observations revealed in this study, which need to be subjected to further research for confirmation.
Conclusion
This is the first study reporting 16S rRNA gene-based phylogenetic characterization of heterotrophic bacteria from the sediments of potential PMN-mining areas in the CIOB. As evident from the present study and other studies, PMNs and associated sediments support phylogenetically-diverse bacterial diversity, which is reportedly distinct from the deep water column32 (Supplementary Table S1). A comprehensive understanding of microbial diversity present in nodule-rich areas of the CIOB is required to assess the impact of large scale removal of nodules and sediments from the deep sea-bed for industrial mining of the PMNs, as suggested Schulse et al.32 in the context of Pacific Ocean. Microbial abundance, diversity and functions can vary across the seabed33, therefore studies employing culture-dependant and -independent methods are required to improve our understanding of microbial diversity and their role in deep-sea ecology.
Methods
Sampling
Sampling was carried out during July–August 2015 with Research Vessel Sindhu Sadhana (SSD-013), as part of ongoing Polymetallic Nodules Program (PMN). The sub-seafloor sediment samples were collected from 13 locations in the CIOB (Fig. 1; Supplementary Table S4), using USNEL-type box-corer of 50 cm × 50 cm × 50 cm dimension.
The box-corer was partitioned into four parts by fixing metallic plates. After siphoning the overlying water, sub-samples were collected using clean 6.3 mm inner diameter plexiglass tubes. On an average, 45 cm bsf long cores were obtained. The cores were sliced at 2 cm intervals up to 10 cm depth and at 5 cm intervals thereafter and extruded aseptically to sterile polythene bags on board. The bags with sediment samples were preserved at 4 °C until further processing.
Abundance of bacteria in deep sub-seafloor sediment samples
For TBC, approximately 1 g of sediment sample was suspended in 9 ml of sterile seawater and processed as detailed in Das et al.34. The DAPI method was used to count the cells. Total bacterial counts were expressed as cells g−1 of dry sediment35.
For retrievable counts of heterotrophic bacteria, each sub-sample was serially diluted to 10–2 with filtered sterile seawater as diluent on-board. Zobell marine agar (ZMA, 12.5%) medium (HiMedia, India) prepared with 50% seawater was spread-plated with 100 μl of original and 10−2 inocula, in duplicates, and incubated at 4 °C for a month and later transferred to 25 °C for another 15 days. After incubation, the plates were examined for retrievable CFUs and calculated per gram of dry sediment.
Bacterial colonies with apparent unique morphology were selected and purified by quadrant streaking method. All the purified bacterial cultures were maintained on ZMA by sub-culturing after every four months as recommended by Bowman36 for further studies. The bacterial cultures were also preserved in 80% glycerol stocks and stored -80 °C for long-term.
DNA extraction and PCR amplification of 16S rRNA gene
Select bacterial cultures were cultivated in Zobell marine broth (ZMB, 12.5%) for 3–7 days at 25 °C, followed by DNA extraction using ZM Fungal/ Bacterial DNA MiniPrep Kit (Zymo Research, D6005). 16S rRNA gene was amplified by PCR using the primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′)37. PCR amplification was performed in a reaction volume of 50 μl, including 5 μl of 10X concentrated buffer (with 15 mM MgCl2) (Genei, Bangalore, India), 1 μl of 10 mM dNTP mix (Genei), 1 μl each of 20 pm/µl 27F and 1492R primers, 1.5 μl of Taq polymerase (1 unit/ μl) (Chromous Biotech, India), 3 μl 20–50 ng template DNA and 37.5 μl nuclease free water.
PCR conditions used were: initial denaturation of 5 min at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 50 °C and 2 min at 72 °C, and a final extension of 72 °C for 10 min. Purification of PCR product was carried out using QIAquick PCR purification columns (QIAGEN, Catalogue number 28106), following the manufacturer’s instructions. The PCR amplicons (DNA) were stored at -20 °C.
Gene sequencing and phylogenetic analysis
The purified PCR products were sequenced using Genetic Analyzer 3130xl (ABI) based on the Big Dye termination v 3.1 (Chain termination) chemistry using the same primer set. For each isolate, a consensus sequence was generated in DNA Dragon (SequentiX, Germany) using both forward and reverse sequences, where possible.
A multiple alignment of 16S rRNA gene sequences, including newly generated sequences from this study and homologous sequences retrieved from NCBI GenBank (Fig. 2), was prepared in MEGA. The 16S rRNA gene sequences generated in this study have been deposited in the NCBI GenBank under accession numbers MH605383–MH605425 (Table 1). Phylogenetic relationships of the bacteria from deep-sea sediments were evaluated in MEGA.
Screening for extracellular enzyme activities
The 43 bacterial cultures were screened for extracellular hydrolytic enzyme activities, including amylases, proteases, lipases, DNases and ureases on agar medium having 1% substrates, i.e. starch, casein, tween 80, toluidine blue and urea, respectively27. Alkaline phosphatase38 and Nitrate reductase39 were also checked for these cultures. After spot inoculation, the plates were incubated for 4 days at 25 ± 1 °C.
Statistical analysis
Two-way analysis of variance (ANOVA) was carried out in Statistica ver. 6 (Statsoft) to find out significant variations in bacterial abundance within the depths and between the cores.
References
Sharma, R. Development of environmental management plan for deep-sea mining. In: Deep-Sea Mining: Resource Potential, Technical And Environmental Considerations, (ed. Sharma, R.), 483–505 (Springer, Gewerbestrasse, Switzerland, https://doi.org/10.1007/978-3-319-52557-0_17 2017).
International Seabed Authority. Polymetallic nodules, https://www.isa.org.jm/files/documents/EN/Brochures/ENG7.pdf (2018).
Banakar, V. K. & Kodagali, V. N. Nodules of the Central Indian Ocean Basin. Deep Ocean Dev. 7‒13 (1988).
Kuhn, T., Wegorzewski, A. Rühlemann, C. & Vink, A. Composition, formation, and occurrence of polymetallic nodules. In: Deep-Sea Mining: Resource Potential, Technical And Environmental Considerations, (ed. Sharma, R.) 23–3 (Springer, Gewerbestrasse, Switzerland, https://doi.org/10.1007/978-3-319-52557-0_2 2017).
Zhang, J., Sun, Q., Zeng, Z., Chen, S. & Sun, L. Microbial diversity in the deep-sea sediments of Iheya North and Iheya Ridge, Okinawa Trough. Microbiological Res. 177, 43–52, https://doi.org/10.1016/j.micres.2015.05.006 (2015).
Sujith, P. P. & LokaBharathi, P. A. Manganese oxidation by bacteria: Biogeochemical aspects. In: Molecular Biomineralization, (ed. Muller WEG), 49‒76 (Prog. Mol. Subcell. Biol. 52, https://doi.org/10.1007/978-3-642-21230-7_3 2011).
Danovaro, R., Molari, M., Corinaldesi, C. & Dell’Anno, A. Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems. Sci. Adv. 2, 1–11, https://doi.org/10.1126/sciadv.1500961 (2016).
Danovaro, R., Dell’Anno, A. & Fabiano, M. Bioavailability of organic matter in the sediments of the Porcupine Abyssal Plain, northeastern Atlantic. Mar. Ecol. Prog. Ser. 220, 25–32, https://doi.org/10.3354/meps220025 (2001).
Chandramohan, D., LokaBharathi, P. A., Nair, S. & Matondkar, S. G. P. Bacteriology of ferromanganese nodules from the Indian Ocean. Geomicrobiol. J. 5, 17–31, https://doi.org/10.1080/01490458709385954 (1987).
LokaBharathi, P. A. & Nair, S. Rise of the dormant: Simulated disturbance improves culturable abundance, diversity and functions of deep-sea bacteria of Central Indian Ocean Basin. Mar. Georesour. Geotechnol. 23, 419–428, https://doi.org/10.1080/10641190500446805 (2005).
Naik, S. S. et al. Relationship of sediment-biochemistry, bacterial morphology, and activity to geotechnical properties in the Central Indian Basin. Mar. Georesour. Geotechnol. 34, 21–32, https://doi.org/10.1080/1064119X.2014.954680 (2016).
Das, A. et al. Astrobiological implications of dim light phototrophy in deep-sea red clays. Life Sci. Space Res. 12, 39–50, https://doi.org/10.1016/j.lssr.2017.01.002 (2017).
Sujith, P. P., Gonsalves, M. J. B. D., Bhonsle, S., Shaikh, S. & LokaBharathi, P. A. Bacterial activity in hydrogenetic ferromanganese crust from the Indian Ocean: a combined geochemical, experimental and pyrosequencing study. Environ. Earth Sci. 76, 191, https://doi.org/10.1007/s12665-017-6495-y (2017).
Wang, S. et al. Microbial community composition and diversity in the Indian Ocean deep sea REY rich muds. PLoS ONE. 13, e0208230, https://doi.org/10.1371/journal.pone.0208230 (2018).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874, https://doi.org/10.1093/molbev/msw054 (2016).
Khandeparker, R., Meena, R. M. & Deobagkar, D. Bacterial diversity in deep-sea sediments from AfanasyNikitin Seamount, Equatorial Indian Ocean. Geomicrobiol. J. 31, 942–949, https://doi.org/10.1080/01490451.2014.918214 (2014).
Sujith, P. P., Mouryaa, B. S., Krishnamurthia, S., Meenaa, R. M. & LokaBharathia, P. A. Mobilization of manganese by basalt associated Mn(II)-oxidizing bacteria from the Indian Ridge System. Chemosphere. 95, 486–495, https://doi.org/10.1016/j.chemosphere.2013.09.103 (2014).
Zhao, B., Liao, L., Yu, Y. & Chen, B. Complete genome of Brachybacterium sp. P6-10-X1 isolated from deep sea sediments of the Southern Ocean. Mar. Genomics. 35, 27–29, https://doi.org/10.1016/j.margen.2017.04.001 (2017).
Wang, W., Shao, Z., Liu, Y. & Wang, G. Removal of multi-heavy metals using biogenic manganese oxides generated by a deep-sea sedimentary bacterium - Brachybacterium sp. strain Mn32. Microbiology. 155, 1989–1996, https://doi.org/10.1099/mic.0.024141-0 (2009).
Abraham, W. R., Lunsdorf, H., Vancanneyt, M. & Smit, J. Cauliform bacteria lacking phospholipids from an abyssal hydrothermal vent: proposal of Glycocaulis abyssi gen. nov., sp. nov., belonging to the family Hyphomonadaceae. Int. J. Syst. Evol. Microbiol. 63, 2207–2215, https://doi.org/10.1099/ijs.0.047894-0 (2013).
Tanaka, T., Burgess, J. W. & Wright, P. C. High-pressure adaptation by salt stress in a moderately halophilic bacterium obtained from open seawater. Appl. Microbiol. Biotechnol. 57, 1–2, https://doi.org/10.1007/s002530100759 (2001).
Tortorella, E. et al. Antibiotics from deep-sea microorganisms: current discoveries and perspectives. Mar. Drugs. 16, 355, https://doi.org/10.3390/md16100355 (2018).
Lindh, M. V. et al. From the surface to the deep-sea: bacterial distributions across polymetallic nodule fields in the Clarion-Clipperton Zone of the Pacific Ocean. Front. Microbiol. 8, 1698, https://doi.org/10.3389/fmicb.2017.01696 (2017).
Russell, J. A., Zayas, R. S., Wrighton, K. & Biddle, J. F. Deep Subsurface Life from North Pond: Enrichment, Isolation, Characterization and Genomes of Heterotrophic Bacteria. Front. Microbiol. 7, 678, https://doi.org/10.3389/fmicb.2016.00678 (2016).
Wang, X. et al. Complete genome sequence of the highly Mn(II) tolerant Staphylococcus sp. AntiMn-1 isolated from deep-sea sediment in the Clarion-Clipperton Zone. J. Biotechnol. 266, 34–38, https://doi.org/10.1016/j.jbiotec.2017.12.004 (2018).
Rodriguez, V. et al. Anthracimycin B, a potent antibiotic against Gram-positive bacteria isolated from cultures of the deep-sea actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs. 16, 406, https://doi.org/10.3390/md16110406 (2018).
Raghukumar, C., Sheelu, G., LokaBharathi, P. A., Nair, S. & Mohandass, C. Microbial biomass and organic nutrients in the deep-sea sediments of the Central Indian Ocean Basin. Mar.Georesour.Geotechnol. 19, 1–16, https://doi.org/10.1080/10641190109353801 (2001).
Mukhopadhyay, R., Ghosh, A. K. & Iyer, S. D. Handbook of Exploration and Environmental Geochemistry- The Indian Ocean Nodule Field: Geology and Resource Potential. Elsevier: Amsterdam (2008).
Cho, H., Kim, K. H., Son, S. K. & Hyun, J. H. Fine-scale microbial communities associated with manganese nodules in deep-sea sediment of the Korea Deep Ocean Study Area in the Northeast Equatorial Pacific. Ocean Sci. J. 53, 337–353, https://doi.org/10.1007/s12601-018-0032-0 (2018).
Nair, S., Mohandass, C., LokaBharathi, P. A., Sheelu, G. & Raghukumar, C. Microscale response of sediment variables to benthic disturbance in the Central Indian Ocean Basin. Mar. Georesour. Geotechnol. 18, 273–283, https://doi.org/10.1080/10641190009353795 (1999).
Biche, S. U. et al. Alkaline Phosphatase: An appraisal of its critical role in C-limited deep-sea sediments of Central Indian Basin. Geomicrobiol. J. 34, 274–288, https://doi.org/10.1080/01490451.2016.1190804 (2016).
Shulse, C. N., Maillot, B., Smith, C. R. & Church, M. J. Polymetallic nodules, sediments, and deep waters in the equatorial North Pacific exhibit highly diverse and distinct bacterial, archaeal, and microeukaryotic communities. Microbiol Open. 6(e428), 1–16, https://doi.org/10.1002/mbo3.428 (2016).
D’Hondt, S. et al. Distributions of microbial activities in deep subseafloor sediments. Science. 306, 2216–2221, https://doi.org/10.1126/science.1101155 (2004).
Das, A., Sujith, P. P., Mourya, B. S., Biche, S. U. & LokaBharathi, P. A. Chemosynthetic activity prevails in deep-sea sediments of the Central Indian Basin. Extremophile. 15, 77–189, https://doi.org/10.1007/s00792-010-0346-z (2011).
Parsons, T. R., Maita, Y. & Lalli, C. M. A manual of chemical and biological methods for seawater analysis. Pergamon Press, Oxford (1984).
Bowman, J. P. Methods for psychrophilic bacteria. Methods in Microbiology. 30, 591–614, https://doi.org/10.1016/S0580-9517(01)30064-8 (2001).
Shinde, V. L., Meena, R. M. & Shenoy, B. D. Phylogenetic characterization of culturable bacteria and fungi associated with tarballs from Betul beach, Goa, India. Mar. Pollut. Bull. 128, 593–600, https://doi.org/10.1016/j.marpolbul.2018.01.064 (2018).
Kannapiran, E. & Ravindran, J. Dynamics and diversity of phosphate mineralizing bacteria in the coral reefs of Gulf of Mannar. J. Basic Microbiol. 52, 91–98, https://doi.org/10.1002/jobm.201100095 (2012).
Smibert, R. M. & Krieg, N. R. Phenotypic Characterization. In: Methods for General and Molecular Bacteriology. (eds Gerhardt, P., Murray, R. G. E., Wood, W. A. & Krieg, N. R.) 607–649 (American Society for Microbiology, Washington DC 1994).
Schlitzer, R. Interactive analysis and visualization of geoscience data with Ocean Data View. Comput. Geosci. 28, 1211–1218, https://doi.org/10.1016/S0098-3004(02)00040-7 (2002).
Acknowledgements
The authors thank the following: Director, CSIR-NIO; Head, Biological Oceanography Division; Ministry of Earth Sciences (MoES), Govt. of India for funding the project GAP 2128; CSIR India for funding under project PSC0206 for DNA sequencing; Drs Rahul Sharma, LokaBharathi, H.N. Khadge, T.N.R. Srinivas and Moturi Shri Ramakrishna for their support and encouragement; and the Master and crew who provided support in scientific operations, during the cruise SSD013. This manuscript has NIO Communication Number 6395.
Author information
Authors and Affiliations
Contributions
M.S.S., S.R.D. and B.D.S. conceived the idea, All authors (V.S.G., M.S.S., S.R.D., D.P., R.M.M. and B.D.S.) conducted experiments; V.S.G., M.S.S. and B.D.S. analysed the results. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gawas, V.S., Shivaramu, M.S., Damare, S.R. et al. Diversity and extracellular enzyme activities of heterotrophic bacteria from sediments of the Central Indian Ocean Basin. Sci Rep 9, 9403 (2019). https://doi.org/10.1038/s41598-019-45792-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45792-x
This article is cited by
-
Microbial Diversity in the Indian Ocean Sediments: An Insight into the Distribution and Associated Factors
Current Microbiology (2022)
-
Bacterial diversity of a floating vegetation (Phumdi) of Loktak Lake and its extracellular enzymes and bacterial antagonistic property
Archives of Microbiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.