Abstract
Patients with inborn errors of amino acid metabolism frequently show neuropsychiatric symptoms despite accurate metabolic control. This study aimed to gain insight into the underlying mechanisms of neural dysfunction. Here we analyzed the expression of brain-derived neurotrophic factor (BDNF) and 10 genes required for correct brain functioning in plasma and blood of patients with Urea Cycle Disorders (UCD), Maple Syrup Urine Disease (MSUD) and controls. Receiver-operating characteristic (ROC) analysis was used to evaluate sensitivity and specificity of potential biomarkers. CACNA2D2 (α2δ2 subunit of voltage-gated calcium channels) and MECP2 (methyl-CpG binding protein 2) mRNA and protein showed an excellent neural function biomarker signature (AUC ≥ 0,925) for recognition of MSUD. THBS3 (thrombospondin 3) mRNA and AABA gave a very good biomarker signature (AUC 0,911) for executive-attention deficits. THBS3, LIN28A mRNA, and alanine showed a perfect biomarker signature (AUC 1) for behavioral and mood disorders. Finally, a panel of BDNF protein and at least two large neural AAs showed a perfect biomarker signature (AUC 1) for recognition of psychomotor delay, pointing to excessive protein restriction as central causative of psychomotor delay. To conclude, our study has identified promising biomarker panels for neural function evaluation, providing a base for future studies with larger samples.
Similar content being viewed by others
Introduction
Although inborn errors of amino acid metabolism (IEM) are treatable disorders, long-term cognitive and behavioral problems are almost constant in patients despite strict dietary management and other treatment approaches. Factors underlying brain dysfunction are not fully understood.
Urea cycle disorders (UCDs, OMIM #311250) are IEM characterized by recurrent hyperammonemic episodes, due to dysfunctions in any of the urea cycle pathway enzymes, a detoxification system that converts ammonia into urea. In the brain, astrocytes are responsible for detoxifying ammonia through glutamine synthetase (GS) and the amidation of glutamate to glutamine1,2. Severe enzyme defects usually manifest symptoms during the first days of life, while symptoms of partial defects tend to appear later in life. Hyperammonemia can damage cerebral tissue by altering cerebral energy metabolism, glutamine/glutamate levels, neurotransmission, and signal transduction of pathways related to neuronal survival and plasticity3,4. Neurocognitive dysfunctions and behavioral impairment represent common long-term outcomes that are likely to reflect chronic ammonia/glutamine toxicity5,6.
Maple syrup urine disease (MSUD; OMIM #248600) is caused by the deficiency of branched-chain α-keto acid dehydrogenase (BCKD) complex activity, a key enzyme for the amino acid catabolism, leading to increased levels of branched-chain amino acids (BCAAs) and their corresponding α-ketoacids7. The BCKD complex is composed of three catalytic components: E1, E2 and E3 and two regulatory enzymes, the BCKD kinase (BCKDK) and BCKD phosphatase (PP2Cm)8. Mutations in BCKDK (OMIM #614923) are associated with decreased levels of BCAAs, epilepsy, intellectual disability and autism9.
Patients with the most severe form of MSUD develop metabolic decompensation and encephalopathy within the first weeks of life and die if untreated7,10,11. Therapy is based on strict dietary management with low BCAAs (mainly leucine) content and/or liver transplantation12,13. Despite treatment, several studies have described small reductions in the intelligence quotient and increased rate of neuropsychiatric conditions, mainly attention deficit hyperactivity disorder (ADHD), depression and anxiety14,15,16,17,18. Neurotoxicity of BCAAs and their α-ketoacids, mainly leucine, might underlie neuropsychiatric disorders, as leucine plasma concentrations are closely correlated with acute symptoms17. Due to blood-brain barrier (BBB) amino acid transporter saturation, increased BCAAs and α-ketoacids in the brain, are concomitant with deficiency of essential amino acids, causing brain-specific deficits in protein synthesis, neurotransmitter depletion and inhibition of mitochondrial enzymes8,19.
Early detection and treatment is the gold standard to avoid long-term disabilities in IEM patients. Therefore, there is a need to find new biomarkers of neural function for the identification and management of those children that, despite metabolic management, are at risk to develop neurologic, cognitive and behavioral problems.
One currently accepted biomarker candidate for brain functioning is brain-derived neurotrophic factor (BDNF), as BDNF signaling is involved in brain development, synaptic function and plasticity20. Activity-dependent regulation of BDNF expression is in part mediated by transcription factor methyl-CpG binding protein 2 (MeCP2), and altered levels of MeCP2, BDNF or their downstream signaling are widely implicated in neuropsychiatric diseases and mood disorders21,22. Altered BDNF levels in plasma have been recently described in MSUD patients and the hippocampus of hyperammonemic rats23,24. BDNF is also involved in glucose and energy homeostasis through control of energy intake and expenditure25, in part by regulation of miRNA and protein synthesis through LIN28A RNA-binding protein26,27. Additional candidate biomarkers are CACNA2D1-2 genes, which codify for α2δ1-2 regulatory subunits of the voltage-gated calcium channels (VGCCs)28. Aside from its modulatory channel function, α2δ1-2 are essential for the formation and stabilization of new synapses by binding to oligomeric extracellular matrix glycoproteins thrombospondins (TSPs), which are codified by THBS genes29. L-leucine and L-isoleucine are also well-known ligands of α2δ1-2 subunits30 and have been postulated as necessary for their correct function31,32.
Therefore, to gain insight into the molecular mechanisms underlying neurological deficits in IEM patients, we analyzed in peripheral whole blood the mRNA expression of a group of genes involved in brain development and synaptic function (ADORA2A, CACNA2D2, FMR1, IRAK1, LIN28A, PTEN, MECP2 E1/E2, THBS1, THBS3) in a cohort patients affected by UCD, MSUD and BCKDK deficiency. Gene expression in blood was correlated with AAs profile, BDNF protein levels in plasma, and neuropsychological symptoms, in an attempt to identify specific molecular pathways altered in each disorder, as well as common mechanisms underlying the pathophysiology of the neurological impairment present in different IEMs.
Results
Clinical information of patients
Table 1 shows a summary of clinical data and Table 2 shows treatment details and historical biochemical parameters of IEM patients included in this study.
UCD patients (7 males and 12 females) include ornithine transcarbamylase deficiency (OTC deficiency) (14), citrullinemia type I (CTLN1) (3), hyperornithinemia-hyperammonemia-homocitrullinuria syndrome, (HHH syndrome) (1) and argininosuccinate lyase deficiency (ASL deficiency) (1), and five of them (26%) were asymptomatic. All UCD patients were treated with L-Arginine +/− L-Citrulline and all but two followed a protein-restricted diet (0.5–1,3 g/Kg/day). Asymptomatic patients showed mean glutamine levels <800 µmol/L; while half of symptomatic patients showed mean glutamine levels ≥800 µmol/L. Eight symptomatic patients (42%) were treated with ammonia scavengers, all with sodium phenylbutyrate (NaPB) and one also with sodium benzoate. Patients without scavengers were those with normal ammonia and amino acids in the follow-up metabolic controls. MSUD patients (3 males and 6 females) include mutations in DBT (2) and BCKDHB (6), one of them asymptomatic. We also included a patient with a genetic diagnose of BCKDK deficiency. MSUD patients followed a low BCAA diet while BCKDK deficiency patient received a high protein diet supplemented with BCAAs. Despite low BCAA diet, mean leucine levels ≥200 µmol/L (normal range in young patients <200 µmol/L) were observed in 78% of MSUD patients.
From the 29 IEM patients in our cohort, 21 symptomatic patients had a neuropsychological evaluation to detect abnormal executive functions and attention deficits (15 patients) cognitive disabilities (7 patients) and other behavioral disorders (7 patients). Three patients of those who fulfilled ADHD criteria and had poor academic results were treated with psychostimulants (methylphenidate). Neuropsychological evaluation was not performed in healthy control subjects (n = 27) and in asymptomatic patients that achieved successfully both academic and personal life events (n = 8).
Analysis of metabolite levels
The 21 principal amino acids and 4 amino acid related-compounds were measured in all plasma samples by ion-exchange chromatography with nynhydrin detection derivatives. Ammonia levels were measured only in OTC patients. Average values for each condition are shown in Table 3.
Despite metabolic management, several mean AAs values were out of normal range both in UCD and MSUD groups (Table 3). The mean ammonia level in the UCD group was in the normal range but nitrogen-rich AAs glutamine (p > 0.0001) and asparagine (p = 0.014) were significantly increased in UCD patients with respect to controls; and as were with glycine (p = 0.014), methionine (p > 0.0001) and histidine (p = 0.000). As expected, citrulline was only increased in patients with ASS deficiency (p = 0.04). When segregating UCD patients by treatment, patients treated with ammonia scavengers showed specific downregulations of phenylalanine (p = 0.045) and BCAAs (Val p = 0.013; Leu p = 0.003; Ile, p = 0.015) with respect to untreated UCD patients and phenylalanine (p = 0.032) and valine (p = 0.041) also with respect to controls.
BCAAs levels were high in most MSUD patients with leucine (p = 0.01) and isoleucine (p = 0.000) over the control group. glycine (p = 0.000), taurine (p = 0.000), serine (p = 0.004), threonine (p = 0.002), glutamate (p = 0.024), asparagine (p = 0.019), histidine (p = 0.017) and ornithine (p = 0.026) were also significantly elevated in MSUD group with respect to controls. The only BCKDK deficiency patient analyzed showed normal BCAAs, a marked decrease in α-Aminobutyric acid (AABA) and levels above the normal range in other twelve AAs (Table 3).
mRNA and protein expression analysis
To ensure the reliability of RT-PCR analysis we set strict sample exclusion criteria (see patients and methods) and as a result, 27 controls, 16 UCD, 7 MSUD, and 1 BCKDK deficiency patients were included in mRNA and further analysis. In addition, outliers were also excluded from the analysis.
A summary of significant differences in mRNA RT-PCR analysis is shown in Table 4. MSUD and UCD groups exhibit significant differences in mRNA gene expression profiles with respect to controls and between them. All genes analyzed in the BCKDK deficiency patient were in the control range. In MSUD patients, CACNA2D2, and MECP2 E1 and E2 isoforms mRNAs were significantly downregulated (p = 0.007; p = 0.027; p = 0.001 respectively), while THBS1 mRNA was upregulated (p = 0.047) with respect to controls. In UCD patients, THBS1 and LIN28A mRNA were upregulated with respect to controls (p = 0.031; p = 0.023 respectively). No differences between groups were found for FMR1, IRAK1, PTEN, and THBS3.
When UCD patients were segregated by ammonia scavenger treatment, ADORA2A mRNA was significantly increased in those treated with scavengers (n = 8) with respect to control group (p = 0.042) and UCD untreated group (n = 8, p = 0.021). In addition, MECP2 E1 (p = 0.047) and THBS1 (p = 0.031), mRNA were significantly increased in those treated with scavengers with respect to control group, but not to untreated UCD patients. Taken together these results suggest that ammonia scavenger treatment might directly increase, THBS1, MECP2 E1, and ADORA2A mRNA expression.
To evaluate the protein expression of the main potential gene biomarkers identified in this study, we performed western blot analysis in purified leukocytes from the same blood samples. We found that protein levels of α2δ2 and MeCP2 in leukocytes from MSUD patients (n = 8) was significantly lower than in controls (n = 5) (p = 0.011, p = 0.048, respectively), corroborating our mRNA results in whole blood (Fig. 1d). Finally, BDNF protein was measured by ELISA in plasma from the same samples included in the mRNA analysis. BDNF levels were highly variable, even in controls, and although no statistically significant differences were found between groups, there was a tendency of lower levels of BDNF in UCD patients (p = 0.061).
ROC curves of different neural function biomarker combinations for diagnosis between MSUD or UCD patients and healthy controls. (a) UCD; (b) MSUD mRNA; (c) MSUD protein; (d) Representative western blot and densitometric analysis showing significant differences for α2δ2 and MeCP2 in MSUD patients (n = 8) and controls (n = 5). *p < 0.05, Mann-Whitney U test.
Spearman correlation between levels of candidate biomarkers and amino acids
We then investigated correlations among potential neural function biomarkers and relevant amino acids using Spearman coefficient of pairwise comparison between samples (Fig. 2). Correlations were performed between each candidate biomarker for the overall population and relevant statistically significant correlations were identified from the correlation matrix heat map obtained. Spearman coefficient (ρ) of 1 or −1 represents perfect positive or negative correlation respectively.
Correlations matrix heat maps. Heat map of Spearman correlation between amino acids and potential gene biomarkers performed between each biomarker for the overall population analyzed. 1 is positive correlation. 0 no correlation and −1 is negative correlation. Statistically significant correlations (p < 0.05) are black boxed.
We found a positive correlation between glutamine levels in plasma and mRNA levels in blood of THBS1 (ρ = 0.342; p = 0.025) and THBS3 (ρ = 0.313; p = 0.041), which were also positively correlated between them (ρ = 0.449; p = 0.005). In addition, THBS1 mRNA positively correlated with LIN28A mRNA (ρ = 0.312; p = 0.047), and negatively with BDNF protein levels (ρ = −0.348; p = 0.019); while THBS3 mRNA positively correlated with mRNA levels of MECP2 e1 and e2 isoforms (ρ = 0.353/0.433; p = 0.032/0.007 respectively), PTEN (ρ = 0.450; p = 0.006) and FMR1 (ρ = 0.447; p = 0.033). We also found a significant strong negative correlation between mean historical leucine levels and BCAAs receptor CACNA2D2 mRNA (ρ = −0.857; p = 0.014); and a significant positive correlation between mean historical leucine levels and taurine (ρ = 0.648; p = 0.043). Consistently, CACNA2D2 mRNA also had a significant negative correlation with taurine (ρ = −0.420; p = 0.006), isoleucine (ρ = −0.377; p = 0.015) and glutamate (ρ = −0.378; p = 0.015), three AAs that are significantly increased in MSUD patients.
Identification of UCD and MSUD gene biomarker signature
To validate the potential diagnostic and recognition effectiveness of neural function biomarker panels, ROC analysis was applied and the area under the ROC curve (AUC) was calculated for each candidate biomarker. AUC value varies from 0 to 1, where values 0.8 > AUC < 0.9 and 0.9 > AUC < 1 reflect good and very good biomarker performance respectively.
We first tested sensibility and specificity of candidate biomarkers altered in MSUD or UCD patients with respect to control group, and a summary of relevant potential gene biomarkers is shown in Table 5 and Fig. 1. From BDNF and the 10 genes analyzed in this study only LIN28A (AUC = 0.712) and THBS1 (AUC = 0.707) exhibit fair biomarker accuracy for UCD. On the contrary, in the case of MSUD, good or very good biomarker performance was found for MECP2 (AUC E2 mRNA = 0.942, E1 mRNA = 0.802, MeCP2 protein = 0.857), CACNA2D2 (AUC CACNA2D2 mRNA = 0.865, α2δ2 protein = 0.925) and with less accuracy THBS1 (AUC mRNA = 0.791). Biomarker specificity and sensitivity are maximal when MECP2 E2 and CACNA2D2 mRNA are simultaneously considered (AUC mRNA = 1, protein = 0.95). Taking together, these results suggest that altered α2δ2 and MeCP2 signaling might be involved in the neural function deficits present in MSUD patients.
Identification of gene biomarker signature of neural function in IEM patients
We used the Kruskal Wallis test to identify significant correlations between candidate biomarkers and the presence of neuropsychological symptoms. No significant correlations were found between individual candidate mRNA biomarkers or BDNF and neuropsychological symptoms within MSUD or UCD patients, and the only BCKDK deficiency patient included in the study precludes this type of analysis.
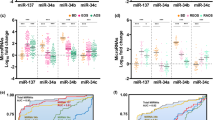
To identify common traits for neuropsychological deficits in IEMs we then analyzed all patient samples as a whole. Low BDNF and threonine levels in plasma, a clinical indicator of protein restriction, have a statistically significant correlation with impaired psychomotor development (BDNF p = 0.035; Thr p = 0.009) (Fig. 3a). On the other hand, increased LIN28A mRNA expression in blood and increased levels of alanine and cysteine in plasma significantly correlated with the presence of other behavioral symptoms (LIN28A p = 0.032; Ala p = 0.025; Cys p = 0.027) (Fig. 3c). Nevertheless, we could not detect LIN28A protein expression in purified leukocytes samples.
Correlation graphs and ROC curves for candidate biomarkers of neural dysfunction in IEM patients. (a) Graphs showing changes in BDNF and threonine in plasma of patients with deficits in psychomotor development and the ROC curves for different biomarker combinations calculated comparing affected with non-affected IEM patients. (b) ROC curves for THBS3 mRNA in blood and AABA in plasma and its combination for the detection of deficits in attention and/or executive functions calculated comparing affected with non-affected IEM patients. (c) Graphs showing changes in LIN28A mRNA in blood and alanine and cysteine in plasma of patients with other behavioral symptoms and the ROC curves for LIN28A. THBS3 and alanine biomarker combinations calculated comparing affected with non-affected IEM patients.
We then calculated AUC coefficients and a summary of the relevant potential effectiveness of candidate neural function biomarkers is shown in Table 6. For psychomotor delay, the combination of BDNF in plasma and MECP2 E1 mRNA in whole blood exhibited very good biomarker accuracy (AUC = 0.909). However, accuracy improved when BDNF was combined with serine or threonine in plasma, reaching maximal accuracy (AUC = 1) when a third amino acid between leucine, isoleucine or citrulline was added (Fig. 3a). Unexpectedly, although no significant differences were found in mRNA in whole blood or in protein expression in leukocytes (data not shown), THBS3 mRNA exhibited good biomarker performance for executive functions and attention deficits (AUC = 0.857). Biomarker performance increased when THBS3 was combined with AABA in plasma (AUC = 0.911) (Fig. 3b). Finally, the combination of THBS3 and LIN28A mRNA exhibited good biomarker performance to detect other behavioral symptoms (AUC = 0.870) in IEM patients, reaching maximal accuracy when was added alanine levels in plasma (AUC = 1) (Fig. 3c).
Taking together, these data point to excessive protein restriction and reduced BDNF levels as hallmarks of psychomotor delay in IEM patients, and point to THBS3 signaling as involved in executive and attention functions, and THBS3 with LIN28A in behavior control.
Discussion
UCD and MSUD are intoxication-type IEM while BCKDK deficiency results in BCAAs deficits, and despite correct metabolic management, affected patients frequently suffer psychomotor delay, cognitive, behavioral and psychiatric conditions. To date, early detection and rigorous medical follow-up is the gold standard to minimize neuropsychiatric sequelae19,33,34.
Several studies have assessed amino acid expression in plasma samples of UCD and MSUD patients. However, no biomarkers directly correlated with the neuropsychological features, and the molecular mechanisms involved are still poorly understood35,36. To our knowledge, this is the first study where the mRNA expression of genes involved in brain development and synaptic function were assessed in blood from IEM patients and correlated with BDNF and metabolite profiles in plasma.
The main limitation of this study is the reduced sample size (UCD n = 19, MSUD n = 9, and BCKDK deficiency n = 1), as IEMs are rare disorders. The use of blood is also challenging, as biomarker levels in blood can be highly variable and do not directly correlate with levels in brain, especially in diseases with preserved BBB integrity as IEMs. Even with these limitations, we found specific biomarker signatures for MSUD patients and neuropsychiatric dysfunctions in IEMs patients, that pointed to CACNA2D2, THBS1,3, MECP2, and LIN28A as new potential phenotype modifiers, after further investigation and verification with larger samples.
The first conclusion of this study is that despite metabolic management, most of symptomatic UCD and MSUD patients in our cohort still had glutamine and leucine/isoleucine respectively in the upper limits or above the normal range, and general AAs plasma profile in patients significantly differed from healthy controls.
In the case of MSUD, our data corroborate previous studies conducted in Brazilian and Filipino patients affected with MSUD which reported altered biochemical profile but no correlation between biochemical markers in serum and neuropsychological features23,35. However, we did not find a significant reduction in plasma BDNF levels as reported in Brazilian patients23, which might reflect different metabolic management and nutritional status in the two MSUD cohorts. In our cohort, none of the MSUD patients reported psychomotor delay while it was highly prevalent in the Brazilian cohort (40%).
BDNF levels are highly variable and directly related to food intake and energy metabolism37 and a reduction in BDNF expression was found in brains of rat offspring after maternal low-protein diet38. In this sense, 42% of our IEM cohort presented some degree of psychomotor delay, mostly corresponding to UCD patients treated with sodium phenylbutyrate, an ammonia scavenger known to produce a selective reduction in BCAAs39. We have found a significant inverse correlation between psychomotor delay and levels in plasma of BDNF and essential neutral AAs (mainly threonine, serine, phenylalanine, and BCAAs). These results highlight the necessity of careful metabolic surveillance and diet formulation based on the changing energy requirements of growing patients, as excessive essential neutral amino acid restriction and low BDNF seems to be a hallmark of psychomotor delay.
Nevertheless, we cannot rule out a role for altered BDNF in MSUD neurocognitive and behavioral sequelae, as we have found statistically significant reductions in the expression of MECP2 mRNA, an epigenetic regulator of BDNF transcription40,41. In addition, we have found statistically significant reductions in CACNA2D2 mRNA in MSUD patients, which was inversely correlated with chronically elevated BCAAs levels. We also found increased levels of THBS1 in MSUD and UCD patients, which expression was unrelated to BCAAs levels but correlated positively with glutamine and negatively with BDNF plasma levels. Results that are in accordance with published reports showing TSP1 upregulation in mice brains with decreased levels of BDNF42. TSP1, the protein codified by THBS1, is an extracellular matrix (ECM) glycoprotein with antiangiogenic properties that exerts multifunctional effects by binding cell-surface receptors and other proteins in the ECM. TSP1 is up-regulated with injury, chronic pathologies and in response to glucose and AAs, being considered a potent modulator of human diseases43,44,45.
One of the most important findings of this work is the identification, at the level of mRNA and protein, of a biomarker signature specific at for MSUD that includes α2δ2 (AUC 0.865/0.925) and MeCP2 (AUC 0.942/0.857), as the combination of both yielded the excellent biomarker accuracy (AUC 1/0.95). No specific biomarker signature was found for UCD.
VGCCs auxiliary subunits α2δ1-2, codified by CACNA2D1-2 genes, regulate calcium ion entry in response to electrical activity in excitable cells46,47. In neurons, α2δ1-2 expression modulates neurotransmitter release probability, trafficking and gating properties of AMPA-selective glutamate receptors (AMPARs)48,49, and have been involved in learning, memory, anxiety-related behaviors and epilepsy in rats and humans50,51,52,53,54. α2δ1-2 subunits are also essential for the formation and stabilization of new synapses by binding to TSP1-4 secreted by astrocytes29,55. It is assumed that BCAAs can induce conformational changes that indirectly affect TSPs binding to α2δ1-2 [29, 30, 31]. Moreover, α2δ subunits are the main targets of gabapentinoid drugs (gabapentin and pregabalin), which compete for the same binding site than BCAAs, blocking α2δ synaptogenic functions29, and biochemically mimicking the action of BCAAs56.
MeCP2 is an epigenetic regulator that preferentially binds to methylated CpG sites in promoter regions of DNA and is crucial for the correct brain development and the stability control of the neural network in response to activity57,58,59. Excessive or defective MECP2 function causes neurodevelopmental disorders associated with mental retardation, epilepsy, loss of speech and anxiety, among others60,61,62,63,64,65.
In addition to the above described, glycine and taurine were significantly increased in MSUD patients, data that is coincidental with previous studies23,35, and they stand out for its very good MSUD biomarker performance (AUC glycine 0.909 and AUC taurine 0.889). Glycine and taurine are abundant in brain, where they are involved in several aspects of normal development including neurogenesis, neuronal migration and differentiation66,67. Both AAs act as neurotransmitters by binding to glycine receptors, to NMDA receptors (glycine) and to GABA receptors (taurine). In addition, taurine inhibits K+-Cl− cotransporter KCC2, modulating Cl− homeostasis, the functionality of inhibitory neurotransmission and neuronal excitability68,69. Thus, the increase in glycine and taurine together with the reduced levels of α2δ2 and MeCP2 observed in MSUD patients might reflect an altered synaptogenesis and misbalanced excitatory-inhibitory neural network.
Unexpectedly, although no significant differences were found, THBS3 mRNA together with AABA in plasma has revealed a good biomarker performance for executive function, attention deficits (AUC 0,911). Reduced AABA in plasma is a frequent consequence of low protein diets and it has been recently associated with depression in older Japanese patients70. TSP3 is developmentally regulated44 and few is known about functions. It has been involved in modulating integrin membrane expression and function71, which is also essential for the formation of correct synaptic structures72. As TSP3 is a secreted protein, ELISA analysis in plasma would be required to gain insight into their potential biomarker significance.
Finally, increased levels of LIN28A mRNA, alanine, and cysteine correlated with behavioral dysfunctions, which include autism spectrum disorder, depression or aggressiveness, and the combination of THBS3 with LIN28A and alanine gave a perfect biomarker signature (AUC 1). LIN28 RNA binding proteins inhibit the biogenesis of let-7 family of miRNAs, a group of miRNAs crucial for embryonic and postnatal development, being at least the 50% of miRNAs present in mature neurons73,74. LIN28/let-7 pathway has been involved in controlling cell growth and energy metabolism26, and regulate dendritic growth and cell survival in response to BDNF75. Elevated levels of LIN28 and disruption of let-7 biogenesis have been described in animal models and in blood of patients with major depression76,77. Previous studies described that elevated levels of cysteine in infants derived in behavioral deficits in adults78 and plasma levels of alanine have been proposed as a marker of depression severity79. Taking together, our results are in concordance of what is observed in depression, which is commonly described in patients with inborn errors of metabolism, including UCD and MSUD80,81.
Our study, though preliminary, has identified several potential biomarker panels for neural function evaluation, providing a base for future studies. Most importantly, α2δ2 and MeCP2 showed an excellent neural function biomarker signature for MSUD. In addition, THBS3 mRNA and AABA gave a very good biomarker signature for executive and attention deficits. THBS3 and LIN28A mRNA and alanine showed a perfect biomarker signature for behavioral and mood disorders. Finally, a panel of BDNF and large neutral AAs showed a perfect biomarker signature for psychomotor delay, pointing to excessive protein restriction as potential causatives of psychomotor delay in diet-treated IEM patients. Although these results are very promising, a large number of clinical samples should be collected and the potential biomarkers in each panel quantified at the protein level for the ultimate goal to translate these neural function biomarkers to the clinical practice.
Material and Methods
Ethical statement
Research Ethics Committee of both hospitals, Sant Joan de Déu and Santiago de Compostela Hospitals, approved the study and informed consent was subscribed by patients and controls (when >18 years old) or by their parents (when <18 years old) prior to the collection of data. All methods were performed in accordance with the relevant guidelines and regulations.
Subjects and assessment
19 patients with a diagnosis of UCDs, 9 patients diagnosed with MSUD, 1 patient with BCKDK (branched-chain ketoacid dehydrogenase kinase deficiency) and 27 healthy age-matched control subjects were recruited for the study at Sant Joan de Déu Hospital in Barcelona and University Hospital of Santiago de Compostela, Spain. Patients were followed in their respective center with the same standardized protocol (Haeberle et al. 2012) from the date of diagnosis up to current date. Controls were healthy age-matched individuals with no history of learning difficulties, psychiatric and behavioral problems, who underwent blood analysis in the context of minor surgical interventions.
Inclusion criteria were patients with genetic and/or enzymatic diagnosis of UCDs, MSUD, and BCKDK; asymptomatic and female carriers of OTC deficiency were also included because of abnormal metabolic profile that required a protein-restricted diet. Table 1 summarizes the clinical characteristics of the patients. We evaluated the following parameters: age of symptoms onset, psychomotor development and number of decompensations (defined as symptomatic hyperammonemia episode with plasma ammonia greater than 100 μmol/L or amino acid decompensation with leucine greater than 1000 µmol/L). Cognitive functions were assessed by Wechsler Intelligence Scale for Children (WISC)-IV or Kaufman Brief Intelligence Test, Second Edition (K-BIT), considering five severity levels for intellectual disability: Borderline = IQ 70–85, Mild = IQ 55–70; Moderate = IQ 40–55; Severe IQ 25–40; Profound IQ < 25. Behavioral disorders, attention, and executive functions were assessed by NEPSY-II subtests, Behavior Rating Inventory for Executive Functions (BRIEF) and Conners’ Continuous Performance Test II (CPT II). ADHD rating scale –IV (Dupaul) was used for behavioral characterization associated with attention-deficit hyperactivity disorder.
Peripheral blood sample collection
Venous blood samples were collected in an anticoagulation EDTA tube after overnight fasting. Biochemical measurements of plasma ammonia, and amino acids were assessed by spectrophotometric technique and ion-exchange chromatography respectively.
Gene expression assays
RNA from 500 µL of whole blood was extracted using miRCURY™ RNA Isolation Kit - Cell & Plant (300110 Exiqon) following supplier instructions. RNA concentration and purity were analyzed in IMPLEN NanoPhotometer® P-Class (Implen). RNA samples (0.5–1 µg) were reverse transcribed to cDNA (High Capacity cDNA Reverse Transcription Kit, 4368814, Applied Biosystems) and Real-Time PCR was performed using TaqMan PCR Assays (4331182, Applied Biosystems) with TaqMan Universal PCR Master Mix (4324018, Applied Biosystems) in the 7900HT Real-Time PCR System (Applied Biosystems). Assays analyzed were: ADORA2A (Hs00169123_m1), CACNA2D2 (Hs01021049_m1), FMR1 (Hs00924547_m1), IRAK1 (Hs01018347_m1), LIN28A (Hs00702808_s1), PTEN (Hs02621230_s1), MECP2 E1 (Hs01598237_m1), MECP2 E2 (Hs00172845_m1), THBS1 (Hs00962908_m1), THBS3 (Hs00938498_m1). GUSB (Hs00939627_m1) and GAPDH (Hs99999905_m1) were used as endogenous controls. Data analysis was performed with Expression Suite Software (Life Technologies) and data expressed as Relative Quantification (RQ) normalized with respect to gusb. Sample exclusion criteria were: threshold cycles (Cts) of more than 35, replicates with a standard deviation greater than 0.25 and Cts of GAPDH and GUSB not consistent. Outliers (identified using interquartile ranges) were removed from the analysis.
BDNF protein detection
The remaining whole blood was centrifuged at 2000 rpm 10 minutes and the plasma obtained was frozen at −80 °C until use. BDNF was measured in duplicates using RayBio® Human BDNF ELISA Kit following supplier instructions.
Western Blot analysis
Leukocytes were obtained from pelleted blood cells after red blood cells lysis with RBC Lysis Buffer (21205, Norgen Biotek corp) following supplier instructions. Proteins were extracted, separated by SDS-page in polyacrylamide gels, and transferred to nitrocellulose membranes (1620112, Bio-Rad). Membranes were blocked for 1 hour at room temperature, primary antibodies were incubated overnight at 4 °C and then with their corresponding secondary HRP-conjugated antibodies for 1 hour (1:5000, Invitrogen). Protein signal was detected with ECL chemiluminiscent system (Amersham, GE Healthcare) in ImageQuant LAS 500 (GE Healthcare). Images were processed with Image Studio Lite 5.2 (LI-COR). Densitometry analysis was performed using gapdh as loading control and ImageJ software (National Institutes of Health). Primary antibodies: rabbit anti- α2δ2 (1:500, Abgent, AP13380C), rabbit anti-MeCP2 (1:1500, Millipore, ABE333), rabbit anti-TSP3 (1:1000, Abgent, AP18972a), rabbit anti-LIN28A (1:200, #8641, Cell Signaling) and mouse anti-Vinculin (1:750, SCBT, sc-59803). Full length gels are shown in Supplementary info file.
Data analysis
SPSS program (IBM SPSS Statistics 24.0) was used to analyze data.
Two tailored non-parametric Kruskal Wallis and Mann-Whitney U tests were used to establish metabolic and gene expression differences between conditions and to correlate candidate biomarkers with clinical variables respectively, as the sample size was not enough to presume normality within the data. Signification values in multiple comparisons were adjusted by Bonferroni correction to reduce Type 1 Error. A bivariate correlation test was used to correlate quantitative variables while Pearson product-moment correlation was used if both variables were qualitative.
To assess biomarker feasibility Receiver Operating Characteristic (ROC) curves and the area under the ROC curve (AUC) were performed. AUC vary from 0 to 1, where values 0.8 ≤ AUC < 0.9 reflect good and 0.9 ≤ AUC ≤ 1 very good biomarker performance. Statistical significance was set at p values *p < 0.05, **p < 0.01 and ***p < 0.001.
References
Martinez-Hernandez, A., Bell, K. P. & Norenberg, M. D. Glutamine synthetase: glial localization in brain. Science 195, 1356–1358 (1977).
Hertz, L. & Zielke, H. R. Astrocytic control of glutamatergic activity: astrocytes as stars of the show. Trends in neurosciences 27, 735–743, https://doi.org/10.1016/j.tins.2004.10.008 (2004).
Cagnon, L. & Braissant, O. Hyperammonemia-induced toxicity for the developing central nervous system. Brain research reviews 56, 183–197, https://doi.org/10.1016/j.brainresrev.2007.06.026 (2007).
Rangroo Thrane, V. et al. Ammonia triggers neuronal disinhibition and seizures by impairing astrocyte potassium buffering. Nature medicine 19, 1643–1648, https://doi.org/10.1038/nm.3400 (2013).
Kolker, S. et al. The phenotypic spectrum of organic acidurias and urea cycle disorders. Part 1: the initial presentation. Journal of inherited metabolic disease 38, 1041–1057, https://doi.org/10.1007/s10545-015-9839-3 (2015).
Jamiolkowski, D. et al. Behavioural and emotional problems, intellectual impairment and health-related quality of life in patients with organic acidurias and urea cycle disorders. Journal of inherited metabolic disease 39, 231–241, https://doi.org/10.1007/s10545-015-9887-8 (2016).
Chuang, D. T. Maple syrup urine disease: it has come a long way. The Journal of pediatrics 132, S17–23 (1998).
Carpenter, K. In Branched Chain Amino Acids in Clinical Nutrition (eds Preedy, V. Rajendram, R. & Patel, V.) 145–156 (Humana Press, 2015).
Novarino, G. et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 338, 394–397, https://doi.org/10.1126/science.1224631 (2012).
Simon, E. et al. Maple syrup urine disease: favourable effect of early diagnosis by newborn screening on the neonatal course of the disease. Journal of inherited metabolic disease 29, 532–537, https://doi.org/10.1007/s10545-006-0315-y (2006).
Lee, W. T. Disorders of amino acid metabolism associated with epilepsy. Brain & development 33, 745–752, https://doi.org/10.1016/j.braindev.2011.06.014 (2011).
Strauss, K. A. et al. Elective liver transplantation for the treatment of classical maple syrup urine disease. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons 6, 557–564, https://doi.org/10.1111/j.1600-6143.2005.01209.x (2006).
Strauss, K. A. et al. Classical maple syrup urine disease and brain development: principles of management and formula design. Molecular genetics and metabolism 99, 333–345, https://doi.org/10.1016/j.ymgme.2009.12.007 (2010).
Simon, E., Schwarz, M. & Wendel, U. Social outcome in adults with maple syrup urine disease (MSUD). Journal of inherited metabolic disease 30, 264, https://doi.org/10.1007/s10545-007-0475-4 (2007).
Shellmer, D. A. et al. Cognitive and adaptive functioning after liver transplantation for maple syrup urine disease: a case series. Pediatric transplantation 15, 58–64, https://doi.org/10.1111/j.1399-3046.2010.01411.x (2011).
Mazariegos, G. V. et al. Liver transplantation for classical maple syrup urine disease: long-term follow-up in 37 patients and comparative United Network for Organ Sharing experience. The Journal of pediatrics 160, 116–121 e111, https://doi.org/10.1016/j.jpeds.2011.06.033 (2012).
Strauss, K. A., Puffenberger, E. G. & Morton, D. H. In GeneReviews (eds Ardinger, H. H. et al.) (2013).
Bouchereau, J. et al. Neurocognitive profiles in MSUD school-age patients. Journal of inherited metabolic disease 40, 377–383, https://doi.org/10.1007/s10545-017-0033-7 (2017).
Muelly, E. R. et al. Biochemical correlates of neuropsychiatric illness in maple syrup urine disease. The Journal of clinical investigation 123, 1809–1820, https://doi.org/10.1172/JCI67217 (2013).
Kowianski, P. et al. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cellular and molecular neurobiology 38, 579–593, https://doi.org/10.1007/s10571-017-0510-4 (2018).
Autry, A. E. & Monteggia, L. M. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacological reviews 64, 238–258, https://doi.org/10.1124/pr.111.005108 (2012).
Chahrour, M. et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229, https://doi.org/10.1126/science.1153252 (2008).
Scaini, G. et al. Serum Markers of Neurodegeneration in Maple Syrup Urine Disease. Molecular neurobiology 54, 5709–5719, https://doi.org/10.1007/s12035-016-0116-8 (2017).
Galland, F. et al. Hyperammonemia compromises glutamate metabolism and reduces BDNF in the rat hippocampus. Neurotoxicology 62, 46–55, https://doi.org/10.1016/j.neuro.2017.05.006 (2017).
Motamedi, S., Karimi, I. & Jafari, F. The interrelationship of metabolic syndrome and neurodegenerative diseases with focus on brain-derived neurotrophic factor (BDNF): Kill two birds with one stone. Metabolic brain disease 32, 651–665, https://doi.org/10.1007/s11011-017-9997-0 (2017).
Shyh-Chang, N. & Daley, G. Q. Lin28: primal regulator of growth and metabolism in stem cells. Cell stem cell 12, 395–406, https://doi.org/10.1016/j.stem.2013.03.005 (2013).
Leal, G., Comprido, D. & Duarte, C. B. BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology 76 Pt C, 639–656, https://doi.org/10.1016/j.neuropharm.2013.04.005 (2014).
Catterall, W. A., Perez-Reyes, E., Snutch, T. P. & Striessnig, J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacological reviews 57, 411–425, https://doi.org/10.1124/pr.57.4.5 (2005).
Eroglu, C. et al. Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell 139, 380–392, https://doi.org/10.1016/j.cell.2009.09.025 (2009).
Brown, J. P., Dissanayake, V. U., Briggs, A. R., Milic, M. R. & Gee, N. S. Isolation of the [3H]gabapentin-binding protein/alpha 2 delta Ca2+ channel subunit from porcine brain: development of a radioligand binding assay for alpha 2 delta subunits using [3H]leucine. Analytical biochemistry 255, 236–243, https://doi.org/10.1006/abio.1997.2447 (1998).
Davies, A. et al. Functional biology of the alpha(2)delta subunits of voltage-gated calcium channels. Trends in pharmacological sciences 28, 220–228, https://doi.org/10.1016/j.tips.2007.03.005 (2007).
Zamponi, G. W., Striessnig, J., Koschak, A. & Dolphin, A. C. The Physiology, Pathology, and Pharmacology of Voltage-Gated Calcium Channels and Their Future Therapeutic Potential. Pharmacological reviews 67, 821–870, https://doi.org/10.1124/pr.114.009654 (2015).
Manoli, I. & Venditti, C. P. Disorders of branched chain amino acid metabolism. Translational science of rare diseases 1, 91–110, https://doi.org/10.3233/TRD-160009 (2016).
Summar, M. L. & Mew, N. A. Inborn Errors of Metabolism with Hyperammonemia: Urea Cycle Defects and Related Disorders. Pediatric clinics of North America 65, 231–246, https://doi.org/10.1016/j.pcl.2017.11.004 (2018).
Chiong, M. A. et al. Plasma amino acid and urine organic acid profiles of Filipino patients with maple syrup urine disease (MSUD) and correlation with their neurologic features. Molecular genetics and metabolism reports 9, 46–53, https://doi.org/10.1016/j.ymgmr.2016.10.004 (2016).
Waisbren, S. E. et al. Biochemical markers and neuropsychological functioning in distal urea cycle disorders. Journal of inherited metabolic disease. https://doi.org/10.1007/s10545-017-0132-5 (2018).
Marosi, K. & Mattson, M. P. BDNF mediates adaptive brain and body responses to energetic challenges. Trends in endocrinology and metabolism: TEM 25, 89–98, https://doi.org/10.1016/j.tem.2013.10.006 (2014).
Marwarha, G., Claycombe-Larson, K., Schommer, J. & Ghribi, O. Maternal low-protein diet decreases brain-derived neurotrophic factor expression in the brains of the neonatal rat offspring. The Journal of nutritional biochemistry 45, 54–66, https://doi.org/10.1016/j.jnutbio.2017.03.005 (2017).
Tuchman, M. et al. Cross-sectional multicenter study of patients with urea cycle disorders in the United States. Molecular genetics and metabolism 94, 397–402, https://doi.org/10.1016/j.ymgme.2008.05.004 (2008).
Chen, W. G. et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889, https://doi.org/10.1126/science.1086446 (2003).
Martinowich, K. et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302, 890–893, https://doi.org/10.1126/science.1090842 (2003).
Qin, L., Kim, E., Ratan, R., Lee, F. S. & Cho, S. Genetic variant of BDNF (Val66Met) polymorphism attenuates stroke-induced angiogenic responses by enhancing anti-angiogenic mediator CD36 expression. The Journal of neuroscience: the official journal of the Society for Neuroscience 31, 775–783, https://doi.org/10.1523/JNEUROSCI.4547-10.2011 (2011).
Zhao, C., Isenberg, J. S. & Popel, A. S. Transcriptional and Post-Transcriptional Regulation of Thrombospondin-1 Expression: A Computational Model. PLoS computational biology 13, e1005272, https://doi.org/10.1371/journal.pcbi.1005272 (2017).
Adams, J. C. & Lawler, J. The thrombospondins. Cold Spring Harbor perspectives in biology 3, a009712, https://doi.org/10.1101/cshperspect.a009712 (2011).
Matsuo, Y. et al. Thrombospondin 1 as a novel biological marker of obesity and metabolic syndrome. Metabolism: clinical and experimental 64, 1490–1499, https://doi.org/10.1016/j.metabol.2015.07.016 (2015).
Dooley, D. J., Taylor, C. P., Donevan, S. & Feltner, D. Ca2+ channel alpha2delta ligands: novel modulators of neurotransmission. Trends in pharmacological sciences 28, 75–82, https://doi.org/10.1016/j.tips.2006.12.006 (2007).
Andrade, A. et al. The alpha(2)delta subunit augments functional expression and modifies the pharmacology of Ca(V)1.3 L-type channels. Cell calcium 46, 282–292, https://doi.org/10.1016/j.ceca.2009.08.006 (2009).
Dolphin, A. C. Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nature reviews. Neuroscience 13, 542–555, https://doi.org/10.1038/nrn3311 (2012).
Hoppa, M. B., Lana, B., Margas, W., Dolphin, A. C. & Ryan, T. A. alpha2delta expression sets presynaptic calcium channel abundance and release probability. Nature 486, 122–125, https://doi.org/10.1038/nature11033 (2012).
Zhou, J. J., Li, D. P., Chen, S. R., Luo, Y. & Pan, H. L. The alpha2delta-1-NMDA receptor coupling is essential for corticostriatal long-term potentiation and is involved in learning and memory. The Journal of biological chemistry 293, 19354–19364, https://doi.org/10.1074/jbc.RA118.003977 (2018).
Lotarski, S. M. et al. Anxiolytic-like activity of pregabalin in the Vogel conflict test in alpha2delta-1 (R217A) and alpha2delta-2 (R279A) mouse mutants. The Journal of pharmacology and experimental therapeutics 338, 615–621, https://doi.org/10.1124/jpet.111.180976 (2011).
Cioli, C., Abdi, H., Beaton, D., Burnod, Y. & Mesmoudi, S. Differences in human cortical gene expression match the temporal properties of large-scale functional networks. PloS one 9, e115913, https://doi.org/10.1371/journal.pone.0115913 (2014).
Barclay, J. et al. Ducky mouse phenotype of epilepsy and ataxia is associated with mutations in the Cacna2d2 gene and decreased calcium channel current in cerebellar Purkinje cells. The Journal of neuroscience: the official journal of the Society for Neuroscience 21, 6095–6104 (2001).
Edvardson, S. et al. Early infantile epileptic encephalopathy associated with a high voltage gated calcium channelopathy. Journal of medical genetics 50, 118–123, https://doi.org/10.1136/jmedgenet-2012-101223 (2013).
Christopherson, K. S. et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433, https://doi.org/10.1016/j.cell.2004.12.020 (2005).
Goldlust, A., Su, T. Z., Welty, D. F., Taylor, C. P. & Oxender, D. L. Effects of anticonvulsant drug gabapentin on the enzymes in metabolic pathways of glutamate and GABA. Epilepsy research 22, 1–11 (1995).
Guy, J., Cheval, H., Selfridge, J. & Bird, A. The role of MeCP2 in the brain. Annual review of cell and developmental biology 27, 631–652, https://doi.org/10.1146/annurev-cellbio-092910-154121 (2011).
Qiu, Z. et al. The Rett syndrome protein MeCP2 regulates synaptic scaling. The Journal of neuroscience: the official journal of the Society for Neuroscience 32, 989–994, https://doi.org/10.1523/JNEUROSCI.0175-11.2012 (2012).
Lewis, J. D. et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69, 905–914 (1992).
Ramocki, M. B., Tavyev, Y. J. & Peters, S. U. The MECP2 duplication syndrome. American journal of medical genetics. Part A 152 A, 1079–1088, https://doi.org/10.1002/ajmg.a.33184 (2010).
Ehrhart, F. et al. Rett syndrome - biological pathways leading from MECP2 to disorder phenotypes. Orphanet journal of rare diseases 11, 158, https://doi.org/10.1186/s13023-016-0545-5 (2016).
Ramocki, M. B. et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Annals of neurology 66, 771–782, https://doi.org/10.1002/ana.21715 (2009).
Barnes, K. V. et al. Anxiety-like behavior in Rett syndrome: characteristics and assessment by anxiety scales. Journal of neurodevelopmental disorders 7, 30, https://doi.org/10.1186/s11689-015-9127-4 (2015).
Ausio, J. MeCP2 and the enigmatic organization of brain chromatin. Implications for depression and cocaine addiction. Clinical epigenetics 8, 58, https://doi.org/10.1186/s13148-016-0214-5 (2016).
Panayotis, N. et al. Importin alpha5 Regulates Anxiety through MeCP2 and Sphingosine Kinase 1. Cell reports 25, 3169–3179 e3167, https://doi.org/10.1016/j.celrep.2018.11.066 (2018).
Ascenzi, M. & Bony, G. The building of the neocortex with non-hyperpolarizing neurotransmitters. Developmental neurobiology 77, 1023–1037, https://doi.org/10.1002/dneu.22495 (2017).
Kilb, W. & Fukuda, A. Taurine as an Essential Neuromodulator during Perinatal Cortical Development. Frontiers in cellular neuroscience 11, 328, https://doi.org/10.3389/fncel.2017.00328 (2017).
El Idrissi, A., El Hilali, F., Rotondo, S. & Sidime, F. Effects of Taurine Supplementation on Neuronal Excitability and Glucose Homeostasis. Advances in experimental medicine and biology 975 Pt 1, 271–279, https://doi.org/10.1007/978-94-024-1079-2_24 (2017).
Inoue, K. et al. Taurine inhibits K+-Cl- cotransporter KCC2 to regulate embryonic Cl- homeostasis via with-no-lysine (WNK) protein kinase signaling pathway. The Journal of biological chemistry 287, 20839–20850, https://doi.org/10.1074/jbc.M111.319418 (2012).
Adachi, Y. et al. Association between plasma alpha-aminobutyric acid and depressive symptoms in older community-dwelling adults in Japan. Geriatrics & gerontology international, https://doi.org/10.1111/ggi.13585 (2018).
Schips, T. G. et al. Thrombospondin-3 augments injury-induced cardiomyopathy by intracellular integrin inhibition and sarcolemmal instability. Nature communications 10, 76, https://doi.org/10.1038/s41467-018-08026-8 (2019).
Lilja, J. & Ivaska, J. Integrin activity in neuronal connectivity. Journal of cell science 131, https://doi.org/10.1242/jcs.212803 (2018).
Juhila, J. et al. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PloS one 6, e21495, https://doi.org/10.1371/journal.pone.0021495 (2011).
Shinohara, Y. et al. miRNA profiling of bilateral rat hippocampal CA3 by deep sequencing. Biochemical and biophysical research communications 409, 293–298, https://doi.org/10.1016/j.bbrc.2011.05.004 (2011).
Amen, A. M. et al. A Rapid Induction Mechanism for Lin28a in Trophic Responses. Molecular cell 65, 490–503 e497, https://doi.org/10.1016/j.molcel.2016.12.025 (2017).
Wei, Y. B. et al. Elevation of Il6 is associated with disturbed let-7 biogenesis in a genetic model of depression. Translational psychiatry 6, e869, https://doi.org/10.1038/tp.2016.136 (2016).
Maffioletti, E. et al. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. Journal of affective disorders 200, 250–258, https://doi.org/10.1016/j.jad.2016.04.021 (2016).
Shapre, L. G. et al. Brain damage and associated behavioral deficits following the administration of L-cysteine to infant rats. Pharmacology, biochemistry, and behavior 3, 291–298 (1975).
Mitani, H. et al. Correlation between plasma levels of glutamate, alanine and serine with severity of depression. Progress in neuro-psychopharmacology & biological psychiatry 30, 1155–1158, https://doi.org/10.1016/j.pnpbp.2006.03.036 (2006).
Gropman, A. L., Summar, M. & Leonard, J. V. Neurological implications of urea cycle disorders. Journal of inherited metabolic disease 30, 865–879, https://doi.org/10.1007/s10545-007-0709-5 (2007).
Walterfang, M., Bonnot, O., Mocellin, R. & Velakoulis, D. The neuropsychiatry of inborn errors of metabolism. Journal of inherited metabolic disease 36, 687–702, https://doi.org/10.1007/s10545-013-9618-y (2013).
Acknowledgements
This work was supported by FIS: PI15/01082 (Instituto de Salud Carlos III: ISCIII and “Fondo Europeo de desarrollo regional” FEDER).
Author information
Authors and Affiliations
Contributions
A.G.C. and S.A. conceived and supervised the study. A.A.C., R.B., A.T.N., A.O. and M.B. and C.S. performed the experiments and collected the data. D.G. collected and processed clinical data. A.A.C. and D.G. analyzed the results. E.C.S., F.R., S.M., D.L.S.M., R.A. M.D.S., C.G.V., R.C., M.L.C. and J.A. provided samples and clinical data of the patients. A.A.C. and D.G. wrote and S.A. and A.G.C. critically revised the manuscript. All authors reviewed the article for intellectual content and approved the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Castells, AA., Gueraldi, D., Balada, R. et al. Discovery of Biomarker Panels for Neural Dysfunction in Inborn Errors of Amino Acid Metabolism. Sci Rep 9, 9128 (2019). https://doi.org/10.1038/s41598-019-45674-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45674-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.