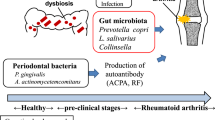
Abstract
Rheumatoid arthritis (RA) is an autoimmune disease characterized by joint inflammation. Individuals with RA have a higher risk of periodontitis and periodontitis has been linked to RA through the production of enzymes by periodontal pathogens that citrullinate proteins. This linkage is supported by findings that periodontitis is associated with increased RA severity and treatment of periodontitis can improve the symptoms of RA. The possible mechanism for this association is through dysbiosis of the oral microbiota triggered by RA-induced systemic inflammation. We examined the RA status of subjects by measuring the number of tender and swollen joints, anti-citrullinated protein antibody and rheumatoid factor. Periodontal disease status and salivary cytokine levels were measured, and dental plaque analyzed by 16S rRNA high throughput sequencing. RA patients had a higher bacterial load, a more diverse microbiota, an increase in bacterial species associated with periodontal disease, more clinical attachment loss, and increased production of inflammatory mediators including IL-17, IL-2, TNF, and IFN-γ. Furthermore, changes in the oral microbiota were linked to worse RA conditions. Our study provides new insights into the bi-directional relationship between periodontitis and RA and suggest that monitoring the periodontal health of RA patients is particularly important.
Similar content being viewed by others
Introduction
The oral cavity is the second largest microbial niche after the gastrointestinal tract with over 700 bacterial species1. In periodontally healthy individuals, microbial populations co-exist in equilibrium with the host. The change in this equilibrium is linked to the pathogenesis of oral diseases such as periodontitis1,2. Oral bacteria, which exist as a biofilm on the tooth surface, can induce inflammation in the adjacent gingiva, leading to osteoclast formation and bone loss which, in severe cases causes tooth loss3. Systemic inflammatory diseases may contribute to disrupting the balance between host and oral microbiota4.
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by chronic inflammation and damage to articular tissues5. An increased incidence of periodontitis has been reported in patients with RA6. Furthermore, treatment of periodontitis has been shown to reduce RA activity7. Both conditions exhibit similar pathological features, including bone resorption8,9,10. It has been proposed that periodontal disease and RA are linked by periodontal pathogens that produce enzymes capable of modifying proteins to enhance their antigenicity through the addition of malondialdehyde-acetaldehyde, citrullination and carbamylation8,10,11. These anti-cyclic citrullinated peptide (CCP) antibodies can be detected before RA onset and have been identified as an etiologic factor in the disease process12. Furthermore, RA enhance systemic inflammation which can amplify the local inflammatory response in the periodontium, increasing periodontal destruction13.
Few studies have described the composition of the oral microbiota in patients with RA14,15,16,17. Zhang examined dental and salivary microbiome but the periodontal status of RA subjects was not defined15. Another study evaluated subgingival microbiota and the periodontal condition of RA subjects, but did not evaluate the impact of RA and periodontitis independently14. Mikuls compared RA to osteoarthritis patients16 while Lopez-Oliva analyzed only RA patients without periodontitis17. Furthermore, neither of these studies assessed inflammatory parameters in the oral cavity. Our study characterized the subgingival microbiome of RA patients and its association with periodontal status, inflammatory markers and RA scores to establish links between these parameters.
Results
Periodontal destruction and RA outcomes
Table 1 shows that of the 42 patients with RA included in the study, 21 (50%) had periodontitis compared to 20 (42.6%) of 47 non-RA subjects (P > 0.05). The mean duration of RA was similar for patients without periodontitis (16.18 ± 8.2 years) and for those with periodontitis (12.46 ± 9.7years) (P > 0.05). RA activity parameters (number of tender and swollen joints, DAS-28) and medications in use were not different between RA patients with or without periodontitis (Table 1). Interesting, the majority of RA subjects (85.7%) with periodontitis were positive for the presence of autoantibodies (ACPA) compared to only 33% in RA patients without periodontitis (P < 0.05).
The presence of RA was associated with worse periodontal parameters compared with control subjects: probing depth (Controls 3.0 × RA 3.8 mm) and clinical attachment loss (Controls 3.0 × RA 4 mm) (Table 1). These data indicate severe periodontitis in RA patients. However, self-reported hygiene habits, plaque index and bleeding on probing did not differ among RA and control subjects (P > 0.05) (Table 1).
RA affects subgingival microbial load, richness and diversity
We investigated the total microbial biomass in RA subjects and found that non-periodontitis RA patients had a significantly ~1 log higher bacterial burden than did control individuals without periodontitis (Fig. 1A). A total of 779 OTUS were found and the impact of RA status on microbial diversity and richness was examined by assessing the number of observed OTUs, Chao1 and Shannon indexes. RA patients had increased microbial diversity compared to controls. In subjects with periodontitis, RA was associated with increased diversity assessed by number of OTUs (Fig. 1C) and Shannon Index (Fig. 1D). In patients without periodontitis, RA caused a change in microbial diversity assessed by Chao1 index (Fig. 1B).
Bacterial load and microbial diversity in subgingival biofilm samples. (A) Bacterial load. (B–D) Metrics of alpha diversity in control subjects and RA patients with or without periodontitis. *Statistically different compared to non-periodontitis subjects within the same group. #Statistically different compared to matched controls. p < 0.05, Kruskal-Wallis test.
To analyze whether the subgingival microbial communities in patients with RA were distinct from that of controls, we performed unweighted UniFrac distance analysis (Fig. 2). Microbial communities in patients with RA had distinct clusters compared to control patients without the complicating factor of periodontitis (Fig. 2A, PERMANOVA, p < 0.01). The presence of periodontitis in patients with RA obviated the difference between the RA and control group (P > 0.05, Fig. 2B). However, RA patients with periodontitis clustered separately from RA patients without periodontitis (P < 0.05, Fig. 2C).
Principal coordinates analysis plots comparing subgingival microbial community compositions. Each point represents a subject. (A) Microbial communities in control and RA subjects without periodontitis. (w/o P). (B) Microbial communities in RA patients versus control subjects with periodontitis (w/P). (C) Microbial communities in RA patients without periodontitis versus with periodontitis.
Subgingival RA and control group sites harbor distinct bacterial communities
We performed LEFSE (linear discriminant analysis coupled with effect size measurements) for analysis of the relative abundance of microbial taxonomic groups. A number of pathogenic bacteria were significantly elevated in the RA group that are associated with worse periodontal status1. RA patients without periodontitis had enrichment in periodontitis-associated bacteria such as Prevotella species (P. melaninogenica, P. denticola, P. histicola, P. nigrescens, P. oulorum, and P. maculosa) and other pathogenic species (Selenomonas noxia, S. sputigena and Anaeroglobus geminatus). In addition, RA subjects presented a reduction of health-associated species (Streptococcus, Rothia aeria, Kingella oralis, Haemophilus, Actinomyces) (Fig. 3A). In the same way, pathogenic species such as Prevotella, Aggregaticbacter actinomycetemcomitans and Parvimonas micra were significantly increased in RA patients with periodontitis compared to control subjects with periodontitis (Fig. 3B). We also observe an increased concentration of gram-negative anaerobic species on RA sites compared to control sites with periodontitis (Supplementary Fig. S1).
OTUs with different relative abundance based on LEfSe (linear discriminant analysis coupled with effect size measurements) results in control subjects (red) and RA subjects (green). (A) Subjects without periodontitis. (B) Subjects with periodontitis. Bars represent linear discriminant analysis scores (LDA).
RA parameters and oral bacteria species
Several bacterial taxa were correlated with RA parameters. In RA subjects with periodontitis, bacteria related to periodontal health, such as Actinomyces, were negatively correlated with number of tender joints (rho = −0.36, p < 0.05). On the other hand, in the same subjects, the presence of pathogenic species such as Fretibacterium fastidiosum18, Parvimonas micra19 and Anaeroglobus geminatus1 were correlated with augmented numbers of swollen (rho = 0.35) and tender joints (rho = 0.30), p < 0.05.
Phylogenetic investigation of communities in subgingival microbiota in RA patients
To identify functional gene families in bacteria that may be differentially enriched in the subgingival microbiota of control subjects and RA patients, we applied PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states), a computational algorithm that provides a functional-gene profile for each sample based on a prediction of the number of copies of each gene family found in the 16S rRNA sequence information. Analysis using PICRUSt identified a number of genes involved with energy metabolism, lipopolysaccharide (LPS) biosynthesis, amino acid and carbohydrate metabolism, cell cycle and peptidases that were significantly more abundant in the subgingival bacteria of subjects with RA independent of periodontal status. In controls, bacterial genes involved with amino acid biosynthesis, and carbohydrate metabolism were over-represented in the periodontal microbiota (Supplementary Fig. S2).
Salivary concentration of inflammatory cytokines in RA patients
To investigate whether the above-mentioned dysbiosis in subgingival microbiota could be associated with an altered inflammatory response we measured cytokines in saliva of RA and control subjects (Fig. 4). The levels of IL-2, IL-6 and IFN-γ were increased in saliva from RA patients compared to control subjects, both without periodontitis (P < 0.05). IL-33 and TNF-α were increased in all RA groups independent of periodontal status (P < 0.05). IL-17 was increased in RA subjects with periodontitis compared to control subjects (P < 0.05, Fig. 4).
Levels of inflammatory cytokines in saliva. Control subjects and patients with rheumatoid arthritis (RA) with and without periodontitis. Cytokines were measured by cytometric bead array (CBA) or enzyme-linked immunosorbent assay (ELISA). *Statistically different compared to subjects without periodontitis within the same group. #Statistically different compared to Control. p < 0.05, Kruskal-Wallis test.
The increased levels of the cytokines IL-6, IL-17 and IL-33 were positively correlated with periodontal parameters such as probing depth and number of missing teeth as demonstrated in Table 2 (P < 0.05). IL-33 levels were positively correlated with RA parameters such as rheumatoid factor and c-reactive protein (CRP), while IL-6 was positively correlated with CRP and ESR. Furthermore, the presence of healthy-related species including Streptococcus, Rothia aeria, Actinomyces was negatively correlated with cytokines IL-17 and TNF-α. In contrast, the presence of pathogenic species, such as Selenomas and Prevotella, were correlated with increased levels of inflammatory cytokines (IL-2, IL-6, IL-17, IL-33 and TNF-α) (Table 2).
Subgingival biofilm of RA patients elicited high IFN-γ production from PBMCs
To evaluate the potential of dysbiotic subgingival biofilm of RA patients to stimulate inflammatory response, we exposed PBMCs from control individuals to inactivated bacterial plaque samples from RA or healthy donors, neither of which had periodontitis. Plaque samples from RA subjects’ stimulated significantly higher production of IFN-γ compared to plaque from control subjects (Supplementary Fig. S3).
Discussion
A relationship between RA and periodontitis has previously been reported, but the impact of RA on the subgingival microbiota linked to periodontal disease has not been thoroughly investigated and mechanisms for the potential impact have not been addressed20,21,22,23,24,25. In the present study, we showed significant differences in the subgingival bacterial community between RA patients and controls. RA patients had a higher bacterial load, a more diverse microbiota and increased abundance of pathogenic species compared to controls, even in periodontally healthy individuals. Accordingly, periodontal destruction (probing depth and clinical attachment loss) was significantly greater in RA subjects.
Microbiota homeostasis can be modulated by a number of parameters including genetic, environmental and inflammatory factors26. Chronic systemic inflammation, as observed in RA, may affect the levels of inflammation in periodontal tissues, which in turn may alter the bacterial community. IL-2, IFN-γ, TNF and IL-33 exhibited an approximately two-fold increase in RA subjects without periodontitis compared to matched controls. IL-6 had a significant although less robust change. Taken together these results indicate that RA impacts the cytokine levels in periodontal tissues reflecting increased inflammation. Similar changes in periodontal ifnlamation have been reported in subjects with systemic lupus erythematosus27 and RA28. Several studies report a positive correlation between salivary cytokine levels and periodontitis29,30 while some have not31. The discrepancy may be due to different stages of disease, different disease classification systems, methodological differences and that whole saliva does not simply represent health versus diseased sites but reflects the aggregate of healthy, gingivitis and periodontitis sites making the analysis more complex. IFN-γ is a Th1 cytokine that enhance cell-mediate responses32. TNF, IL-6 and IL-17 have multiple overlapping functions that contribute to RA and periodontal disease by stimulating leukocyte activation and migration, chemokine expression and bone resorption23,33. IL-33 stimulates Th2 responses34, while IFN-γ and IL-2 regulate several aspects of the immune response35.
The increased inflammatory cytokine levels in RA subjects was reflected by increased inflammatory potential of the bacteria from these individuals as reflected by greater stimulation of IFN-γ in PBMCs from inactivated dental plaque compared to stimulation by dental plaque from matched control subjects. Previous studies have suggested that periodontitis and RA are interdependent with elevated levels of pro-inflammatory molecules in both36. Inflammatory mediators found in the subgingival microenvironment may change the ecological conditions to favor the outgrowth of pathogenic bacteria, leading to periodontal destruction37 as shown for diabetes38. The increased inflammation caused by RA coupled with microbial changes may amplify periodontal inflammation and explain the greater su sceptibility to periodontitis in RA subjects that we and others have observed39,40. Studies in mice showed that RA is linked to greater periodontal bone loss and changes in the oral microbiota24. Our results provide evidence that similar events occur in humans.
We found that RA was associated with increased microbial load and diversity, which is consistent with previous reports that periodontitis, unlike most polymicrobial infections, is associated with increased bacterial diversity41. In addition, we observed that the severity of RA, reflected by the number of tender and swollen joints, was significantly correlated with the presence of pathogenic oral bacteria (i.e. Fusobacterium nucleatum and Treponema socransky). RA patients without periodontal disease have previously been reported to have increased bacterial biomass in their dental plaque, consistent with our results17 and bacterial changes in individuals with RA have been linked to parameters of RA severity. We examined samples that reflected similar sites within the same individual and our analysis focused on differences between individuals. However, this does not rule out the possibility that within an individual there are differences based on the periodontal status of a specific tooth.
Increased numbers of gram-negative anaerobic species are associated with proteolysis, inflammation and periodontal destruction42. Our analysis of predicted functions using PICRUSt identified an enrichment of several pathways linked to amino acid metabolism and peptidases in the microbiota from RA subjects. We also observed an abundance of genes linked to bacterial virulence such as LPS and sporulation, with the caveat that PICRUST analysis is descriptive, predicting genes that are present in the biofilm, but not necessarily genes that are expressed. These peptidases and virulence factors may increase inflammation and contribute to periodontal breakdown43.
Local changes in the periodontal tissues associated with enhanced inflammation can alter the environment and promote the growth of pathogenic bacteria, amplifying inflammation40,44. Subgingival sites in RA patients had an increase in gram-negative anaerobic species in dental plaque that are strongly associated with periodontal destruction and inflammation. We found higher levels of Selenomonas noxia and Parvimonas micra in RA subjects in agreement with previous findings in animal model of RA24 as well as elevated levels of Prevotella. A recent study found an association between anti-citrullinated peptide antibody (ACPA) titers and evidence of greater exposure to Prevotella45. ACPAs may be regarded as etiologic factors and diagnostic markers as they are detected in the serum before the disease onset. It is also possible that ACPAs represent different forms of RA46 and that ACPA-positive patients have a worse clinical prognosis with higher rates of erosive damage47. Here we observed that most of the RA patients with periodontitis (85%) were ACPA-positive. In agreement with this finding, ACPA-positive patients have a higher incidence of periodontitis than patients with osteoarthritis48 and RA patients with periodontitis exhibit high titers of ACPAs and greater periodontal inflammation49,50.
Recently our group reported the impact of another systemic autoimmune disease, systemic lupus erythematosus (SLE) on periodontal status and subgingival microbiota51. Patients with SLE exhibit a higher prevalence of periodontitis, which occurs at a younger age when compared to healthy individuals. Like RA, SLE caused an increased bacterial load in subgingival sites and induced changes in the microbial composition and diversity that were linked to increased cytokines concentration on saliva (IL-6, IL-17 and IL-33), similar to the changes observed for RA patients. Together these results corroborate the hypothesis that systemic inflammatory conditions are associated with dysbiosis of subgingival microbiota and increased risk of periodontitis.
A limitation of our study is its cross-sectional design with a single time measurement. Therefore, we are unable to answer the question whether changes in the microbiota are a cause and/or effect of the RA. In addition, the use of different medications to treat RA may affect periodontal inflammation and the microbiota. Despite these limitations that data presented is significant. Our study is the first to demonstrate the influence of RA on subgingival microbiota taking into account the periodontal status of the subjects. The results support the concept that RA is an inflammatory disease that can modulate the subgingival biofilm and thus increase susceptibility to periodontal diseases.
Methods
Subjects
During one year we evaluated patients from the Rheumatology Outpatient Clinic of Clinics Hospital of Federal University of Minas Gerais (UFMG), Belo Horizonte, Brazil that were diagnosed with RA based on the 2010 American College of Rheumatology and EULAR classification criteria52. Two hundred thirty-nine patients agreed to participate, however, the study group consisted of forty-two patients based on the following inclusion criteria: no other rheumatic disease, no treatment for periodontal disease within the last 6 months, no use of orthodontic appliances, no use of antibiotics within the last 3 months, no pregnancy or lactation and the presence of at least 8 teeth. The control group consisted of 47 subjects without RA or other rheumatic diseases, that were age and gender matched with the RA group, randomly assigned from a population with demographic, social, and educational backgrounds similar to RA patients. Their medical history was obtained from an interview. Patients’ medical history and medications were determined by review of medical charts. Each patient had laboratory assessments that were taken from the medical record which included systemic measures of RA including blood levels of IgM rheumatoid factor (RF) and anti–citrullinated protein antibody (ACPA). As a measure of systemic inflammation C-reactive protein (CRP) was assessed as well as erythrocyte sedimentation rate (ESR). Periodontal status was assessed by two calibrated examiners (JDC and SMM) and the following parameters were recorded: plaque index, probing depth, clinical attachment level and bleeding on probing. Periodontitis was defined as the presence of two or more interproximal sites with probing depth ≥4 mm or one site with probing depth ≥5 mm53.
Ethics statement
The subjects gave written informed consent, and the study protocol was approved by the Federal University of Minas Gerais Ethics Committee (CAAE: 03128012.0.0000.5149/2012). All patient data and subgingival samples were anonymized. For sampling of PBMCs from the blood of healthy subjects they also gave written informed consent and their data were anonymized. All methods used here were carried out in accordance with the relevant guidelines and regulations.
Subgingival samples collection
Subgingival samples were collected using endodontic paper points (ISO40) (Tanariman, Manacaparu, AM, Brazil) that were inserted in 5 sites with deepest probing depth for one minute. After removal, the material was pooled together and stored in a sterile tube containing 500 µL of sterile distilled water and centrifuged at 3,000 g for 5 minutes. The paper points were discharged, and the pellet was kept at −80 °C until DNA extraction.
Saliva collection
Saliva was collected by continuous drooling into a sterile 50 mL tube for 5 minutes. The salivary flow was measured in milliliters per minute (ml/min). The saliva samples were diluted (1:1) in a phosphate-buffered saline (PBS) solution containing protease inhibitors and subsequently frozen at −80 °C until analysis.
Cytokines measurement
Concentrations of interleukin-2 (IL-2), IL-6, IL-17, tumor necrosis factor-α (TNF-α and interferon-γ (IFN-γ) in saliva were determined using a Cytokine Bead Array (CBA) Human Th1/Th2/Th17 Kit (BD Biosciences, San Diego, CA) and analyzed on a BD FACS Calibur flow cytometer (BD Biosciences). The concentration of IL-33 was measured by enzyme-linked immunosorbent assay (ELISA) (R&D Systems, Minneapolis, MN, USA). Assays were performed according to the manufacturer’s instructions. The results were expressed as picograms of cytokine (pg/ml) adjusted according to salivary flow.
DNA extraction and sequencing
DNA was extracted from plaque samples using the Quick-g DNA MicroPrep kit (Zymo Research, Irvine, CA, USA) and 50 µL (10 mg/ml) of lysozyme per sample to enhance bacterial lysis as described54. The quantity and quality of DNA was measured spectrophotometrically (Tecan, Männedorf, Switzerland). The primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) which target the hypervariable V4 region of the 16S rRNA gene were used for amplification55. After, agarose gel electrophoresis was performed to check size integrity. All amplicons were subjected to Illumina MiSeq Platform at the Next-Generation Sequencing Core of University of Pennsylvania and sequenced together at the same run. All Illumina sequence data were submitted to the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA325500.
Microbiota analysis
The raw reads were trimmed to remove regions with a low Phred score. Trimmomatic56 was the tool used with the TRAILING:5 and SLIDINGWINDOW:4:15 parameters. The trimmed reads were merged using FLASH57 tool requiring 30 reads overlap. The assembled amplicons were mapped to the CORE database using the Qiime’s pick_closed_reference_otus and 97% identity threshold. The representative set of sequences had their taxonomic classification using the same database and the Qiime’s assign_taxonomy script. The alpha diversity indexes were assessed using the Vegan R package58. Beta diversity was calculated using Unique Fraction metric (UNIFRAC) unweighted. Quantification of total bacterial load was determined by real-time PCR using universal primers for 16S rRNA gene. (F: AGAGTTTGATCCTGGCTCAG; R: ACGGCTACCTTGTTACGACTT) (IDT, Coralville, Iowa, USA) based on a a standard curve prepared using DNA extracted from a known number of Porphyromonas gingivalis (colony forming units) separated with flow cytometry and amplified with the same qPCR protocol. Samples were assayed in duplicate in a 25 μl reaction mixture containing 2.5 μl of template DNA, 2.5 μl of 10x TaqMan Universal PCR Master Mix, 1.5 μl of MgCl2,1 μl dNTP, 12.5 pmol of forward primer and reverse primer. The standard curve was used to derive the Cq (quantification cycle value) vs log CFUs linear equation. The cycling conditions used were as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min each.
To provide an inference of the functional profile of the microbial community based on 16 S rRNA gene sequence results we assigned taxonomy to the representative set of sequences using the GreenGenes 13.5 database. R. We utilized PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states)59, a computational approach to predict the functional composition of a microbial community using marker 16S sequencing data and a database of reference genomes, to identify and quantify the Pathways and KEGG Orthology Groups.
Blood collection and in vitro induction of PBMCs with oral biofilm
Blood was obtained from 5 systemically healthy individuals that did not have periodontitis or current use of immunosuppressive or anti-inflammatory drugs. Peripheral blood mononuclear cells (PBMCs) isolated by Ficoll–Paque gradient (Amersham Biosciences, Uppsala, Sweden) for 40 minutes at 20 °C. PBMCs were washed twice with PBS and counted in a hemocytometer chamber. The PBMCs of each patient (106 cells/ml) were incubated in a complete RPMI medium of 2 mM L-glutamine, 5% normal human serum, 100 l g/mL streptomycin, and 100 UI/mL penicillin G potassium with subgingival plaque in 96-well plates. For these assays total subgingival plaque was removed from a given site, placed in 100 ul of TE buffer, homogenized by vortexing and heat/freeze inactivated as described by Moutsopoulos and colleagues60. PBMCs were exposed to a standardized amount of inactivated bacteria (1 × 108 CFU) for 24 h and then supernatants were processed to evaluate cytokine concentration.
Statistics
After evaluating the normality of distribution by Kolmogorov-Smirnov tests, clinical, demographic, alpha diversity, cytokine levels and bacterial load data were compared using One Way ANOVA or Kruskal-Wallis test. Correlations between relative abundance of taxa and clinical parameters of periodontal disease and RA were calculated using Spearman correlation coefficients (SPSS software, version 20). PERMANOVA was performed to compare beta diversity (QIIME software, version 1.9). In case of multiple comparisons, the p-value was corrected using Bonferroni correction. The predicted functional groups, and Operational Taxonomic Units (OTU), were compared among RA and control subjects and tested for statistical significance using DESeq261 (R statistical software, version 3.5) and LEFSE62, respectively. P values < 0.05 were statistically significant.
Data Availability
All Illumina sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under BioProject Accession Number PRJNA325500.
References
Diaz, P. I., Hoare, A. & Hong, B. Subgingival Microbiome Shifts and Community Dynamics in Periodontal Diseases. J. Calif. Dent. Assoc. 44, 397–472 (2016).
Graves, D. T., Corrêa, J. D. & Silva, T. A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 1–9 (2018).
Graves, D. T., Jiang, Y. & Genco, C. Periodontal disease: bacterial virulence factors, host response and impact on systemic health. Curr. Opin. Infect. Dis. 13, 227–232 (2000).
Hajishengallis, G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44 (2015).
Koch, A. E. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 36, 5–8 (2007).
Demmer, R. T., Molitor, J. A., Jacobs, D. R. Jr. & Michalowicz, B. S. Periodontal disease, tooth loss and incident rheumatoid arthritis: results from the First National Health and Nutrition Examination Survey and its epidemiological follow-up study. J. Clin. Periodontol. 38, 998–1006 (2011).
Okada, M. et al. Periodontal Treatment Decreases Levels of Antibodies to Porphyromonas Gingivalis and Citrulline in Patients With Rheumatoid Arthritis and Periodontitis. J. Periodontol. 84, 74–84 (2013).
Araújo, V. M. A., Melo, I. M. & Lima, V. Relationship between Periodontitis and Rheumatoid Arthritis: Review of the Literature. Mediators Inflamm. 2015, 1–15 (2015).
Fuggle, N. R., Smith, T. O., Kaul, A. & Sofat, N. Hand to mouth: A systematic review and meta-analysis of the association between rheumatoid arthritis and periodontitis. Front. Immunol. 7, 1–10 (2016).
de Smit, M. J. et al. Periodontitis and Rheumatoid Arthritis: What do we Know? J. Periodontol. 86, 1013–1019 (2015).
Bright, R. et al. Gingival tissue, an extrasynovial source of malondialdehyde-acetaldehyde adducts, citrullinated and carbamylated proteins. J. Periodontal Res. 53, 139–143 (2018).
Brusca, S., Abramson, S. B. & Scher, J. U. Microbiome and mucosal inflammation as extra-articular triggers for rheumatoid arthritis and autoimmunity. Curr. Opin. Rheumatol. 26, 101–107 (2014).
Queiroz-Junior, C. M. et al. Experimental arthritis exacerbates Aggregatibacter actinomycetemcomitans-induced periodontitis in mice. J. Clin. Periodontol. 39, 608–616 (2012).
Scher, J. U. et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 64, 3083–94 (2012).
Zhang, X. et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015).
Mikuls, T. R. et al. The subgingival microbiome in patients with established rheumatoid arthritis. Rheumatology (2018).
Lopez-Oliva, I. et al. Dysbiotic Subgingival Microbial Communities in Periodontally Healthy Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 70, 1008–1013 (2018).
Deng, Z. L., Szafrański, S. P., Jarek, M., Bhuju, S. & Wagner-Döbler, I. Dysbiosis in chronic periodontitis: Key microbial players and interactions with the human host. Sci. Rep. 7, 1–13 (2017).
Colombo, A. P. V. et al. Comparisons of Subgingival Microbial Profiles of Refractory Periodontitis, Severe Periodontitis, and Periodontal Health Using the Human Oral Microbe Identification Microarray. J. Periodontol. 80, 1421–1432 (2009).
Mercado, F. B., Marshall, R. I. & Bartold, P. Inter-relationships between rheumatoid arthritis and periodontal disease. A review. J Clin Periodontol 30, 761–772 (2003).
de Pablo, P. et al. Periodontitis in systemic rheumatic diseases. Nat. Rev. Rheumatol. 5, 218–224 (2009).
Routsias, J., Goules, J., Goules, A., Charalampakis, G. & Pikazis, D. Autopathogenic correlation of periodontitis and rheumatoid arthritis. Rheumatol. 50, 1189–1193 (2011).
Queiroz-Junior, C. M. et al. Experimental arthritis triggers periodontal disease in mice: involvement of TNF-α and the oral Microbiota. J. Immunol. 187, 3821–30 (2011).
Corrêa, J. D. et al. Arthritis-induced alveolar bone loss is associated with changes in the composition of oral microbiota. Anaerobe 39, 91–96 (2016).
Scher, J. U. & Abramson, S. B. The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol. 7, 569–78 (2011).
Molloy, S. Microbiome: Tipping the balance. Nat. Rev. Microbiol. 10, 3 (2012).
Marques, C. P. C. et al. Salivary levels of inflammatory cytokines and their association to periodontal disease in systemic lupus erythematosus patients. A case-control study. Cytokine 85, 165–170 (2016).
Mirrielees, J. et al. Rheumatoid arthritis and salivary biomarkers of periodontal disease. J. Clin. Periodontol. 37, 1068–1074 (2010).
Isaza-Guzman, D. M. et al. Association study between salivary levels of interferon (IFN)-gamma, interleukin (IL)-17, IL-21, and IL-22 with chronic periodontitis. Arch. Oral Biol. 60, 91–99 (2015).
Liukkonen, J., Gursoy, U. K., Pussinen, P. J., Suominen, A. L. & Kononen, E. Salivary Concentrations of Interleukin (IL)-1beta, IL-17A, and IL-23 Vary in Relation to Periodontal Status. J. Periodontol. 87, 1484–1491 (2016).
Teles, R. P., Likhari, V., Socransky, S. S. & Haffajee, A. D. Salivary Cytokine Levels in Chronic Periodontitis and Periodontally Healthy Subjects. A cross-sectional Study. J. Periodontal Res. 44, 411–417 (2009).
Gemmell, E., Marshall, R. I. & Seymour, G. J. Cytokines and prostaglandins in immune homeostasis and tissue destruction in periodontal disease. Periodontol. 2000 14, 112–43 (1997).
McInnes, I. B., Buckley, C. D. & Isaacs, J. D. Cytokines in rheumatoid arthritis - shaping the immunological landscape. Nat. Rev. Rheumatol. 12, 63–8 (2016).
Köseoğlu, S., Hatipoğlu, M., Sağlam, M., Enhoş, Ş. & Esen, H. H. Interleukin-33 could play an important role in the pathogenesis of periodontitis. J. Periodontal Res. 50, 525–534 (2015).
Ebersole, J. L. et al. Cytokine gene expression profiles during initiation, progression and resolution of periodontitis. J. Clin. Periodontol. 41, 853–861 (2014).
Biyikoglu, B. et al. Periodontal therapy in chronic periodontitis lowers gingival crevicular fluid interleukin-1beta and DAS28 in rheumatoid arthritis patients. Rheumatol. Int. 33, 2607–2616 (2013).
Hajishengallis, G. The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral Microbiol. 29, 248–257 (2014).
Xiao, E. et al. Diabetes Enhances IL-17 Expression and Alters the Oral Microbiome to Increase Its Pathogenicity. Cell Host Microbe 22, 120–128 (2017).
Smit, M. D. et al. Periodontitis in established rheumatoid arthritis patients: a cross-sectional clinical, microbiological and serological study. Arthritis Res. Ther. 14, R222 (2012).
Bingham, C. O. III. & Moni, M. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr. Opin. Rheumatol. 25, 345–353 (2013).
Diaz, P. I. et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol Oral Microbiol 27, 180–201 (2012).
Marsh, P. D., Head, D. A. & Devine, D. A. Ecological approaches to oral biofilms: Control without killing. Caries Res. 49, 46–54 (2015).
Kurita-Ochiai, T. et al. Butyric Acid Induces Apoptosis in Inflamed Fibroblasts. J Dent Res 87, 51–55 (2008).
Takahashi, N. Oral Microbiome Metabolism: From ‘Who Are They?’ to ‘What Are They Doing?’. J. Dent. Res. 94, 1628–37 (2015).
Schwenzer, A. et al. Association of Distinct Fine Specificities of Anti–Citrullinated Peptide Antibodies With Elevated Immune Responses to Prevotella intermedia in a Subgroup of Patients With Rheumatoid Arthritis and Periodontitis. Arthritis Rheumatol. 69, 2303–2313 (2017).
Daha, N. A. & Toes, R. E. M. Rheumatoid arthritis: Are ACPA-positive and ACPA-negative RA the same disease? Nat. Publ. Gr. 7, 202–203 (2011).
Seegobin, S. D. et al. ACPA-positive and ACPA-negative rheumatoid arthritis differ in their requirements for combination DMARDs and corticosteroids: secondary analysis of a randomized controlled trial. Arthritis Res. Ther. 16, 1–12 (2014).
Gonzalez, S. M. et al. Alveolar bone loss is associated with circulating anti-citrullinated protein antibody (ACPA) in rheumatoid arthritis patients. J. Periodontol. 86, 222–231 (2015).
Potikuri, D. et al. Periodontal disease is signifi cantly higher in non-smoking treatment-naive rheumatoid arthritis patients: results from a case-control study. 1541–1544, https://doi.org/10.1136/annrheumdis-2011-200380 (2012).
Nesse, W. et al. The periodontium of periodontitis patients contains citrullinated proteins which may play a role in ACPA (anti-citrullinated protein antibody) formation. J Clin Periodontol 39, 599–607 (2012).
Corrêa, J. D. et al. Subgingival microbiota dysbiosis in systemic lupus erythematosus: association with periodontal status. Microbiome 5, 34 (2017).
Aletaha, D. et al. Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 62, 2569–2581 (2010).
Eke, P. I., Page, R. C., Wei, L., Thornton-Evans, G. & Genco, R. J. Update of the Case Definitions for Population-Based Surveillance of Periodontitis. J. Periodontol. 83, 1449–1454 (2012).
Grice, E. A. et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc. Natl. Acad. Sci. USA 107, 14799–804 (2010).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–4 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
Moutsopoulos, N. M. et al. Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology. PLoS Pathog. 11, 1–19 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Acknowledgements
We thank Professor Dr. Paulo Eduardo Alencar de Souza for his contribution with in vitro experiment.
Author information
Authors and Affiliations
Contributions
The project was conceived by T.A.S., D.T.G., G.A.F. and A.L.T. The clinical material was collected by J.D.C., D.C.C. and S.M.M. All laboratory work was conducted by J.D.C. and T.A.S. with the help of S.M.M., J.M.S., M.L.A. and E.X. Bioinformatics analysis was conducted by E.J.L., C.M., G.R.F., E.X. and J.D.C. J.D.C., D.T.G., F.Q.C. and T.A.S. interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Corrêa, J.D., Fernandes, G.R., Calderaro, D.C. et al. Oral microbial dysbiosis linked to worsened periodontal condition in rheumatoid arthritis patients. Sci Rep 9, 8379 (2019). https://doi.org/10.1038/s41598-019-44674-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-44674-6
This article is cited by
-
BugSigDB captures patterns of differential abundance across a broad range of host-associated microbial signatures
Nature Biotechnology (2023)
-
Nitrate and a nitrate-reducing Rothia aeria strain as potential prebiotic or synbiotic treatments for periodontitis
npj Biofilms and Microbiomes (2023)
-
Influence of Environmental Factors on Salivary Microbiota and Their Metabolic Pathway: Next-Generation Sequencing Approach
Microbial Ecology (2023)
-
Associations between inflammation-related LL-37 with subgingival microbial dysbiosis in rheumatoid arthritis patients
Clinical Oral Investigations (2022)
-
The facultative human oral pathogen Prevotella histicola in equine cheek tooth apical/ periapical infection: a case report
BMC Veterinary Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.