Abstract
Pyrrolone-fused benzosuberene (PBS) compounds were semi-synthesized from α,β,γ-Himachalenes extracted from the essential oil of Cedrus deodara following amino-vinyl-bromide substituted benzosuberenes as intermediates. These PBSs compounds classified as an attractive source of therapeutics. The α-isoform of PI3K which is a pivotal modulator of PI3K/AKT/mTOR signaling pathway, responsible for neurological disorders like epilepsy, found as a potential target molecule against these 17 semi-synthesized PBS compounds using in silico ligand-based pharmacophore mapping and target screening. The compounds screened using binding affinities, ADMET properties, and toxicity that were accessed by in silico docking simulations and pharmacokinetics profiling. Ultimately two compounds viz., PBS-8 and PBS-9 were selected for further in vivo evaluation using a zebrafish (Danio rerio) model of pentylenetetrazol (PTZ)-induced clonic convulsions. Additionally, gene expression studies performed for the genes of the PI3K/AKT/mTOR pathway which further validated our results. In conclusion, these findings suggested that PBS-8 is a promising candidate that could bedeveloped as a potential antiepileptic.
Similar content being viewed by others
Introduction
Himachalenes a mixture of sesquiterpenes extracted from Cedrus deodara oil containing hexahydrobenzocycloheptene as basic skeleton can be easily converted to benzocycloheptene/benzosuberene, a core structure of several natural products like colchicine, allocolchicine, demethylsalvicanol and feveline etc. which are clinically reported bioactive compounds1,2. In earlier developments, α, β, γ-himachalenes as a mixture were applied through sequential and consecutive approaches for the synthesis of substituted benzosuberenes as a reactive and bio-active precursor3,4. The present study aimed to search target molecules from benzosuberene classes of compounds which could serve as promising therapeutics for treating the target disease. After successful in-silico ligand-based pharmacophore mapping and target identification5, we found phosphoinositide-3-kinase-α (PI3K-α) as a potential target against the selected molecules. PI3Ks are lipid kinases that control mTOR (mammalian target of rapamycin) signaling pathway which is responsible for cell proliferation, cell invasion, cell migration and cell death6. The mTOR pathway is a frequent target of epilepsy treatment. mTOR hyperactivation has been found to be active in many types of human cancers and neurological disorders. mTOR is a serine/threonine protein kinase that belongs to the PI3K family and is encoded by the MTOR gene7,8. PI3K consist of three classes: Class I, Class II and Class III, in which Class I is divided into Class IA and Class IB. PI3K-α falls under the Class IA. It catalyze the phosphorylation of 3′-hydroxyl group of the inositol ring of phosphatidylinositol and also activated by cell surface receptors such as receptor tyrosine kinases (RTKs), G-protein coupled receptors (GPCRs) and small G-protein oncogenes (Ras)9,10. They are heterodimers of catalytic and regulatory subunits, such as p110 (catalytic) and p85 (regulatory)11,12. Human cells contain the PIK3CA gene that encodes catalytic subunit such as p110α of class I PI3K13. Phosphorylation of tyrosine kinase receptor results in the activation of PI3K which activates cascading steps of phosphorylation. PI3K further activates AKT, which in turn, phosphorylates mTOR, that has downstream regulatory effects on genes such as ribosomal protein S6 kinase (Rps6kb1) and ribosomal protein S6 (Rps6). PI3K inhibition counteracts the downstream activation of mTOR14,15. PI3K/AKT/mTOR signaling pathway has been proven to be involved in neurogenesis and dysregulation, or hyperactivation of this signaling pathway strongly associated with many severe brain disorders, including epilepsy16. Moreover, mutations in this signaling pathway have been found in a large number of human patients with epilepsy17.
Epilepsy is the fourth most common neurological disorder that affects people of all ages. It is characterised by recurrent neuronal seizures with or without loss of consciousness15. If not treated well, it might result in brain damage which could even lead to death18. Many anti-epileptic drugs are available in the market which possess significant adverse effects on the health of the individual19. This necessitates an urgent need for an effective therapeutic drug against epilepsy with minimal or no side effects.
These demands initiated an enormous interest in screening the active therapeutic agents for the treatment of epilepsy. Our present study suggested a potential lead compound that could be used to treat epilepsy by compressing PI3K-α activation. In this direction, we screened and evaluated benzosuberene classes of compounds by in silico approaches. Further, to validate the activity of the computationally suggested compound(s) against epilepsy, we tested these compounds in a Zebrafish (Danio rerio) larvae model of pentylenetetrazol (PTZ)-induced convulsions. The study identified a potential lead compound against epilepsy, and this workflow is presented in Fig. 1. As this compound is naturally derived, therefore it could probably be a better candidate for the therapeutic purpose with more bio- compatibility and less toxicity.
Results
Semi-synthesis of PBS compounds
The pyrrolone fused benzosuberenes (PBSs) used under this study were semi-synthesized from α,β,γ-himachalenes following the intermediates of amino-vinyl-bromide substituted benzosuberenes. Further, these intermediates in the presence of oxalic acid as in situ CO source under palladium catalyzed condition gave pyrrolone-fused benzosuberenes (PBSs) (Fig. 2, ligand 1–17). Under this study, several functional groups were found to be toleratnt and ended with good yields20.
Identification of a target molecule
Further, to identify the target molecule against 17 PBS compounds, we used a ligand-based virtual screening approach21 with the help of Accelrys Discovery studio package. The 3D pharmacophore model against these PBS ligands were mapped using the interaction pattern of cations, anions, aromatic, aliphatic, hydrophobic and hydrogen bond donors/acceptors5. The pharmacophore model thus generated was then used to search the pre-existing structured databases to identify the molecular structure that best matches with the pattern of that pharmacophore map. This similarity search unearths PI3K (α-isoform) as the biological target against our PBS compounds.
Analyses of binding energies and binding interactions
For enumeration of specific inhibitors against α isoform of PI3K lipid kinase, we docked our 17 naturally originated compounds with this isoform. We calculated the energy of interaction between PI3K-α and 17 PBS ligands. Docking with Autodock 4.2.622 exhibited different binding energies of 17 docked ligands with PI3K, ranging from −8 to −10 kcal/mol (Fig. 3). Lowest binding energies of our 17 PBS compounds docked with α isoform following the ligand order of PBS-9, PBS-12 (−9.35 kcal/mol) < PBS-2 (−9.28 kcal/mol) < PBS-5 (−9.25 kcal/mol) < PBS-3 (−9.22 kcal/mol) < PBS-10 (−9.17 kcal/mol) < PBS-11 (−9.16 kcal/mol) < PBS-6 (−9.13 v) < PBS-8 (−8.99 kcal/mol) < PBS-13, PBS-17 (−8.96 kcal/mol) < PBS-7 (−8.86 kcal/mol) < PBS-16 (−8.83 kcal/mol) < PBS-4 (−8.60 kcal/mol) < PBS-1 (−8.31 kcal/mol) < PBS-14 (−8.26 kcal/mol) < PBS-15 (−8.19 kcal/mol), as shown in Table 1. The atomic interactions were further explored by LigPlot+ v.1.4 software23. This software could plot 2D views of in-depth ligand bonds, non-ligand bonds, hydrogen bonding and hydrophobic interactions pattern between the docked ligands and the active site residues of the corresponding receptor (Fig. 4).
Docked conformations of selected PBS compounds (represented as red color spheres) complexed with PI3K-α (represented as a green color cartoon); that are screened through binding energies obtained by Autodock 4.2.6. An enlarged view of docked PBS ligands and PI3K-α residues shows 2D interactions using LigPlot+. Purple lines represent ligand bonds, and yellow lines represent non-ligand bonds. Hydrogen-bonds are represented by green dotted lines and distances between atoms are expressed in Å. Residues involved in hydrophobic interactions are identified by a red color semicircle surrounding them with corresponding atoms represented by black dots.
Analyses of drug-likeliness
To determine the drug-likeliness of our PBSs compounds, we calculated their ADMET properties. The screened results of ADMET were summarized in Table 2, revealing six descriptors such as absorption, solubility, cytochrome P450 2D6 (CYP2D6), plasma protein binding (PPB), hepatotoxicity, and AlogP98. Easy absorption of the drug through blood brain barrier (BBB) measured by its AlogP98 value which must be less than 5. The obtained absorption levels determine drug absorption and absorption decreases inversely with the level, i.e., level 0 denotes proper absorption, level 1 denotes moderate absorption and so on. To determine the inhibitory effect and toxicity of the drug CYP2D6 and hepatotoxicity descriptors gave two predicted classes, such as class 0 (non-inhibitor and non-toxic) and class 1 (inhibitor and toxic). Another ADMET descriptor PPB has given two results true or false that symbolizes the binding or non-binding of the drug. Moreover, solubility descriptor predicted the molar solubility of the drug, which gives good molar solubility within a range of −2.0 and −4.0. If it is below that range, solubility decreases gradually and becomes extremely low below −8.0, and above that range, it increases gradually to become too soluble at 0.0.
ADMET model also developed using two descriptors such as 2D polar surface area (2D-PSA) and AlogP98 (Fig. 5). This model includes the eclipses of 99% and 95% confidence limits which were used to define the regions with compounds having proper intestinal absorption and penetration across the BBB. The results of the obtained ADMET model showed that all our PBS compounds fell inside all the eclipses expect two of them fell outside the eclipse of 95% confidence limits for intestinal absorption.
A plot of AlogP98 versus 2D Polar Surface Area (PSA) for our PBS compounds. The plot is showing green and blue colored eclipses of 99% confidence limits for intestinal absorption and blood-brain barrier (BBB), as well as red and pink colored eclipses of 95% confidence limits for intestinal absorption and BBB.
Another method used to determine the therapeutic compatibility of the drug is toxicity prediction by komputer assisted technology (TOPKAT), summarized in Table 3. TOPKAT is a useful tool for in-silico prediction of toxicity quantitatively, and it is employed in quantitative structure-activity relationship (QSTR) models. Moreover, with the help of these QSTR models, it calculates probability values and evaluates toxicity through them. It follows the criterion of checking the components in the optimal predictive space (OPS), and when they lie outside then the results were considered as unreliable, i.e., false positives. Obtained Ames probability values determine the level of toxicity, such as, 0.0 to 0.30 (non-toxic), 0.30 to 0.70 (inter vocal), and 0.70 to 1.0 (toxic). Other additional descriptors provided was Ames mutagenicity, Ames enrichment and Ames score to determine the reliability of the predictions. It also provides values of the carcinogenic potency of TD50 mouse, rat oral LD50, rat inhalational LC50, and Daphina EC50. Increase in TD50, LD50, LC50, and EC50 values predicts the decrease in toxicity and increase in safety index of the drug which makes it more potent.
Further analyses of drug-likeliness were performed by Lipinski’s rule-of-five to determine the ability of the drug to diffuse passively through the BBB. Lipinski’s rule-of-five follows the criteria of number of violations listed in Table 4 such as, molecular weight of compound should be less than 500, AlogP value should be less than 5, hydrogen bond donors should be less than 5, hydrogen bond acceptors should be less than 10, and number of rotatable bonds should be less than 5, respectively.
Effect on PTZ-induced clonic seizures
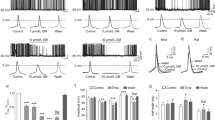
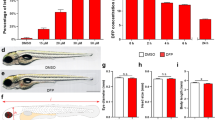
The exposure with 8 mM PTZ showed the appearance of clonic seizure in vehicle control larvae with a latency of 4.42 ± 0.15 min. Pre-incubation with 1 µM concentration of PBS-9 (P = 0.002) and PBS-8 (P < 0.001) resulted in a marked increase in latency to clonic-like seizures in comparison to vehicle control. However insignificant difference in latency to first clonic-like seizure was observed at 1 µM concentration of PBS-9 and PBS-8 (P = 0.670). The group pre-incubated with sodium valproate showed a significant (P < 0.001) increase in latency to clonic-like seizures. The effect of both PBS-8 (P = 1.00) and PBS-9 (P = 0.639) at 1 µM concentration was found to be equipotent when compared to sodium valproate. Both test compounds were found to be ineffective at 0.25 µM and 0.5 µM concentrations as compared to vehicle control (Fig. 6).
Effect on mRNA levels
Larvae exposure to PTZ resulted in a significant increase in mRNA levels of c-fos (P = 0.003), PIK3CA (P < 0.001), PIK3R1 (P < 0.001), AKT1 (P < 0.001), mTOR (P < 0.001), Rps6 (P 0.003) and Rps6kb1 (P < 0.001) as compared to naive. The level of c-fos mRNA was found to be significantly decreased in PBS-8 (P = 0.004), and PBS-9 (P = 0.005) exposed larvae in contrast to vehicle control. Furthermore, pre-incubation with a 1 µM concentration of PBS-8 and PBS-9 showed a significant (P < 0.001) decrease in mRNA levels of AKT, PIK3CA, PIK3R1, mTOR, and Rps6kb1 as compared to the vehicle control larvae. A marked decrease in mRNA level of Rps6 was also observed in the larvae exposed to PBS-8 (P = 0.049) and PBS-9 (P = 0.005) and as that of vehicle control (Fig. 7).
Discussion
For identification of the biological target against ligand 1–17, they were undergone through in silico studies. The procedures of virtual screening were performed to identify the biological target using Discovery studio package. Virtual screening is an efficient approach that is widely used for the discovery of novel compounds24. In the absence of target molecule information ligand-based virtual screening approach had been successfully applied, such as pharmacophore mapping and similarity searching5.
PI3K-α was a resulted target of ligand-based virtual screening against these 17 PBSs compounds. PI3K as reported plays an essential role in the activation of mTOR signaling pathway. The role of the PI3K/AKT/mTOR pathway has been widely deciphered in epilepsy14,25. Studies suggested that injury due to seizures led to activation of the pathway which further propagates seizure progression and related pathogenic changes26. Furthermore, the constitutive stimulation of the pathway has been well explored in various in vitro27 and in vivo models28. Studies conducted on PI3K suggested its phosphatidylinositol group to be responsible for the emergence of a large number of mitotic factors, thus resulting in the proliferation of cells29 through phosphorylating AKT and activation of its downstream genes such as mTOR, Rps6, and Rps6kb1. It has been found that all mammalian cells when activated by receptor tyrosine kinases, expresses at least one of the isoforms of PI3K. Moreover, the PI3K (class IA) is functional when the catalytic subunit p110α binds with its regulatory adapter protein p85α to form a dimer30. Thus this target is beneficial for the development of new treatment avenues in epilepsy.
In this study, in silico docking and pharmacokinetic profiling were performed for screening of more drug-likely PBSs ligands against the PI3K-α protein. Molecular docking is a computational approach widely used in the drug discovery process31,32. These PBSs compounds were then screened by obtaining binding energies using docking simulations. Smaller the binding energy, more potential it is. Based on binding energies nine compounds were screened viz., PBS-2, PBS-3, PBS-5, PBS-6, PBS-8, PBS-9, PBS-10, PBS-11 and PBS-12, and based on the torsional free energy they were screened down to eight viz., PBS-3, PBS-5, PBS-6, PBS-8, PBS-9, PBS-10, PBS-11, and PBS-12. Consequently, the screening of these remaining eight PBS compounds were done using their ADMET properties. PBS-12 was screened out based on ADMET adsorption level descriptor as well as PBS-5 was screened out based on its inhibition effect and toxicity showed by CYP2D6 and hepatotoxicity descriptors. After ADMET screening six compounds were selected viz., PBS-3, PBS-6, PBS-8, PBS-9, PBS-10, and PBS-11. Further screening of PBSs compounds were done based on TOPKAT results. PBS-3 and PBS-11 compounds were screened out based on their low potency values obtained by TOPKAT carcinogenic potency of TD50 mouse descriptor. Moreover, PBS-6 and PBS-10 were screened out based on the applicability of rat oral LD50, rat inhalational LC50, and Daphina EC50. Finally,in silico docking studies and pharmacokinetic profiling suggested that PBS-8 and PBS-9 compounds were found suitable inhibitor for PI3K-α. Finally, they were evaluated through in vivo studies using zebrafish as a model organism for human epilepsy.
Since the past few decades, zebrafish has gained popularity as a developed disease model. The foremost criterion for using zebrafish in epilepsy research is to ascertain seizure like clonic convulsions as depicted by the Racine scale in rodent model33. Accordingly, in the present study, seizure-like behavior in 7 dpf (days post fertilization) zebrafish larvae were established using PTZ, and latency to first clonic seizure was recorded. The study found that, exposure to the test compounds (PBS-8 and PBS-9) depicted a considerable increase in PTZ-mediated clonic seizure latency at 1 µM concentration. Epileptic studies conducted on rodent models have revealed that seizure generation leads to immediate early genes expression, particularly c-fos upregulation in the brain34, a similar observation has also been made earlier in the zebrafish larvae model33. In line with this observation, the present study showed increased c-fos expression in PTZ exposed vehicle control larvae, and subsequent decrease upon pre-incubation with PBS-8 and PBS-9, supporting its anti seizure effect. Our findings were further supported by previous work showing c-fos downregulation in larvae treated with an antiseizure compound35.
Our study revealed that the genes encoding these units, i.e., p110α (PIK3CA gene) and p85α (PIK3R1 gene) were upregulated in the larvae exposed to PTZ. Pre-incubation with PBS-8 and PBS-9 showed a reduction in their expression. The fact that the intricate mechanism of PI3K/AKT activation leading to mTOR hyperactivation in rodent models of epilepsy36 can be corroborated with our findings, which showed that the expression of all the downstream genes, i.e.,AKT, mTOR, Rps6, and Rps6kb1 were reduced dramatically following drug treatment on PTZ treated larvae.
The study identified few potential lead compounds against epilepsy. Our adopted approaches addresse the complexity in searching enormous natural bioactive space. Moreover cellular targets of a natural lead is crucial for the process of drug discovery. We strongly recommand computational exploration in target identification and screening of lead before going for an in-vivo analysis. It helps to reduce the effort and time of a researcher.
Materials and Methods
Synthesis of chemical compounds
All the 1–17 pyrrolone-fused benzosuberene (PBS) compounds used under this study were synthesized following our earlier developed protocols20. These compounds (ligand 1–17) were synthesized with different functional groups at the specific position denoted by R, as shown in Fig. 2. Further, these molecules were investigated for therapeutic applications.
Ligand-based Virtual screening to natural analogs
Accelrys Discovery studio package (Dassault Systèmes BIOVIA, 2017R2, San Diego) was used for deriving pharmacophore mapping5, that is a type of ligand-based virtual screening. Naturally extracted seventeen ligands were fed in the Accelrys Discovery studio package to generate pharmacophore model, with default parameters. The pharmacophore model was selected and then used to search the 3D structure database to identify the appropriate receptor structure.
Protein dataset
The crystallographic structure of PI3K (α-isoform) class I lipid kinase was achieved from Brookhaven PDB (Protein Data Bank; www.rcsb.org) server37. We selected a catalytic subunit of α isoform (PDB ID: 4JPS) of Homo sapiens organism, solved by X-ray diffraction method at a resolution of 2.2 Å13.
Pharmacokinetic properties
Drug-likeliness of 17 PBS compounds were analyzed by assessing Lipinski’s rules, Absorption, distribution, metabolism, excretion, and toxicity (ADMET) descriptors, and TOPKAT descriptors using Accelrys Discovery studio package. ADMET analyses were performed using sixdescriptors, such as absorption, solubility, CYP2D6, Plasma Protein Binding (PPB), hepatotoxicity, and AlogP98. Also, Toxicity Prediction by Komputer Assisted Technology (TOPKAT) analyses were performed using carcinogenic potency of LD50 mouse, Ames mutagenicity, Ames probability, Ames enrichment, Ames score, rat oral LD50, rat inhalational LC50, and Daphina EC50.
Docking simulations
Binding affinities of our 17 PBS compounds against an alpha isoform of PI3-Kinase were computed using an open source software Autodock 4.2.6 version22. These compounds were used as ligands against PI3K- α protein receptor.
Protein structure of PI3K-α was preprocessed for docking by computing Gasteiger charges, adding hydrogens and removing water. The receptor was kept rigid while the ligands were kept flexible by setting torsion angles. Precise binding complexes of ligands and receptor molecule were obtained by setting grid maps. The grid points per map in X, Y, Z dimensions: 60 × 60 × 60 Å with 0.375 Å spacing and grid center X: −1.318, Y: −9.512, Z: 16.948, were used to cover catalytic pockets of PI3K-α. Binding conformations were estimated as per default docking parameters and Lamarckian Genetic Algorithm for ligands. For further interaction analysis, lowest binding energy conformations were plotted by LigPlot + v.1.4.523.
Zebrafish maintenance
Adult wild-type zebrafish of 4–5 months were housed in a ZebTEC Stand-Alone system (Tecniplast, Buguggiate, Varese, Italy) set at temperature 26–28 °C, pH 7.0–7.5 and conductivity 400 - 600 µS. The room photo period cycle was maintained at 14:10 h light: dark and fish were fed twice a day with freshly hatched live Artemia (Inve Aquaculture, Inc., Salt Lake City, USA). The experimental protocol was duly approved by the Institutional Animal Ethics Committee of CSIR-IHBT and was performed in accordance with the approved guidelines.
Egg collection
The eggs were obtained from the induced spawning of healthy adults in separate breeding tanks. Briefly, at the end of the light cycle of the day prior to collection of eggs, two males and four females (1:2 ratio) were transferred to a breeding tank (Tecniplast, Buguggiate, Varese, Italy) separated by a transparent divider, containing system water maintained at 28.5 °C temperature. The breeding setup comprised of an internal grid bottom tank (Model: ZB10BTI) with a sliding transparent divider (Model: ZB10BTD), fitted into the external solid bottom tank (Model: ZB10BTE) covered with a transparent lid (Model: ZB10BTL). On the day of egg collection, the lid was removed after 30 min of the start of the light cycle. Healthy fertilized eggs were collected in sterile petri dishes using a pipette and were cleaned with 2–3 rounds of washing with system water. Around 50 eggs per plate were kept in a BOD incubator (Relitech, Ambala, India) at 28.5 °C. Regular water changes (twice a day) were done till 7 dpf.
PTZ-induced epileptic seizures
The larvae at 7 dpf in different groups (n = 6) were pre-incubation with 3 different concentrations [0.25, 0.5 and 1 μM (selected on the basis of pilot studies)] of PBS-8 and PBS-9 for 1 h at 28.5 °C. The stock solutions of the compounds were made in pure dimethyl sulfoxide, and working dilutions were made with system water (0.01% concentration of DMSO in final solution). Following pretreatment, each larva was exposed to 8 mM of PTZ at 28.5 °C, and the induced seizures were recorded with upper cut off time of 15 min. Two separate groups of larvae (n = 6) were also exposed to PTZ after pre-incubation in system water and valproic acid sodium salt (3 mM in system water) that served as vehicle control and standard, respectively. Three different stages appeared in larva exposed to PTZ as, Stage I: enhanced swimming activity or hyperactivity; Stage II: circular whirlpool-like movements and; Stage III: clonic seizures with loss of posture and falling33. Increase in latency to clonic seizures was recorded as a parameter for anticonvulsant effect.
Analysis of target gene expression
For gene expression studies healthy zebrafish larvae of 7 dpf were pre-incubated with test compounds (1 µM) for 1 h and later exposed to 8 mM PTZ solution for 15 min (3 set of larvae for each concentration, n = 20/set). Similar, separate sets were made that served as vehicle control (pre-incubated in system water and exposed to PTZ) and naïve control (not exposed to PTZ). At 1 h, post-PTZ exposure, total RNA was extracted from whole larvae (n = 20/group) using Trizol reagent (Sigma Aldrich, USA). The homogenate formed after Trizol exposure was treated with chloroform and centrifuged at 12,000 g for 15 min at 4 °C following incubation at room temperature for 5 min. The aqueous layer obtained after chloroform treatment was further subjected to 100% isopropanol to precipitate the RNA and centrifuged at 12,000 g for 10 min at 4 °C. The pellet obtained was repeatedly washed in 75% ethanol, centrifuged at 7500 g for 5 min at 4 °C, dissolved in nuclease-free water and quantified using Nanodrop ND-1000 (Thermo Scientific, USA). The total RNA obtained was treated with RNase-free DNase kit (Promega, Madison, USA) for removal of trace amounts of DNA, following which cDNA synthesis was done using high capacity cDNA-RT kit (Applied Biosystems, USA), as per manufacturers guidelines. The quantitative real-time polymerase chain reaction (qRT-PCR) analysis was done by SYBR Green Jump start Taq Ready Mix (Sigma Aldrich, USA) on Step One Plus Real-Time PCR system (Applied Biosystems, USA) using elongation factor-1-α (elf1α) of zebrafish as the reference standard. The target-specific primers were designed (Table 5) using Primer Express Software 3.0 (Applied Biosystems, USA). Moreover, to reduce sampling error, the mean of each sample was considered after performing the reaction in triplicate. The annealing temperature of each gene was standardized at 55 °C, and gene expression was denoted by fold change using comparative 2^ddCT method38.
Statistical analysis
The results of latency to clonic seizures were shown as mean ± standard error. The gene expression study results were presented as mean ± standard deviation. The statistical significance among different groups was analyzed using one-way analysis of variance followed by Tukey’s test. The results were regarded as significant at P < 0.05.
References
Williams, M., Kowaluk, E. A. & Arneric, S. P. Emerging molecular approaches to pain therapy. Journal Medicinal Chemistry 42(9), 1481–500 (1999).
Brossi, A. Bioactive alkaloids. 4. Results of recent investigations with colchicine and physostigmine. Journal of Medicinal Chemistry 33, 2311–2319 (1990).
Chaudhary, A. et al. Naturally occurring himachalenes to benzocycloheptene amino vinyl bromide derivatives: as antidepressant molecules. Molecular Diversity. 357–366 (2012).
Reddy, C. B., Bharti, R. & Das, P. ‘RSC Advances Supported palladium nanoparticles-catalyzed decarboxylative coupling approaches to aryl alkynes, indoles and pyrrolines synthesis †. Royal Society of Chemistry. 6, 71117–71121 (2016).
Markt, P. et al. Discovery of novel CB2 receptor ligands by a pharmacophore-based virtual screening workflow. Journal of medicinal chemistry. 52(2), 369–378 (2008).
Shayesteh, L. et al. PlK3CA is implicated as an oncogene in ovarian cancer. Nature Genetics. 21(1), 99–102 (1999).
Guertin, D. A. & Sabatini, D. M. Defining the Role of mTOR in Cancer. Cancer Cell. 12(1), 9–22 (2007).
Bellacosa, A. et al. Activation of AKT kinases in cancer: Implications for therapeutic targeting’. Advances in Cancer Research. 94(04), 29–86 (2005).
Katso, R. et al. 3-K INASES: Implications for Development, Class I PI3Ks. Annu. Rev. Cell Dev. Biol. 17, 615–75 (2001).
Samuels, Y. & Ericson, K. Oncogenic PI3K and its role in cancer. Current Opinion in Oncology. 18(1), 77–82 (2006).
Vivanco, I. & Sawyers, C. L. The phosphatidylinositol 3-Kinase–AKT pathway in human cancer. Nature Reviews Cancer. 2(7), 489–501 (2002).
Liu, P. et al. Targeting the phosphoinositide 3-kinase (PI3K) pathway in cancer. Nat. Rev. Drug. Discov. 8(8), 627–644 (2009).
Furet, P. et al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorganic and Medicinal Chemistry Letters. Elsevier Ltd. 23(13), 3741–3748 (2013).
Mazumder, A. G., Padwad, Y. S. & Singh, D. Anticancer Mammalian target of rapamycin (mTOR) signaling pathway inhibitors: Current status, challenges and future prospects in management of epilepsy. CNS and Neurological Disorders - Drug Targets. 15(8), 1–11 (2016).
Wei, H. et al. Geniposide attenuates epilepsy symptoms in a mouse model through the PI3K/Akt/GSK - 3 β signaling pathway. Experimental and therapeutic medicine. 15(1), 1136–1142 (2018).
Brandt, C. et al. The novel, catalytic mTORC1/2 inhibitor PQR620 and the PI3K/mTORC1/2 inhibitor PQR530 effectively cross the blood-brain barrier and increase seizure threshold in a mouse model of chronic epilepsy. Neuropharmacology. 40, 107–120 (2018).
Cheng, X. et al. The effect of P85 on neuronal proliferation and di ff erentiation during development of mouse cerebral cortex. Developmental biology. 441(1), 95–103 (2018).
Xia, J. et al. Biomedicine & Pharmacotherapy Therapeutic effects of scoparone on pilocarpine (Pilo) -induced seizures in mice. Biomedicine & Pharmacotherapy. Elsevier. 97(June 2017), 1501–1513 (2018).
Wu, Q. Down-regulation of Long Noncoding RNA MALAT1 Protects Hippocampal Neurons Against Excessive Autophagy and Apoptosis via the PI3K/Akt Signaling Pathway in Rats with Epilepsy. Journal of Molecular Neuroscience, 1–12 (2018).
Bharti, R., Reddy, C. B. & Das, P. Oxalic Acid as Sustainable CO Source for Pyrrolone-Fused Benzosuberenes Synthesis through Palladium Catalyzed Carbonylative Cyclization. ChemistrySelect. 2, 4626–4629 (2017).
Hamza, A., Wei, N. & Zhan, C. Ligand-Based Virtual Screening Approach Using a New Scoring Function. Frontiers in Pharmacology. 9, 11 (2012).
Morris, G. & Huey, R. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry. 30(16), 2785–2791 (2009).
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. Ligplot - a Program To Generate Schematic Diagrams of Protein Ligand Interactions. Protein Engineering. 8(2), 127–134 (1995).
Drwal, M. N. & Griffith, R. Combination of ligand- and structure-based methods in virtual screening. Drug Discovery Today: Technologies. Elsevier Ltd. 10(3), e395–e401 (2013).
Mazumder, A. G., Patial, V. & Singh, D. Mycophenolate mofetil contributes to downregulation of hippocampal interleukin type 2 and 1β mediated PI3K/AKT/mTOR pathway hyperactivation and attenuates neurobehavioral comorbidities in rat model of temporal lobe epilepsy. Brain, Behavior and Immunity. 75, 84–93 (2018).
Xiao, Z. et al. Interleukin-1 β plays a role in the pathogenesis of mesial temporal lobe epilepsy through the PI3K/Akt/mTOR signaling pathway in hippocampal neurons. Journal of Neuroimmunology. Elsevier. 282, 110–117 (2015).
Claessens, Y. et al. Role of the phosphatidylinositol 3-kinase/Akt and mTOR/P70S6-kinase pathways in the proliferation and apoptosis in multiple myeloma. Molecular cancer therapeutics. 6587–6597 (2002).
Dai, R. et al. Involvement of PI3K/Akt Pathway in the Neuroprotective Effect of Sonic Hedgehog on Cortical Neurons under Oxidative Stress. Journal of Huazhong University of Science and Technology [Medical Sciences]. 32(6), 856–860 (2012).
Mahoss, C. J. et al. A Specific Inhibitor of Phosphatidylinositol 3-Kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). 269(7), 5241–5248 (1994).
Vanhaesebroeck, B. & Waterfield, M. D. Signaling by Distinct Classes of Phosphoinositide 3-Kinases. 254, 239–254 (1999).
Chen, Y. C. Beware of docking! Trends in Pharmacological Sciences. Elsevier Ltd. 36(2), 78–95 (2015).
Dhanaraj, P., Devadas, A. & Muthiah, I. A comparative meta-genomic analysis of HPV strains: A step towards the design, synthesis and characterization of noval quenazoline derivative for antiviral activity. Computational Biology and Chemistry. Elsevier Ltd 73, (213–220 (2018).
Baraban, S. C., Taylor, M. R. & Castro, P. A. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience. 131, 759–768 (2005).
Morgan, J. I. et al. Mapping Patterns of c-fos Expression in the Central Nervous System After Seizure. Science. 237(4811), 192–197 (1983).
Baxendale, S. et al. Identification of compounds with anti-convulsant properties in a zebrafish model of epileptic seizures. 784, 773–784 (2012).
Meng, X. et al. Journal of the Neurological Sciences Role of the mTOR signaling pathway in epilepsy. Journal of the Neurological Sciences. Elsevier B.V. 332(1–2), 4–15 (2013).
Berman, H. M. et al. The protein data bank. Nucleic acids research. 28(1), 235–242 (2000).
Livak, K. J. & Schmittgen, T. D. Analysis of Relative Gene Expression Data Using Real- Time Quantitative PCR and the 2 Ϫ ⌬⌬ C T Method. 408, 402–408 (2001).
Acknowledgements
The authors are thankful to the Director, CSIR-IHBT for providing necessary facilities and support. GT and VB acknowledges the Department of Science and Technology, New Delhi, India for providing junior research fellowship SERB File No: ECR/2016/000031. AGM acknowledges the Department of Science and Technology, New Delhi, India for providing DST-INSPIRE fellowship vide letter no. DST/INSPIRE Fellowship/2013/820. The authors are also grateful to Dr. Rajesh Ramachandran of Indian Institute of Science Education and Research, Mohali, Punjab, India for providing initial stock of adult zebrafish. This manuscript represents CSIR-IHBT Communication No. 4330. The CSIR support in the form of projects MLP:0201 for bioinformatics studies and MLP: 0204 for zebrafish studies is highly acknowledged.
Author information
Authors and Affiliations
Contributions
R.P. designed the study and formatted the manuscript. R.P., G.T. and V.B. performed and interpreted the in-silico data. A.G.M., S.K. and D.S. performed, evaluated and interpreted zebrafish experimentation part. R.B., Y. and P.D. synthesized and provided in-house active compounds for in-silico and in-vitro experimentation. All authors contributed in the preparation of manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tanwar, G., Mazumder, A.G., Bhardwaj, V. et al. Target identification, screening and in vivo evaluation of pyrrolone-fused benzosuberene compounds against human epilepsy using Zebrafish model of pentylenetetrazol-induced seizures. Sci Rep 9, 7904 (2019). https://doi.org/10.1038/s41598-019-44264-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-44264-6
This article is cited by
-
Systematic Profiling of Mitogen-Inducible Gene 6 and Its Derived Peptides Binding to Receptor Tyrosine Kinases in Bone Cancers at Molecular and Cellular Levels
International Journal of Peptide Research and Therapeutics (2024)
-
Exosomes derived from human umbilical cord mesenchymal stem cells decrease neuroinflammation and facilitate the restoration of nerve function in rats suffering from intracerebral hemorrhage
Molecular and Cellular Biochemistry (2024)
-
Discovery of Palbociclib as a potent c-Myc G4 stabilizer for lung cancer treatment using molecular docking, molecular dynamics simulation, and in vitro activity evaluation
Molecular Diversity (2024)
-
Computational insights into NIMA-related kinase 6: unraveling mutational effects on structure and function
Molecular and Cellular Biochemistry (2023)
-
High-throughput virtual screening of potential inhibitors of GPR52 using docking and biased sampling method for Huntington’s disease therapy
Molecular Diversity (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.