Abstract
Gastric adenocarcinoma (GAC), also known as stomach adenocarcinoma (STAD), is one of the most lethal malignancies in the world. It is vital to classify and detect the hub genes and key pathways participated in the initiation and progression of GAC. In this study, we collected and sequenced 15 pairs of GAC tumor tissues and the adjacent normal tissues. Differentially expressed genes (DEGs) were analyzed and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analysis were used to annotate the unique biological significance and important pathways of enriched DEGs. Moreover, we constructed the protein-protein interaction (PPI) network by Cytoscape and conducted KEGG enrichment analysis of the prime module. We further applied the TCGA database to start the survival analysis of these hub genes by Kaplan-Meier estimates. Finally, we obtained total 233 DEGs consisted of 64 up-regulated genes and 169 down-regulated genes. GO enrichment analysis found that DEGs most significantly enriched in single organism process, extracellular region, and extracellular region part. KEGG pathway enrichment analysis suggested that DEGs most significantly enriched in Protein digestion and absorption, Gastric acid secretion, and ECM-receptor interaction. Furthermore, the PPI network showed that the top 10 hub genes in GAC were IL8, COL1A1, MMP9, SST, COL1A2, TIMP1, FN1, SPARC, ALDH1A1, and SERPINE1 respectively. The prime gene interaction module in PPI network was enriched in protein digestion and absorption, ECM receptor interaction, the PI3K-Akt signaling pathway, and pathway in cancer. Survival analysis based on the TCGA database found that the expression of the FN1, SERPINE1, and SPARC significantly predicted poor prognosis of GAC. Collectively, we identified several hub genes and key pathways associated with GAC initiation and progression by analyzing the microarray data on DEGs, which provided a detailed molecular mechanism underlying GAC occurrence and progression.
Similar content being viewed by others
Introduction
Gastric cancers are the fifth commonest cancer after lung, breast, colorectal and prostate cancers1. It imposes a considerable health burden worldwide. Gastric adenocarcinoma (GAC), also known as stomach adenocarcinoma (STAD) is the commonest histological type (~90–95%)2. In 2015, GAC was expected to be diagnosed nearly 777,000 new cases and led to deaths of 350,000 people worldwide3. Although advances have been made for the diagnostic and therapeutic techniques for decades, the mortality rate of GAC is still high and the global 5-year survival rates remain unsatisfactory4.
It is well known that cancer is usually characterized by abnormally cell cycle activity, which generally results from either mutation in the up- or down-stream signaling pathways or genetic lesions in protein-encoding genes involved in cell cycle. The highly organized and regulated mammalian cell cycle ensures normal and accurate gene duplication, cell division and cell apoptosis5.
Microarray could be used to probe the key biomarkers and provide a better understanding of the molecular mechanisms involved in GAC. Until now, clinically applicable biomarkers are still lacking. Therefore, exploring novel and effective molecular biomarkers to elucidate effective therapeutic targets for GAC patients is still imperative. In this study, we focused on the different expression pattern between the GAC tumor tissues and matched normal tissues. To discover the hub genes and key pathways associated with the initiation and progression of GAC, we applied differential gene expression analysis and functional enrichment analysis. In conclusion, we identified a set of hub genes that participated in several cancer-relevant pathways and their abnormal expression are correlated with the clinical prognosis of GAC people by overall survival analysis.
Methods and Materials
Patients and samples
Tumor and matched normal tissues samples were obtained from the GAC patients at the Affiliated Hospital of Xuzhou Medical University in 2014. These tissues were stored in RNAlater (Ambion, Life Technologies, ThermoFisher Scientific, Waltham, MA, USA) at 4 °C until full penetration of RNAlater into the tissues and transferred to −80 °C for storage. The selection criteria were as follows: (1) the subject presented was diagnosed as GAC and no history of other tumors; (2) Complete demographic and clinical data including age, gender, clinical manifestations, tumor size, the extent of resection, and date of relapse and/or death have been collected. In order to get the formal permission of surgical procedures and the intelligent use of the resected tissues, the legal surrogates of those participants provided their Written informed consent. The National Regulations on the Use of Clinical Samples in China is as a guideline for human tissue acquisition and legitimate use. This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Xuzhou Medical University. The demographic and clinical features of the patient were summarized in Table 1.
Profiling of gene expression
Total RNA was extracted from frozen tissues separately using EZNA ® HP Tissue RNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA) according to the manufacturer’s recommended procedure. Bioanalyzer 2100 (Agilent Technologies, Palo Alto, Calif.) was used to assess the quality and quantity of these total RNAs. Affymetrix microarray was used for mRNA profiling, which was performed by GeneChem (Shanghai Genechem Co., Ltd.). Briefly, after rRNA removal, biotinylated aRNA (cRNA) was prepared according to the manufacturer’s protocol (3′ IVT Express Kit, Affymetrix 901228). PrimeView Human Gene Expression Array (cat. no. 901838; Affymetrix; Thermo Fisher Scientific, Inc.) were hybridized and scanned according to standard Affymetrix protocols. All samples were processed in technical duplicate. GeneChip Scanner 3000 (Affymetrix) was used to scan the completed arrays. Images were extracted with Affymetrix GeneChip Command Console (AGACC) and analyzed by using Expression Console Software (Affymetrix, CA, USA). Data were deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118916, GSE118916).
Data preprocess and differentially expressed genes identification
Probes were converted to gene symbols according to the platform annotation information of the raw data. The expression value for a gene, which was mapped by multiple probes, was acquired by selecting the max value among those probes. Those invalided probes without any gene information were removed. The original CEL data was then started background correction, normalization, and expression calculation by using the R package “affy”. The R package limma (http://www.R-project.org) was used to conduct data normality by log2 transformation. We applied the R package “limma” to identify the differentially expressed genes (DEGs) following the following criteria: (I) |logFC| > 2; (II) P-value < 0.05 and (III) false discovery rate (FDR) < 0.05.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis
Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were conducted by using clusterProfiler6 to reveal the unique biological significance and key pathways associated with GAC of the DEGs (criteria: p-value < 0.05, significantly enriched). Fisher’s exact test7 was used to identify the significant GO terms and pathways and corrected P-value was obtained by Benjamini and Hochberg (BH) false discovery rate (FDR) algorithm. Cytoscape8, Enrichment Map9, and Gephi10 were used for visualization of the network.
The protein-protein interaction network construction
The Retrieval of Interacting Genes (STRING v10)11 (http://string-db.org/) was used to analyze the interactive relationships among DEGs to construct protein-protein interaction (PPI) network and only experimentally validated interactions with a combined score >0.4 were selected as significant. Cytoscape was used to construct the PPI network and Gephi was used to network visualization. The plug-in Molecular Complex Detection (MCODE) was used to select the prime module from the PPI network. The criteria were set as follows: MCODE scores >2 and number of nodes >5. Then the KEGG pathway enrichment analysis of the DEGs from the module was conducted. P < 0.05 was considered to be significant.
TCGA data acquisition and processing
We searched the GAC cases with both clinical information and gene expression profile from The Cancer Genome Atlas (TCGA) database12 by using the R package “OIsurv”. The expression value of each hub genes was defined as either high (expression value >median value) or low (expression value <median value). Survival curves were analyzed by the Kaplan-Meier method13, and univariate survival analysis was performed using a log-rank test. Overall survival (OS) was defined as the period between the resection date and death for any cause.
TCGA data analysis
We downloaded the RNA-seq data and clinical phenotype information of the GAC-related tumor and the adjacent normal tissues from The Cancer Genome Atlas (TCGA) database (https://xenabrowser.net/datapages/). We calculated the expression level of FN1, SERPINE1, and SPARC and conducted the difference significance test between the GAC tumor group and the normal group. We further did the correlation analysis between the hub gene and the disease stage.
Statistical analysis
The statistical data were processed using SPSS 19.0 software (IBM, SPSS, Chicago, IL, USA). The paired t-test was performed to evaluate the significant difference between tumor tissues and tumor-adjacent normal tissues. The statistical significance of the difference in patients’ overall survival (OS) between the high expression group and low expression group was obtained using the two-sided log-rank test. All p-values were two-sided, and generally, p-values < 0.05 were considered to have statistical significance.
Results
Differentially expressed genes in gastric adenocarcinoma
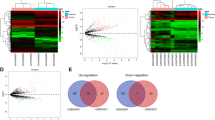
Total 15 pairs of GAC tumor and normal samples were analyzed (Fig. 1A) and the basic global gene expression pattern among all samples was denoted by Pearson correlation matrix for calculation of pairwise correlation coefficient (Fig. 1B). Then the first step was to identify the DEGs between these two groups. The classical Bayesian algorithm was applied to identify the DEGs and volcano plots were used for visualizing the DGEs variation between tumor and normal tissues (Fig. 1C). In total, 233 mRNAs displayed differential expression in GAC, including 64 up-regulated mRNAs and 169 down-regulated mRNAs. Each of the top 10 upregulated DGEs and downregulated DGEs were listed in Table 2. Hierarchical clustering analysis showed the expression pattern of DEGs among samples, which suggested that the expression of genes in GAC tumor tissues significantly differ from those in adjacent-normal tissues (Fig. 1D). Detailed information of these up- and down-regulated probes is shown in Supplementary Tables 1 and 2.
Overview of differentially expressed genes (DEGs) in gastric adenocarcinoma. (A) Principal components analysis (PCA) plot for total of 30 samples. (B) the pearson correlation matrix among all samples by calculating pairwise correlation coefficient. The redder the color, the higher the correlation coefficient, while the bluer the color, the lower the correlation coefficient. (C) Volcano plot of the DEGs. Each circle represents a gene, and red circle represents. (D) Heatmap of the 233 DEGs. Each column in the heatmap represents a sample, and each row represents the expression level of a gene. The color scale beside the heatmap represents the raw Z-score ranging from green (low expression) to red (high expression).
Differentially expressed genes enrichment analysis
Gene enrichment analysis uses predefined gene sets and ranks of genes to identify significant biological changes or gene co-expression pattern, which could assess the functional associations of target gene set derived from the set of experiments. KEGG analysis results showed that DEGs were mostly enriched in Protein digestion and absorption, Gastric acid secretion, and ECM-receptor interaction (Table 3). Figure 2A shows the TOP20 enriched KEGG pathways. The five top mostly enriched pathways were got to construct a network (Fig. 3A). From the network, the up-regulated genes Human fibronectin gene (FN1), collagen α1 (I) gene (COL1A1) and collagen α2 (I) gene (COL1A2) were supposed to have a direct relationship with ECM-receptor interaction, while the down-regulated gene Somatostatin (SST) was supposed to be associated with Gastric acid secretion. GO analysis results showed that DEGs were mostly enriched in the single-organism process (belonging to a biological process), cellular process (belonging to a biological process), and cell (belonging to cellular component) (Table 4). Figure 2B shows the TOP10 enriched function of molecular function (MF), biological process (BP) and cellular component (CC) respectively. The five top mostly enriched functions were also got to construct a network (Fig. 3B). The detailed information of KEGG and GO enrichment results are listed in Supplementary Tables 3 and 4 separately.
PPI network analysis
Based on the STRING database, a co-occurrence network of total 135 nodes and 280 edges was constructed. From the PPI network, the top 10 hub genes with higher degrees were filtered, including Interleukin-8 (IL8, up-regulated), Collagen α1 (I) gene (COL1A1, up-regulated), Matrix metalloproteinase-9 (MMP-9, up-regulated), Somatostatin (SST, down-regulated), Collagen α2 (I) gene (COL1A2, up-regulated), Metalloproteinases 1 (TIMP1, up-regulated), Human fibronectin gene (FN1, up-regulated), Secreted protein, acidic, and rich in cysteine (SPARC, up-regulated), Aldehyde dehydrogenase 1 (ALDH1A1, down-regulated) and Serpin peptidase inhibitor, clade E (SERPINE1, up-regulated) (Fig. 4). Among these genes, IL8 had the highest node degree (21). Besides, the prime module was selected (Fig. 5A), and the KEGG pathway enrichment analysis of the genes in the module was analyzed (Fig. 5B). The prime module analysis revealed that the development of GAC was mainly associated with protein digestion and absorption, ECM receptor interaction, the PI3K-Akt signaling pathway and pathway in cancer.
The protein–protein interaction (PPI) network and hub genes. (A) PPI network of DEGs. Only experimentally validated interactions with a combined score >0.4 were selected as significant. The nodes were colored according to whether it belongs to up- or down-regulated genes. The thicknesses of those edges were associated with the combined scored. The size of each node is proportional to the number of connections, that is, the degree. (B) The expression heatmap of TOP10 hub genes.
TCGA dataset analysis
TCGA research analysis could combine extensive genetic studies of human gene expression with the specific disease. GAC cases were divided into high expression (expression value >median value) and low expression (expression value <median value) groups for each key gene. The Kaplan-Meier survival curves of 411 GAC cases showed that FN1, SERPINE1, and SPARC was significantly related to the prognosis of GAC (Fig. 6), while other 7 hub gene was not significantly associated with GAC (Supplementary Fig. 1).
Overall survival analysis in GAC based on the TCGA data as determined by Kaplan-Meier estimates. 411 GAC cases with full data of both clinical and 10 hub gene expression were downloaded from TCGA database. Kaplan-Meier estimates (log-rank test) were made and found the expression of (A), FN1; (B), SERPINE1 and (C), SPARC was significantly affect the prognosis of GAC in overall survival (p < 0.05).
We further calculated the expression level of FN1, SERPINE1, and SPARC. The results showed that the three hub genes were all extremely significant higher in GAC tumor tissues than the normal tissues (Fig. 7), which were in good consistence with our research. Moreover, we conducted statistical analysis between the expression level of hub genes and disease stage. We found that FN1 (r = 0.21, p = 1.57E-05), SERPINE1 (r = 0.22, p = 5.08E-06), and SPARC (r = 0.14, p = 0.006) all had a significant correlation with CRC disease stage, which indicated that these key genes may have crucial role in GAC progression.
Discussion
GAC is one of the lethal of malignancies originating in the stomach, which is a kind of heterogeneous disease and consists of various phenotypes and genotypes5. Of all, cell cycle plays a key important role in cancer originating including mutation of signaling pathways or mutation of protein-encoding gene, and those relevant cell cycle proteins are thought to have a vital association with cancer progression. Making deeper understanding of the role of cell cycle proteins, including revealing their function mechanisms and finding their relevant upstream or downstream targets, could help us to discover more valuable biomarkers. Fortunately, the development of biotechnology, like microarray, transcriptome sequencing, and proteomics, has provided an advanced way to get the nearly all gene expression information during all stages for any human genomes and lots of potential therapeutic targets have been excavated, including AFP14, BCR-ABL15, BRCA1/BRCA216, EGFR17, HER218 and so on.
In this study, we analyzed the gene expression profiles of 15 GAC tumor tissues and 15 adjacent none-tumor tissues by microarray. We finally identified 64 up-regulated DEGs and 169 down-regulated DEGs. The heatmap of all DEGs showed obviously different between the two groups (Fig. 1D), which demonstrated that each group has its specific gene expression pattern and the samples from the same group seemed to share the similar expression pattern and often participate in a parallel biological process (Fig. 1B). To better understand the interactions among DEGs, we further performed functional enrichment analysis. The GO term enrichment analysis shows that DGEs were mainly involved in cell, extracellular region, extracellular region part, extracellular space, and single organism process (Fig. 3B). As previous studies, the structure of cancer-relevant proteins, like HER219, HER320 has a ligand-activated state in the extracellular region and could interact with other same receptors in the absence of direct ligand binding, which maybe indicate that many DEGs of this study might be transmembrane proteins and may involve in some extracellular signal transduction pathways. KEGG pathway enrichment analysis reveals that DEGs were mainly involved in several cancer-related pathways such as ECM-receptor interaction, protein digestion and absorption, gastric acid secretion and retinol metabolism (Fig. 3A). Gastric acid secretion has been reported to have an association with gastric mucosal healing21, which may be involved in the destruction of normal cell proliferation. The previous study pointed out that retinoic acid receptors are frequently having low expression in epithelial gastric cancer22.
Subsequently, in order to find the co-existence pattern among all the DEGs, we constructed the PPI network. From the network, 10 hub genes with high degrees were found (Fig. 4). Moreover, we extracted the main module from the PPI network and carried out KEGG pathway enrichment analysis (Fig. 5). The result showed that the family of collagen genes (CLO1A1, COL1A2, COL2A1, etc.) are tight clustered and have directed association with protein digestion and absorption, ECM-receptor interaction and P13K-Akt signaling pathway. Besides the other two pathways, P13K-Akt signaling pathway has been reported to be vital in cell cycle including cell proliferation and gene mutation and is activated in various cancers23. The FN1 gene, a glycoprotein involved in cellular adhesion and migration processes, was denoted to have an association with the pathway in cancer.
In order to find the relationship between the target genes and the GAC prognosis, we conducted survival analysis based on the TCGA data, which included 411 GAC cases. The result showed that FN1, SPARC, and SERPINE1 expression were significantly (p < 0.05) associated with GAC prognosis (Fig. 6). The high expression of these three genes often indicates a poor prognosis. The expression level of these three hub genes were all remarkably higher in GAC tumor tissues than the normal tissues (Fig. 7), which indicated these hub genes might have potential impact on the GAC cell progression.
Conclusion
We identified the hub genes and key pathways associated with GAC by analysis microarray data and provided more information about molecular mechanisms about GAC occurrence and progression, holding promise for acting as biomarkers and potential therapeutic targets. However, further molecular biological experiments including q-RT PCR, colony-formation assay, and flow cytometry analysis are required to confirm the specific functional role and mechanisms underlying the three crucial genes including FN1, SPARC, and SERPINE1 in GAC.
Human participants and animal rights
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
Tarver, T. American Cancer Society. Cancer facts and figures 2014. J Consumer Health Internet. 16, 366–7 (2012).
Ajani, J. A. et al. Gastric adenocarcinoma. Nature Reviews Disease Primers. 3, 17036 (2017).
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 136, E359–E86 (2015).
Fujitani, K. Overview of adjuvant and neoadjuvant therapy for resectable gastric cancer in the East. Digestive surgery. 30, 119–29 (2013).
Sherr, C. J. & Bartek, J. Cell cycle–targeted cancer therapies. (2017).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: a journal of integrative biology. 16, 284–7 (2012).
Upton, G. J. Fisher’s exact test. Journal of the Royal Statistical Society Series A (Statistics in Society). 395–402 (1992).
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 27, 431–2 (2010).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 5, e13984 (2010).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Icwsm. 8, 361–2 (2009).
Szklarczyk, D. et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids research. 43, D447–D52 (2014).
Tomczak, K., Czerwińska, P. & Wiznerowicz, M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary oncology. 19, A68 (2015).
Guyot, P., Ades, A., Ouwens, M. J. & Welton, N. J. Enhanced secondary analysis of survival data: reconstructing the data from published Kaplan-Meier survival curves. BMC medical research methodology. 12, 9 (2012).
Tateishi, R. et al. Diagnostic accuracy of tumor markers for hepatocellular carcinoma: a systematic review. Hepatology international. 2, 17–30 (2008).
Shinohara, H. et al. Anti-cancer fatty-acid derivative induces autophagic cell death through modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia. Cancer letters. 360, 28–38 (2015).
Pontikakis, S. et al. Predictive value of ATP7b, BRCA1, BRCA2, PARP1, UIMC1 (RAP80), HOXA9, DAXX, TXN (TRX1), THBS1 (TSP1) and PRR13 (TXR1) genes in patients with epithelial ovarian cancer who received platinum-taxane first-line therapy. The pharmacogenomics journal. 17, 506 (2017).
Vizoso, M. et al. Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nature medicine. 21, 741 (2015).
Alix-Panabières, C. & Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer discovery. (2016).
Cho, H.-S. et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 421, 756 (2003).
Cho, H.-S. & Leahy, D. J. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 297, 1330–3 (2002).
Smolka, A. J. & Schubert, M. L. Helicobacter pylori-induced changes in gastric acid secretion and upper gastrointestinal disease. Molecular pathogenesis and signal transduction by helicobacter pylori: Springer, 227–52 (2017).
Hu, K.-W., Chen, F.-H., Ge, J.-F., Cao, L.-Y. & Li, H. Retinoid receptors in gastric cancer: expression and influence on prognosis. Asian Pacific Journal of Cancer Prevention. 13, 1809–17 (2012).
Hennessy, B. T., Smith, D. L., Ram, P. T., Lu, Y. & Mills, G. B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nature reviews Drug discovery. 4, 988 (2005).
Acknowledgements
This work was financially supported by grants from the Jiangsu Province Social Development Key Projects (BE2017639), Jiangsu Province Key medical talents (ZDRCC2016010), China Postdoctoral Science Foundation funded project (2016M591928), Scientific research project of the health planning committee of Jiangsu (H2017082), Xuzhou Science and Technology Plan Projects (KC16SY148, KC17166).
Author information
Authors and Affiliations
Contributions
S.F. and J.C. conceived and instructed the work. L.L. and Z.Z. performed the analysis and constructed the model. Y.Z. collected the samples. Q.Z., X.W. and B.M. checked the associated database and raw data. L.L. and Z.Z. wrote and revised the manuscript. All of the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, L., Zhu, Z., Zhao, Y. et al. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep 9, 7827 (2019). https://doi.org/10.1038/s41598-019-43924-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-43924-x
This article is cited by
-
Quantified pathway mutations associate epithelial-mesenchymal transition and immune escape with poor prognosis and immunotherapy resistance of head and neck squamous cell carcinoma
BMC Medical Genomics (2024)
-
Developing a prognosis and chemotherapy evaluating model for colon adenocarcinoma based on mitotic catastrophe-related genes
Scientific Reports (2024)
-
Helicobacter pylori-induced fibroblast-derived Serpin E1 promotes gastric cancer growth and peritoneal dissemination through p38 MAPK/VEGFA-mediated angiogenesis
Cancer Cell International (2023)
-
The expression of SERPINE1 in colon cancer and its regulatory network and prognostic value
BMC Gastroenterology (2023)
-
Clinicopathological correlation of Cathepsin K expression in salivary gland carcinomas; relation to patients` outcome
Diagnostic Pathology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.