Abstract
The archipelago of Lakshadweep is considered as a stopover to the maritime route since ancient time. It is not very clear when the human first occupied these islands, however in the long history of the islands, the local legends suggest that Lakshadweep has been ruled by different kingdoms. To have a better understanding of peopling of Lakshadweep, we have analysed 557 individuals from eight major islands for mitochondrial DNA and 166 individuals for Y chromosome markers. We found a strong founder effect for both paternal and maternal lineages. Moreover, we report a close genetic link of Lakshadweep islanders with the Maldives, Sri Lanka and India. Most of the Lakshadweep islands share the haplogroups specific to South Asia and West Eurasia, except Minicoy Island that also shares haplogroups of East Eurasia. The paternal and maternal ancestries of the majority of island populations suggest their arrival from distinct sources. We found that the maternal ancestry was closer to South Indian populations, whereas the paternal ancestry was overwhelmed with the haplogroups, more common in the Maldives and North of India. In conclusion, our first genetic data suggest that the majority of human ancestry in Lakshadweep is largely derived from South Asia with minor influences from East and West Eurasia.
Similar content being viewed by others
Introduction
Lakshadweep is an archipelago of about thirty-five islands, scattered over approximately 78,000 square km of the Arabian Sea, 200–440 kms off the south-western coast of India1,2. The literal meaning of Lakshadweep is “one hundred thousand islands” in Sanskrit. The total population size of the archipelago is approximately 65 thousands, however, it lacks aboriginal population1,3. Presently, majority of the populations follow Islam and are ethnically similar to Malayali people of Kerala state2. The first human settlement of this archipelago is not very clear, nevertheless it was under the power of various regime. Amini, Kalpeni Andrott, Kavaratti and and Agatti are the oldest inhabited islands. The islands were known to sailors since ancient time2. Jataka stories of Buddhism has mentioned these islands, which documented the spread of Buddhism to the islands during 6th century BC1,4. The local stories suggest the arrival of Islam in 661 AD by Arabians. Later, Cholas ruled the islands in 11th, Portuguese in 16th, Ali Rajahs in 17th, Tipu Sultan in 18th and finally it was under British Raj in 19th Century1,2,3,4,5,6. Arab traveller Ibn Batuta mentioned about these islands in many of his stories2.
Due to its geographical location, the present day Lakshadweep populations may offer a unique insight into historic migration events. The exposure of these islands to people from pan-world ethnicity might have created a mosaic like pattern in their genomic ancestry, however no genetic study so far has been done on populations living in these islands. Therefore, for the first time, we have done extensive DNA sampling of several major islands of Lakshadweep (Supplementary Fig. 1), and performed a high-resolution analysis of haploid markers. We looked specifically for the composition of various haplogroups (hg) present in these islands and their intra and inter-relations to the surrounding populations.
Methods
Sampling
Approximately, 5–10 ml blood samples, were collected from 557 individuals belonging to eight major islands of Lakshadweep (Agatti, Andorth, Bitra, Chetlat, Kadmat, Kalpeni, Kiltan and Minicoy), with informed written consent (Supplementary Fig. 1). With detailed interview procedure about their family pedigree, we have excluded the samples of the people who are related for minimum of three generations. This project was approved and permitted by the Institutional Human Ethical Committee, Mangalore University, Karnataka, India and Institutional Ethical Committee of the CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India. All methods were performed in accordance with the relevant guidelines and regulations.
Genotyping
We have used Sanger Sequencing method and utilised 23 F and 23 R markers described elsewhere7 to sequence the Hypervariable segment I (HVS-I) of mtDNA. We sequenced both of the strands to minimize errors. Variations were recorded against the r-CRS8. To assign those samples in haplogroups, we subsequently genotyped coding region diagnostic mutations and used these combined information to assign haplogroups (Table 1, Supplementary Tables 1 and 2). They were further verified by consulting the data published till date in PhyloTree build 169. In case of Y chromosome, we genotyped 166 samples for 44 biallelic markers published recently10 to assign the Y chromosomal haplogroups (Supplementary Table 2). For all the Y chromosome markers, we utilised Sanger sequencing (ABI 3700, Applied Biosystems) method and compared it with the reference to note the variations.
Statistical analysis
For statistical analysis, first we plotted the PC1 vs PC2 estimated by using POPSTR (kindly provided by H. Harpending), to infer the inter and intra relationship of populations. We plotted the regional haplogroup affiliation of all the islands over their geographical co-ordinates. The classification of different haplogroups to East Eurasia, West Eurasia and South Asia was done manually considering few major points e.g. their exclusive presence in a region, their high frequency and diversity in a region and based on classifications used in previous publications11,12. In cases where the particular geography was not determined with the current resolution of data, we have put them as haplogroup of ‘unknown’ origin. We also took special care for haplogroup R1a-M17 which harbours substantial amount of Indian paternal ancestry13. It is a haplogroup of debatable origin14,15, we used it as such. The haplotype diversity and AMOVA tests were performed by using Arlequin 3.5 software16 Median-joining and reduced median networks were reconstructed with NETWORK program (version 5)17. Reduced median and median-joining procedures were applied sequentially for the analysis and they showed similar network structure. For comparison, we have used various datasets published elsewhere11,18,19,20,21,22,23,24,25,26,27,28,29.
Results and Discussion
The Lakshadweep islands, were well known to the maritime sailors since ancient times, presumably due to a staging point between different continents1,2. The extensive maritime history facilitated the access of these islands by various ethnic groups, therefore, it is likely that the genetic landscape of these islands might have shaped uniquely. However, this assumption has not been investigated at the molecular level that simultaneously examines the ancestry composition of different islands and their genetic relationships that might also co-vary with distance from neighbouring regions. To validate these assumptions, we have analysed 557 samples for mitochondrial DNA (mtDNA) and 166 samples for Y chromosome markers and compared them with the neighbouring regions.
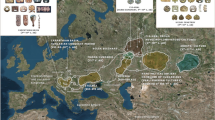
Our analysis on mtDNA has revealed that nearly 56% of the maternal lineages of Lakshadweep islands belong to three major haplogroups (M2b2, M30 and R30) (Table 1 and Supplementary Fig. 2). It is interesting to note that in the background of major haplogroups, all of the individuals carried mostly a single haplotype; therefore we call them as founder haplogroup (Supplementary Table 1). These founder haplogroups are virtually present in all the studied islands. Notably, haplogroup R30, which is also reported previously in the coastal South Asia including Sri Lanka21,30, is the most frequent haplogroup in many of the Lakshadweep islands (Table 1 and Supplementary Table 1). To have a spatial understanding of various maternal lineages present in Lakshadweep, we have grouped haplogroups in to regions and plotted with their geographical co-ordinates (Fig. 1). All the islands (except Minicoy) showed major component of maternal ancestry associated with South Asia. The southern most islands Kalpeni, and Minicoy show substantial West Eurasian-specific maternal ancestry, which is largely similar to the Maldives and Sri Lanka. Additionally, Minicoy island carried East Eurasian-specific haplogroups, which is otherwise virtually absent in this region.
The spatial distribution of regional maternal components in Lakshadweep islanders in comparison with other neighbouring populations. South Asian haplogroups—mtDNA: M2–6, N5, M33–65, R5–8, and R31–32. East Eurasian haplogroups—A–G, M7–12, M66, R22, and N9. West Eurasian haplogroups H-K, U1, U4, U7 and U9 Unresolved haplogroups—M*, R*, N* including other lineages, for example, M31 and M32.
The principle component analysis (PCA) of maternal ancestry including South Asian major regions formed a distinct cline of Lakshadweep populations, which is likely due to drift created by strong founder effect seen in their maternal ancestry (Fig. 2). The closest cluster to many of these islands are the populations of Kerala with whom they share their most common haplogroups i.e. M2 and R30 (Supplementary Table 2).
We measured the haplotype diversity of island populations with respect to the mainland South Asian populations (Supplementary Fig. 2a). Our data indicate a significant (two tailed p value < 0.001) reduction in the haplotype diversity of Lakshadweep populations. The most likely explanation of this reduction is that a few individuals were introduced to these islands, which therefore created a ‘bottleneck followed by founder effect’ like scenario.
In order to have better understanding of founder lineages present in different islands of Lakshadweep we have reconstructed a median-joining network of maternal haplogroups identified for Lakshadweep islands (Fig. 3). The network analysis clearly showed the high prevalence of specific haplotypes present in the islands, likely stemming out from a single founder in each of their haplogroup background. Apart from major common founder maternal lineages, such as hg R30 followed by hg M2 (the top two maternal founder lineages of several islands), some of the islands carry specific founder lineages. For example, island Minicoy carry founder haplogroups M7 and M66 (East Eurasian specific) as well as hg U9 (West Eurasian-specific). Similarly, island Kalpeni harbour hg U4 (West Eurasian-specific) (Fig. 3 and Supplementary Fig. 1). The sharing of many of the maternal lineages among islands of Lakshadweep advocates a high level of intra-island maternal gene flow. At the inter-population level, they are closest to the Kerala populations with whom they share the same tongue (Supplementary Table 2). We have performed AMOVA analysis to compare the genetic relatedness of the Lakshadweep with the mainland South Asian populations. The analysis suggested a close genetic affinity of Lakshadweep populations with the South Indian Kerala populations (Table 2).
The median-joining network of 557 mtDNAs belonging to Lakshadweep islands. Each sample represented on the diagram has been sequenced for the HVS-I region and genotyped for the coding region mutations that are indicated in Supplementary Table 1. Circle sizes are proportional to the number of mtDNAs with that haplotype.
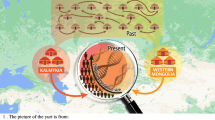
In order to assess the paternal ancestry, we further analysed the Y chromosome markers among 166 individuals of Lakshadweep (Supplementary Tables 2 and 3). Since the sample numbers genotyped for Y chromosomal markers from various islands were uneven and five out of nine islands were genotyped for less than 20 individuals, we have pooled all the Y chromosomal data and represented as single data of Lakshadweep (Supplementary Tables 2 and 3). The paternal affinity illustrates a distinct pattern in comparison with maternal ancestry. In the PCA analysis, Lakshadweep inclines towards Maldives population (Fig. 4), which is likely due to hg J2-M172. In the region wise comparison, the paternal ancestry components of Lakshadweep are more similar to the Maldives, North India and Pakistan rather than Kerala (Fig. 5 and Table 2). This suggests a gene flow to Lakshadweep from all of these regions. However, similar to maternal founders, we also see a reduction in haplotype diversity due to strong founder effect for paternal lineages (Supplementary Fig. 3b). Three haplogroups J2a-M172, Ra1-M17 and R2a-M124 harbour approximately 85% of the paternal ancestry of the Lakshadweep. The most frequent haplogroup was hg R2a-M124 followed by hg R1a1-M17 (Supplementary Table 2). In comparison to the mainland India, the Maldives and Sri Lanka, Lakshadweep show reduced paternal diversity and stronger founder effect (Supplementary Figs 1 and 3b). However, due to absence of Y-STR data, we could not determine if all the persons belonging to one Y chromosomal haplogroup, has been derived from a single founder haplotype or from a multiple founder haplotypes.
The spatial distribution of regional paternal components in Lakshadweep islanders in comparison with other neighbouring populations. South Asian haplogroups—C5, F, H, L, and R2. Southeast Asian haplogroups—C2, C3, D, and M–O. West Eurasian haplogroups G,I, and J. Unresolved haplogroups—K, P, Q and R1a.
In conclusions, the Lakshadweep populations share a large number of ancestries within the islands (except Minicoy). In spite of the fact that these islands served as a maritime crossroad, we have observed a strong founder effect with reduced diversity for both maternal and paternal ancestries. The maternal ancestry of the islands is largely derived from Southern India, whereas the paternal ancestry is showing higher affinity with the populations of the Maldives or Northern or Western part of the South Asia. The most diverse island is Minicoy which might have served as a popular destination for maritime sailors, thus received gene flow from various directions.
References
Saigal, O. Lakshadweep. (National Book Trust, India), 1–219 (2000).
Forbes, A. D. W. Sources towards a history of the Laccadive Islands. South Asia J South Asian Stud. 2, 130–150 (1979).
Vijayakumar, V. Traditional Fortunes: Law and Custom in India’s Lakshadweep Islands (Oxford University Press). 1–351 (2006).
Sharma, A. & Khan, M. Buddha heads and some grave head stones from Lakshadweep. In Rao, S. R. (ed.), The Role of universities and research institutes in marine archaeology, 129–131 (1994).
Tripati, S. Marine investigations in the Lakshadweep Islands, India. Antiquity 73, 827–835 (1999).
Tripati, S. Stone anchors from Minicoy Island, Lakshadweep, India. Int J Naut Archaeol 38, 406–412 (2009).
Rieder, M. J., Taylor, S. L., Tobe, V. O. & Nickerson, D. A. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res 26, 967–973 (1998).
Andrews, R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23, 147 (1999).
van Oven, M. & Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30, E386–94 (2009).
Karmin, M. et al. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 25, 459–466 (2015).
Chaubey, G. et al. Population Genetic Structure in Indian Austroasiatic speakers: The Role of Landscape Barriers and Sex-specific Admixture. Mol Biol Evol 28, 1013–1024 (2011).
Tamang, R. et al. Reconstructing the demographic history of the Himalayan and adjoining populations. Hum Genet 137, 129–139 (2018).
Sahoo, S. et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA 103, 843–848 (2006).
Sharma, S. et al. The Indian origin of paternal haplogroup R1a1* substantiates the autochthonous origin of Brahmins and the caste system. J Hum Genet 54, 47–55 (2009).
Underhill, P. A. et al. The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. Eur J Hum Genet 23, 124–131 (2015).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10, 564–567 (2010).
Bandelt, H.-J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16, 37–48 (1999).
Chaubey, G. et al. Language shift by indigenous population: a model genetic study in South Asia. Int J Hum Genet 8, 41 (2008).
Chaubey, G. et al. Genetic affinities of the Jewish populations of India. Sci Rep 6, 19166 (2016).
Kivisild, T. et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72, 313–32 (2003).
Lanka Ranaweera, L. et al. Mitochondrial DNA history of Sri Lankan ethnic peoples: their relations within the island and with the Indian Sub-continental populations. J Hum Genet 59, 28–36 (2013).
Metspalu, M. et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 5, 26 (2004).
Pijpe, J. et al. Indian Ocean crossroads: human genetic origin and population structure in the Maldives. Am J Phys Anthropol 151, 58–67 (2013).
Qamar, R. et al. Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 70, 1107–24 (2002).
Quintana-Murci, L. et al. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74, 827–45 (2004).
Sharma, G. et al. Genetic affinities of the central Indian tribal populations. PloS One 7, e32546 (2012).
Thangaraj, K. et al. Different population histories of the Mundari- and Mon-Khmer-speaking Austro-Asiatic tribes inferred from the mtDNA 9-bp deletion/insertion polymorphism in Indian populations. Hum Genet 116, 507–17 (2005).
Thanseem, I. et al. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet 7, 42 (2006).
Trivedi, R. et al. Genetic imprints of pleistocene origin of indian populations: a comprehensive Phylogeographic sketch of Indian Y-chromosomes. Int J Hum Genet 8, 97–118 (2008).
Chaubey, G. Language isolates and their genetic identity: a commentary on mitochondrial DNA history of Sri Lankan ethnic people: their relations within the island and with the Indian subcontinental populations. J Hum Genet 59, 61–63 (2013).
Acknowledgements
We thank all the all volunteers, lab technicians and Medical officers from each island of Lakshadweep for their kind participation in this study. We acknowledge Lakshadweep Administration, Department of Science and Technology (DST) and Directorate of health Services Kavaratti island, for an entry related permission to collect samples. This work is supported by Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India (SB/EMEQ-167/2013), New Delhi, India. KT is supported by NCP (MLP0117) fund from the Council of Scientific and Industrial Research (CSIR), Government of India. GC is supported by Estonian Institutional Research grants IUT 24-1 and National Geographic explore grant HJ3-182R-18. AS is supported by the UGC-RET doctoral fellowship and PPS is supported by the CSIR-JRF doctoral fellowship.
Author information
Authors and Affiliations
Contributions
K.T., G.C. and M.S.M. designed and supervised the study. M.S.M., M.R.N., N.P., S.J.C. and I.B. collected the samples and anthropological information. M.S.M., N.R., M.R.N., S.P. and N.P. performed mtDNA and Y chromosomal genotyping. M.S.M., N.R., G.C., A.S., P.P.S. and P.K.D. performed data analysis. K.T. provided laboratory reagents. G.C. and M.S.M. wrote the manuscript with inputs from N.R. and K.T.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mustak, M.S., Rai, N., Naveen, M.R. et al. The peopling of Lakshadweep Archipelago. Sci Rep 9, 6968 (2019). https://doi.org/10.1038/s41598-019-43384-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-43384-3
This article is cited by
-
Genetic evidence for a single founding population of the Lakshadweep Islands
Molecular Genetics and Genomics (2024)
-
The maternal ancestry of the Kavaratti islanders and the last glacial maximum aftermath
Molecular Genetics and Genomics (2023)
-
Island biogeography and human practices drive ecological connectivity in mosquito species richness in the Lakshadweep Archipelago
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.