Abstract
Resistant starch (RS) is a constituent of dietary fibre that has beneficial effects on the intestine physiological function of animals. However, the roles of RS on shrimp intestine health is unknown. In this study, we investigated the the effects of dietary RS on the microbial composition, and digestive and immune-related indices in the intestine of Litopenaeus vannamei. The shrimp were fed with diets containing different levels of RS: 0 g/kg (Control), 10 g/kg (RS1), 30 g/kg (RS2) and 50 g/kg (RS3) for 56 days. The results showed that dietary RS improved the morphology of the intestine mucosa. RS also increased the activity of digestive enzymes (AMS, LPS, Tryp, and Pep) and immune enzymes (PO, T-AOC, T-NOS, and NO), and the expression levels of immune-related genes (proPO, ALF, Lys, HSP70, Trx, Muc-1, Muc-2, Muc-5AC, Muc-5B, and Muc-19). A microbiome analysis indicated that dietary RS increased the short-chain fatty acids (SCFAs) contents and altered the composition of the intestine microbial. Specifically, RS increased the abundances of Proteobacteria and decreased the abundance of Bacteroidetes. At the genus level, the beneficial bacteria (Lutimonas, Ruegeria, Shimia, Mesoflavibacter, and Mameliella) were enriched, which might be involved in degrading toxins and producing beneficial metabolites; while potential pathogens (Formosa and Pseudoalteromonas) were decreased in response to dietary RS. Our results revealed that dietary RS could improve the intestine health of L. vannamei, probably via modulating the intestine microbial composition and SCFAs contents, and enhancing the digestion and immunity of the shrimp.
Similar content being viewed by others
Introduction
The intestine microbial has a mutual relationship with its host which influences the digestibility, metabolism, and immunity of the host1,2. Healthy intestine microbial can protect the host from colonization by pathogenic microbes and produce short-chain fatty acids (SCFAs). Conversely, unbalanced intestine microbial (dysbiosis) may lead to the alteration of immunity, and increase the susceptibility to diseases3,4. SCFAs, including acetate, propionate and butyrate, are shown to improve disease resistance5. SCFAs can provide nutrition for intestine mucosa, and benefit the healthy intestine micro-ecological environment of the host6. The production of SCFAs occurs mainly through the bacterial fermentation of fibre that non-digestible by the gastrointestinal systems of animals7. Therefore, dietary interventions, especially via supplementation with fiber, are efficient for regulation of the intestinal microbial, will be beneficial to the host health.
Different kinds of dietary fibres have unique metabolic effects and produce different kinds and amounts of SCFAs8. As a constituent of dietary fibre, resistant starch (RS) is non-digestible in the intestine of animals but can be fermented by resident microbial9. Diets that are rich in RS have beneficial effects to animal health, especially produce SCFAs by microbial fermentation, which can acidify the intestine environment, modify and stabilize intestine microbial10. Other health benefits associated with dietary RS are the promotion of the growth and proliferation of intestine epithelial cells, and the induction the transcription of genes which are conducive to intestine development11. There are four different types of RS: (1) RS1 is a physically inaccessible starch exist in legumes and grains; (2) RS2 is a raw starch granule, including raw potatoes and green (unripe) bananas; (3) RS3 is a retrograded starch, including potatoes and rice; (4) RS4 is a chemically modified starch, including esters, ethers, and cross-linked starches. Several studies have explored the effects of RS on the intestine health of swine and poultry12,13. However, the roles of RS in shrimp intestine health remain unknown.
The Pacific white shrimp Litopenaeus vannamei is a commercially important marine species in the world. Shrimp farming has suffered from serious problems caused by the various diseases14. High applications of antibiotics and antimicrobial drugs easily pollute the environment, thereby resulting in bacterial resistance; thus, ecofriendly disease preventative approaches must be developed for antibiotics alternative15. RS is rendered as a good alternative to antibiotic. Intestine mucosa is an important barrier for shrimp disease defence, and intestine health clearly affects shrimp health16. Therefore, in this study, we explored the effects of dietary RS on the intestine health of L. vannamei, in terms of intestine histological structure, the digestive and immune indices, and the intestine microbial composition and metabolite SCFAs contents.
Results
Intestine epithelium morphology
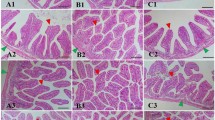
Shrimp fed the RS diet had a better intestine health morphology than those of the control group (Fig. 1). Health characteristics included good epithelial cell morphology, cell density in neat rows, and higher epithelium height (EH) and wall thickness (WT) of the three RS groups than those of the control group (P < 0.05).
Intestine tissue of L. vannamei stained with HE after the shrimp were fed the control and three RS diets for 56 days. (A) Control group, × 400; (B) RS1 group, × 400; (C) RS2 group, × 400; (D) RS3 group, × 400. (a) mucosa brush border, (b) intestine epithelium, (c) cell nuclei, (d) intestine lumen, (e) epithelium height (EH), (f) wall thickness (WT). Bars show the mean ± SE (n = 5). The different letters (a–c) indicate significant differences (P < 0.05) among the groups.
Digestive and immune enzymes activity
Shrimp fed the RS diet had greater digestive and immune enzymes activity than those of the control group (Fig. 2). Amylase (AMS) activity in the RS2 group was highest (P < 0.05). Lipase (Lip) activity in the RS2 and RS3 groups was highest, and there were no differences between them (P > 0.05). The trypsin (Tryp) activity showed no differences in the three RS groups (P > 0.05). The pepsin (Pep) activity was increased in the three RS groups with the increasing doses of RS (P < 0.05). The phenoloxidase (PO) activity was increased in the three RS groups (P < 0.05) with the decreasing doses of RS. The total antioxidant capacity (T-AOC) activity was increased in the three RS groups (P < 0.05) with the increasing doses of RS. The total nitric oxide synthase (T-NOS) activity was highest in the RS3 group (P < 0.05), and there were no differences between the RS1 and RS2 groups. The nitric oxide (NO) content in the three RS groups was statistically the same (P > 0.05).
Digestive and immune enzyme indices in the intestine of L. vannamei that were fed the control and three RS diets for 56 days. (A) AMS; (B) LPS; (C) Tryp; (D) Pep; (E) PO; (F) T-AOC; (G) T-NOS; (H) NO. Bars show the mean ± SE (n = 3). The different letters (a–c) indicate significant differences (P < 0.05) among the groups.
Immune-related genes expression
Shrimp fed the RS diet had a higher immune-related genes expression level than those of the control group, including the antibacterial, antioxidant, and mucus functions (Fig. 3). Antibacterial genes, such as prophenoloxidase (proPO), anti-lipopolysaccharide factor (ALF), and lysozyme (Lys), were the highest in the RS1, RS2, and RS3 groups respectively (P < 0.05). Antioxidant genes, such as the heat shock protein 70 (HSP70) was the highest in the RS1 group (P < 0.05), thioredoxin (Trx) in the three RS groups was statistically the same (P > 0.05). Mucin (Muc) genes, such as Muc-1 and Muc-19, were the highest in the RS3 group (P < 0.05), Muc-2 and Muc-5B were no differences in the three RS groups (P > 0.05), Muc-5AC was the highest in the RS2 and RS3 groups (P < 0.05).
Intestine microbial richness and diversity
Illumina sequencing of the intestine microbial produced 1,587,040 clean reads, and the average sequence length was 316 bp. A rarefaction curve analysis of the observed species per sample was sufficient (Fig. 4A). A total of 564 (97.24%) OTUs were co-owned by all of the groups, and the unique OTUs were higher in the three RS groups compared with those of the control group (Fig. 4B). The largest category of the unique OTUs was the RS3 group (458, 0.4%), the second was the RS1 group (198, 0.4%), and the third was the RS2 group (159, 0.1%). Non-metric multi-dimensional scaling (NMDS) based on the Bray-Curtis distance was performed to detect the relationships among the microbial in different samples and confirmed that the intestine microbial of the four groups were separated (Fig. 4C).
Similar to the OTUs number, the Chao 1, ACE, Simpson and Shannon indices of the three RS groups were all higher than those of the control group (Table 1). The Chao 1 and ACE indices of the RS1 and RS3 groups were higher than those of the RS2 group (P < 0.05). The Shannon index was no difference among the three RS groups (P > 0.05). The Simpson indices of the RS1 and RS2 groups were higher than those of the RS3 group (P < 0.05).
Changes in the intestine bacterial composition
In total, 35 different bacterial phylum were identified; Proteobacteria, Bacteroidetes, and Verrucomicrobia were three of the dominant phylum. Compared with the control group, Proteobacteria, Verrucomicrobia, and Acidobacteria abundance were increased in the three RS groups, while that of Bacteroidetes and Tenericutes were decreased (Fig. 5A). At the class level, the abundances of Alphaproteobacteria, Gammaproteobacteria, and Verrucomicrobiae were increased in the three RS groups, while that of Flavobacteriia was decreased (Fig. 5B). At the genus level, the abundances of certain beneficial bacteria, Lutimonas, Ruegeria, Shimia, Mesoflavibacter, and Mameliella, were increased in the three RS groups, while that of some potential pathogens, such as Formosa and Pseudoalteromonas, were decreased (Fig. 5C).
Intestine microbe composition of L. vannamei that were fed the control and RS diets for 56 days. (A) phylum level; (B) class level. Bars show the mean ± SE (n = 3). The different letters (a–c) indicate significant differences (P < 0.05) among groups. (C) Heatmap analysis of intestine microbial on the top 50 genera. The redder colour show the higher abundance of the genera, and the black colour is the lower abundance.
The linear discriminant analysis (LDA) effect size (Lefse) was applied to detect the differential abundances of bacterial taxa among the four groups. The Lefse LDA scores showed that the abundances of 25, 16 and 5 taxa were increased in the RS1, RS2, and RS3 groups respectively, while that of 17 taxa were all decreased (Fig. 6A). There were 14 bacterial taxa that distinguished the four groups by LDA value, with 5, 3, 5, and 1 taxa in the control, RS1, RS2, and RS3 groups respectively (Fig. 6B). In detail, 2 classes, 4 orders, and 3 families were enriched in the control group, including Micrococcales (from the order level), Lentisphaerae (from the class level), Desulfobacterales (from the order to family level), Alteromonadales (from the order to family level), and Mollicutes (from the class to family level). One phylum, 2 classes, 6 orders, and 7 families were enriched in the RS1 group, including Proteobacteria (from the phylum to family level), Cyclobacteriaceae (from the family level), and LD29 (from the family level). Two orders and 4 families were enriched in the RS2 group, including Propionibacteriales (from the order to family level), Sh765B-TzT-29 (from the family level), Rhizobiales_Incertae_Sedis (from the family level), Bacteriovoracaceae (from the family level), and Oligoflexales (from the order level). One phylum and one order were enriched in the RS3 group, including Acidobacteria (from the phylum to order level).
Changes in the intestine microbial metabolism
Based on random forest KEGG classification, the signalling of “nitrotoluene degradation”, “methane metabolism”, “stilbenoid, diarylheptanoid and gingerol biosynthesis”, “secondary bile acid biosynthesis”, and “starch and sucrose metabolism” were increased in the three RS groups, while that of “retinol metabolism”, “nicotinate and nicotinamide metabolism”, “isoflavonoid biosynthesis” and “beta-lactam resistance” were decreased (Fig. 7).
Shrimp fed the RS diet had a higher SCFAs content, including acetic acid (AA), propionic acid (PA), butyric acid (BA), and valeric acid (VA) than those of the control group (P < 0.05) (Table 2). The AA content was no difference among the three RS groups. The PA content was increased in the three RS groups with the increasing doses of RS. The BA content was highest in the RS3 group, while that in the RS1 and RS2 groups was not significantly different. The VA content was highest in the RS2 and RS3 groups, and they were statistically the same.
The intestine microbial were correlated with biochemical parameters
The correlations between the phylum abundance and the host biochemical parameters are shown in Fig. 8. The proPO, HSP70, Muc-2, and Muc-5B had positive correlations with the Proteobacteria. The PO had a positive correlation with the Proteobacteria, Verrucomicrobia, Actinobacteria, and Chlorobi. The Tyrp, T-NOS, NO, proPO, HSP70, and Muc-5B had positive correlations with the Acidobacteria and Chlorobi. The PA, BA, Pep, T-AOC, Lys, Muc-1, and Muc-19 had positive correlations with the Acidobacteria. The AMS, ALF, Trx, and Muc-2 had positive correlations with the Chlorobi. The T-NOS, proPO, and HSP70 had positive correlations with the Gemmatimonadetes.
Significant correlation between intestine microbial phylum and shrimp health indices. Correlation coefficient is represented by different colours (green: positive correlation; red: negative correlation). *Data represent significantly negative or positive correlations (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
The intestine health influences the shrimp health because intestine mucosa provides an important barrier to pathogens. Intestine health is related to its structural integrity, mucus components and immune molecules2. The intestine mucosa structure of shrimp can be damaged by environmental stress and pathogen challenges16, which may increase the risk of pathogen invasion and impair the host immunity17. The nutrient uptake of the intestine mainly relies on its epithelium and microvilli. Here, we examined the effects of dietary RS on intestine mucosa morphology and the digestive enzymes activity of the shrimp. We found that shrimp fed the RS diet had a higher epithelium height and digestive enzymes activity. Hence, dietary RS improved the intestine histological structure and promoted its digestive capacity, which is likely to benefit nutrient absorption.
Antibacterial molecules play vital roles in the immune homeostasis of shrimp. Mucus is located on the surface of the intestine mucosa, which can lubricate the intestine tract and resist pathogenic infection18. Muc is the main antimicrobial component of intestine mucus19,20. It was reported that dietary RS could increase the Muc-4, Muc-5AC, and Muc-12 genes in the intestine of pigs21. As important immune parameters, ALF, proPO, Lys, NOS and NO function as strong antibacterial forces against pathogenic microbes22,23,24,25. In this study, the PO and T-NOS activity, NO content, and the expression levels of the propO, ALF, Lys, Muc-1, Muc-2, Muc-5AC, Muc-5B and Muc-19 genes were increased in the three RS groups, which suggested that dietary RS could improve the intestine mucin and antibacterial capacity of the host towards resisting pathogenic infection.
The environment induces oxidative stress in aquatic animals26. Organisms have evolved an antioxidant defence system to counteract oxidative stress, including antioxidant enzymes and proteins. The total status of antioxidant enzymes can be reflected by T-AOC activity27. As important antioxidant proteins, HSPs can increase endogenous peroxidase activity to eliminate reactive oxygen species (ROS)28; Trx acts as a peroxiredoxin substrate that functions in cell redox homeostasis29. In this study, dietary RS increased the T-AOC activity and the HSP70 and Trx gene expression in the shrimp intestine. These phenomena revealed that dietary RS could enhance the intestine antioxidant function, which might contribute to the regulation of the redox status of the intestine.
Shrimp microbial are important for its host health, which can be regulated by diet30,31,32. Here, we found that dietary RS improved the composition of intestine microbial of the shrimp and increased the microbial carbon source and lipid metabolism, including starch, sucrose, SCFAs, bile acid, and gingerol. SCFAs can create an acidic environment in the intestine of shrimp, which accelerate increase of beneficial bacteria and reduce the proliferation o pathogenic bacteria9. Thus, dietary RS can increase the diversity and beneficial metabolites of intestine microbial to improve the intestine health of shrimp. After the shrimp fed the RS diets for 56 days, the bacterial phylum Proteobacteria abundance increased, while the Bacteroidetes abundance decreased; these two phylum are the dominant intestine bacterial of the shrimp during its growth stages16,33,34,35. The abundance of Actinobacteria was enriched in the three RS groups. The correlation analysis of the bacterial phylum and its host health indicators showed that Proteobacteria had a positive correlation with immune indices (PO, proPO, HSP70, Muc-2, and Muc-5B). Additionally, the PO had a positive correlation with the Actinobacteria abundance. Actinobacteria are good elaborators of pharmaceutical products, including antibiotics and antimicrobial agents36. Hence, forecasts of these phylum bacterial contributed to the intestine health of the host, particularly in antibacterial, antioxidant, and mucin secretion.
Some beneficial bacterial genera were enriched after the shrimp fed with the RS diets, including Lutimonas, Ruegeria, Shimia, Mesoflavibacter, and Mameliella. Lutimonas is a strictly aerobic heterotrophic nitrifying bacterium for the degradation of ammonia37. Ruegeria and Shimia are both members of the Roseobacter clade, with essential metabolic capabilities, and are involved in protein utilization38. Ruegeria also have high phosphodiesterase and phosphomonoesterase activities that contribute to the degradation of phosphate triester compounds in aquatic environments39,40. Mameliella play important roles in the degradation of aromatic compounds41,42 and can accumulate beneficial poly-β-hydroxybutyrate (PHB) granules in cells43. Mesoflavibacter can produce zeaxanthin that have strong antioxidant and anticancer properties44,45. Interestingly, based on the KEGG pathway analysis, “nitrotoluene degradation” and “methane metabolism” were increased in the three SA groups, which indicated that dietary RS contribute to the enrichment of the beneficial bacteria that are involved in degrading toxins and producing beneficial metabolites. Furthermore, opportunistic pathogens, such as Formosa and Pseudoalteromonas, were absent in response to dietary RS. Formosa was enriched in the intestine of L. vannamei when the shrimp were subjected to ammonia and nitrite stress, respectively33. Pseudoalteromonas can cause high mortality in Portunus pelagicus46. Therefore, this study illuminated that dietary RS could optimize the intestine microbial composition of L. vannamei and decrease the risk of pathogen invasion to the host.
In conclusion, dietary RS improved the intestine health of L. vannamei. Of these, dietary RS improved the intestine mucosa morphology, enhanced the intestine digestive and immune capacity of the shrimp, including mucus, antibacterial, and antioxidant enzyme activity and/or gene expression. Furthermore, dietary RS also regulated the composition and SCFAs contents of the intestine microbial, and there was a positive correlation between some bacterial phylum and the host health indices.
Materials and Methods
Ethics statements
The collection and handling of the animals in this study was approved by the Animal Care and Use Committee at the Chinese Academy of Fishery Sciences (CAFS), and all experimental animal protocols were carried out in accordance with national and institutional guidelines for the care and use of laboratory animals at the CAFS (No. 2016TS07).
Diet preparation
The source of RS in this study was high amylose maize (HI-MAIZE® 260), which was purchased from National Starch Industrial (Shanghai) co., LTD. According to the supplier, this starch was obtained directly from high amylose maize seeds, the RS contents was 80%, and it could be classified as an RS2 type; no other exogenous ingredients were added. Four experimental diets were prepared and differed in RS content: 0 g/kg (Control), 10 g/kg (RS1), 30 g/kg (RS2) and 50 g/kg (RS3), and the feed formula is given in Table S1. In the feed ingredients of the three RS groups, we used RS instead of wheat flour. The diet preparation was referenced to Duan et al.47 Moisture, crude protein, crude lipid and ash of the experimental diets were determined using a standard method of Association of Official Analytical Chemists (AOAC) (1995).
Shrimp and rearing conditions
Healthy juvenile L. vannamei were randomly selected from a local farming pond in Shenzhen, China; the average weight was 3.2 ± 0.3 g. The shrimp were temporarily reared and acclimated for 7 days before the feeding trial experiment. The rearing water of the shrimp was sand-filtered seawater, which was aerated with two stones. The parameters of the water quality were pH 8.4, salinity 30, temperature 30 ± 0.5 °C, and dissolved oxygen 6.0 ± 0.5 mg l–1. The shrimp were fed daily at 5% of body weight. Change one third of the water each day.
After acclimation, the shrimp were divided into four groups: Control, RS1, RS2, and RS3, and each group was fed with the corresponding experimental feed. Each group contained three 500 L replicate tanks, and 40 shrimp per tank. The culture condition was in accord with the acclimation stage. All tanks were fed three times per day at 7:00, 12:00 and 18:00 for 56 days. Uneaten feed and faeces were cleared out from the tanks. At 56 days, the intestine of the shrimp of each tank were sampled individually and were snap-frozen in liquid nitrogen for biochemical, gene expression, and microbiome analysis.
Histological analysis
The intestines from three shrimp per tank were extracted at 56 days and were stored in Davidson solution for 24 h. After being rinsed with running water for 8 h, and the embedded lumps were put into 70% ethanol overnight. Then, the samples were dehydrated in a series of ethanol solutions (80%, 90%, and 100%), followed by acetone and xylene transparent and paraffin embedding. The tissue was sectioned in a microtome (Leica, RM2016, Wetzlar, Germany) to a 4 μm thickness. After staining with haematoxylin and eosin (HE), stained sections were observed and photographed under light microscope (Olympus, Tokyo, Japan). The intestine EH and WT were detected six sites randomly.
Biochemical analysis
The intestine from three shrimp per tank were extracted at 56 days and homogenized to 10% ratio with 0.9% saline solution, then centrifuged (3500 rpm, 4 °C, 10 min). The supernatant was immediately detected for biochemical parameters using a microplate reader (Bio-Rad, USA). AMS, Lip, Tryp, Pep, PO, T-AOC, T-NOS, and NO were measured using related commercial assay kits (Nanjing Jiancheng Institute, China) according to the manufacturer’s protocols. The total protein concentration in tissue homogenates was measured using a Coomassie brilliant-blue protein assay kit (Jiancheng, Nanjing, China). The assays were all run in three replicate samples.
Gene expression analysis
The total RNA from the intestine of three shrimp per tank was extracted at 56 days using TRIzol Reagent (Invitrogen, USA); then, the genome DNA was eliminated using RQ1 RNase-Free DNase (Promega, USA). The total RNA (8 μg) was reverse transcribed to cDNA using PrimeScript™ RT Reagent Kit (Takara, China) according to the manufacturer’s protocol. Real-time RT-qPCR was conducted in a LightCycler480 System using a SYBR® Premix Ex Taq™ II Kit (TaKaRa, Japan). β-actin gene of L. vannamei was chosen as an internal control, and the specific primer sequences of the target genes were designed using Primer Premier 5.0 software and are listed in Table S2. RT-qPCR was carried out using the method of Duan et al.47. The relative gene expression was calculated by the 2−ΔΔCT comparative CT method and is shown as the fold-change in comparison to the control group.
Intestine microbiome analysis
The intestine microbial DNA of six shrimp per tank was extracted at 56 days using a PowerSoil™ DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer’s protocol, and analyzed using 1.0% agarose gel electrophoresis. DNA concentration and quality were checked using a NanoDrop Spectrophotometer. The V4 region of the bacterial 16S rRNA gene was amplified using the barcoded fusion primers 515F and 806R (Table S2). The PCRs were each 20 μL, containing the FastPfu polymerase 0.4 μL, dNTPs (2.5 mM) 2 μL, 5 × FastPfu buffer 4 μL, each primer (5 μM) 0.8 μL, and template DNA 10 ng. The PCR programs were 1 cycle of 95 °C for 5 min, 27 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, followed by 72 °C for 10 min. The PCR products were purified by a PCR purification kit (Qiagen), then were sequenced with an Illumina HiSeq platform. The sequences analysis was referenced to Duan et al.33.
Intestine SCFAs content analysis
SCFAs content, including AA, PA, BA, and VA from the intestine of three shrimp per tank were detected at 56 days using gas chromatography, which was referenced to Weitkunat et al.48.
Statistical analysis
The data were presented as the mean ± SE, and statistically analyzed using one-way ANOVA, followed by Duncan’s multiple range testing (SPSS v22.0). Correlation between the host intestine health indices and the bacterial relative abundance by phylum (top ten) was estimated using the Pearson test (−0.8 < R < 0.8). P < 0.05 was regarded as statistically significant.
References
Brown, K., DeCoffe, D., Molcan, E. & Gibson, D. L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 4, 1095–1119 (2012).
Levy, M., Blacher, E. & Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 35, 8–15 (2017).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
De Vadder, F. & Mithieux, G. Gut-brain signaling in energy homeostasis: the unexpected role of microbiota-derived succinate. J. Endocrinol. 236, R105–R108 (2018).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Backhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Cummings, J. H. & Macfarlane, G. T. Role of intestinal bacteria in nutrient metabolism. Clinical Nutrition 16, 3–11 (1997).
Zhao, L. P. et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018).
Regassa, A. & Nyachoti, C. M. Application of resistant starch in swine and poultry diets with particular reference to gut health and function. Animal Nutrition 4, 305–310 (2018).
Roy, C. C., Kien, C. L., Bouthillier, L. & Levy, E. Short-chain fatty acids: ready for prime time? Nutr. Clin. Pract. 21, 351–366 (2006).
Haenen, D. et al. A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. J. Nutr. 143, 274–283 (2013).
Heo, J. M., Agyekum, A. K., Yin, Y. L., Rideout, T. C. & Nyachoti, C. M. Feeding a diet containing resistant potato starch influences gastrointestinal tract traits and growth performance of weaned pigs. J. Anim. Sci. 92, 3906–3913 (2014).
Krause, D. O., Bhandari, S. K., House, J. D. & Nyachoti, C. M. Response of nursery pigs to a synbiotic preparation of starch and an Anti-Escherichia coli K88 probiotic. Appl. Environ. Microbiol. 76, 8192–8200 (2010).
Thitamadee, S. et al. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452, 69–87 (2016).
Sapkota, A. et al. Aquaculture practices and potential human health risks: current knowledge and future priorities. Environ. Int. 34, 1215–1226 (2008).
Duan, Y. et al. Changes in the intestine microbial, digestive, and immune-related genes of Litopenaeus vannamei in response to dietary probiotic Clostridium butyricum supplementation. Front. Microbiol. 9, 2191 (2018).
Gao, C. et al. Transcriptomic profling revealed the signatures of intestinal barrier alteration and pathogen entry in turbot (Scophthalmus maximus) following Vibrio anguillarum challenge. Dev. Comp. Immunol. 65, 159–168 (2016).
Johansson, M. E. & Hansson, G. C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 16(10), 639–649 (2016).
Derrien, M. et al. Mucin bacterial interactions in the human oral cavity and digestive tract. Gut Microb. 1(4), 254–268 (2010).
Ogata, S., Shimizu, K., Tominaga, S. & Nakanishi, K. Immunohistochemical study of mucins in human intestinal spirochetosis. Hum. Pathol. 62, 126–133 (2017).
Zhou, L., Fang, L., Sun, Y., Su, Y. & Zhu, W. Effects of a diet high in resistant starch on fermentation end-products of protein and mucin secretion in the colons of pigs. Starch Stärke 69, 1600032 (2017).
de la Vega, E. et al. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): a broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection. Mol. Immunol. 45(7), 1916–1925 (2008).
Saurabh, S. & Sahoo, P. K. Lysozyme, an important defence molecule of fish innate immune system. Aquac. Res. 39, 223–39 (2008).
Xie, S. W. et al. Effect of proline supplementation on anti-oxidative capacity, immune response and stress tolerance of juvenile Pacific white shrimp, Litopenaeus vannamei. Aquaculture 448, 105–111 (2015).
Yao, C. L., Ji, P. F., Wang, Z. Y., Li, F. H. & Xiang, J. H. Molecular cloning and expression of NOS in shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 28, 453–460 (2010).
Lushchak, V. I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 101, 13–30 (2011).
Wu, F. J. et al. The protective effect of eicosapentaenoic acid-enriched phospholipids from sea cucumber Cucumaria frondosa on oxidative stress in PC12 cells and SAMP8 mice. Neurochem. Int. 64, 9–17 (2014).
Shi, J. X. et al. Characterization and function analysis of Hsp60 and Hsp10 under different acute stresses in black tiger shrimp, Penaeus monodon. Cell Stress Chaperon. 21, 295–312 (2016).
Ren, Q., Zhang, R. R., Zhao, X. F. & Wang, J. X. A thioredoxin response to the WSSV challenge on the Chinese white shrimp, Fenneropenaeus chinensis. Comp. Biochem. Physiol. C 151, 92–98 (2010).
Turnbaugh, P. J., Bäckhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Wu, G. D. et al. Linking longterm dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Duan, Y. F., Liu, Q. S., Zhang, J. S., Wang, Y. & Xiong, D. L. Impairment of the intestine barrier function in Litopenaeus vannamei exposed to ammonia and nitrite stress. Fish Shellfish Immunol. 78, 279–288 (2018).
Zhang, D. M. et al. Bacterioplankton assemblages as biological indicators of shrimp health status. Ecol. Indic. 38, 218–224 (2014).
Huang, Z. B., Li, X. Y., Wang, L. P. & Shao, Z. Z. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquac. Res. 47(6), 1737–1746 (2016).
Zothanpuia Passari, A. K. et al. Bioprospection of actinobacteria derived from freshwater sediments for their potential to produce antimicrobial compounds. Microb Cell Fact. 17, 68 (2018).
Fu, S. Z., Fan, H. X., Liu, S. J., Liu, Y. & Liu, Z. P. A bioaugmentation failure caused by phage infection and weak biofilm formation ability. J. Environ. Sci. 21, 1153–1161 (2009).
Barreto-Curiel, F. et al. Effects of extruded aquafeed on growth performance and gut microbiome of juvenile Totoaba macdonaldi. Anim. Feed. Sci. Tech. https://doi.org/10.1016/j.anifeedsci.2018.09.002 (2018).
Achbergerová, L. & Nahálka, J. Degradation of polyphosphates by polyphosphate kinases from Ruegeria pomeroyi. Biotechnol. Lett. 36(10), 2029–2035 (2014).
Yamaguchi, H. et al. Phosphotriesterase activity in marine bacteria of the genera Phaeobacter, Ruegeria, and Thalassospira. Int. Biodeter. Biodegr. 115, 186–191 (2016).
Danish-Daniel, M., Han Ming, G., Noor, M. E., Yeong, Y. S. & Usup, G. Draft genome sequence of Mameliella alba strain UMTAT08 isolated from clonal culture of toxic dinoflagellate Alexandrium tamiyavanichii. Genomics Data 10, 12–14 (2016).
Zhao, Q. et al. Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci. Total Environ. 609, 1238–1247 (2017).
Zheng, Q. et al. Mameliella alba gen. nov., sp. nov., a marine bacterium of the Roseobacter clade in the order Rhodobacterales. Int. J. Syst. Evol. Microbiol. 60(4), 953–957 (2010).
Lee, J. H. et al. Mesoflavibacter aestuarii sp. nov., a zeaxanthinproducing marine bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 64, 1932–1937 (2014).
Zhang, Y., Liu, Z., Sun, J., Xue, C. & Mao, X. Biotechnological production of zeaxanthin by microorganisms. Trends Food Sci. Tech. 71, 225–234 (2018).
Talpur, A. D. et al. Pathogenicity and antibiotic sensitivity of pathogenic flora associated with the gut of blue swimming crab, Portunus pelagicus (Linnaeus, 1758). J. Anim. Vet. Adv. 10, 2106–2119 (2011).
Duan, Y. F., Wang, Y., Zhang, J. S., Sun, Y. X. & Wang, J. Dietary effects of succinic acid on the growth, digestive enzymes, immune response and resistance to ammonia stress of Litopenaeus vannamei. Fish Shellfish Immunol. 78, 10–17 (2018).
Weitkunat, K. et al. Effects of dietary inulin on bacterial growth, short-chain fatty acid production and hepatic lipid metabolism in gnotobiotic mice. J. Nutr. Biochem. 26, 929–37 (2015).
Acknowledgements
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2019TS18, 2016TS07, 2017YB12), Central Public-interest Scientific Institution Basal Research Fund, CAFS (2019CY0103), Guangdong Provincial Special Fund for Marine Fisheries Technology (A201701B09), Guangdong Natural Science Foundation (2017A030313147), and Shenzhen Science and Technology Planning Project (JCYJ20170412110605075). We are grateful to the technical assistance of the laboratory staff.
Author information
Authors and Affiliations
Contributions
Y.D. conceived, designed, performed the experiments, and wrote the manuscript. Y.W. and J.Z. assisted in the experimental design, and edited of the manuscript. Y.W., Q.L., H.D., H.L. and D.X. contributed to the sampling and data analyse. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Duan, Y., Wang, Y., Liu, Q. et al. Changes in the intestine microbial, digestion and immunity of Litopenaeus vannamei in response to dietary resistant starch. Sci Rep 9, 6464 (2019). https://doi.org/10.1038/s41598-019-42939-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-42939-8
This article is cited by
-
Unraveling the etiology of shrimp diseases: a review through the perspectives of gut microbial dynamics
Aquaculture International (2024)
-
Microbiome changes of an integrated aquaculture system of shrimp Litopenaeus vannamei and seaweed Ulva lactuca with different water exchanges
Aquaculture International (2024)
-
In silico prediction of potential indigenous microbial biomarkers in Penaeus vannamei identified through meta-analysis and genome-scale metabolic modelling
Environmental Microbiome (2023)
-
Digestive enzyme activities, intestinal histology, and gut microbiota of Pacific white shrimp Litopenaeus vannamei fed with turmeric, curcumin, and nanomicelle curcumin
Aquaculture International (2023)
-
Agavin induces beneficial microbes in the shrimp microbiota under farming conditions
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.