Abstract
Tinea pedis is a type of dermatophytosis caused by anthropophilic keratinolytic fungi such as Trichophyton interdigitale. Quantitative reverse transcription PCR (RT-qPCR) is a reliable and reproducible technique for measuring changes in target gene expression across various biological conditions. A crucial aspect of accurate normalization is the choice of appropriate internal controls. To identify reference genes for accurate evaluation of expression levels in T. interdigitale, the transcription levels of eight candidate reference genes (adp-rf, β-act, ef1-α, gapdh, psm1, sdha, rpl2 and ubc) and one target gene (Tri m4) were analysed by RT-qPCR after growing the dermatophyte under different environmental conditions. The results obtained from expression stability evaluations with NormFinder, geNorm, BestKeeper, and RefFinder software demonstrated that adp-rf and psm1 were the most stable internal control genes across all experimental conditions. The present study constitutes the first report of the identification and validation of reference genes for RT-qPCR normalization for T. interdigitale grown under different environmental conditions resembling the conditions encountered by fungi during invasion of skin.
Similar content being viewed by others
Introduction
Trichophyton interdigitale is a keratinophilic and keratinolytic fungus belonging to the dermatophyte group1, and it is responsible for infections of the feet and toes (tinea pedis)2. Epidemiological studies have indicated that the incidence of dermatophytosis due to this fungus is rising and is not correlated with patient characteristics such as ethnicity or race3. However, tinea pedis is most frequently found in adults between 30 and 60 years old4, and this dermatophytosis is more common in men than in women5 and in developed countries6. Dermatophytes have been recorded worldwide with variations in epidemiology, distribution, incidence and target hosts from one location to another. Different conditions, such as geographic location, climate, health care quality, immigration status, hygiene culture, and socioeconomic status, may influence the development of dermatophyte infections7. Keratin is a structural protein of the stratum corneum, where dermatophytes typically infect, while elastin and collagen are the fibrous proteins of the extracellular matrix in the dermis that dermatophytes can penetrate during infection8. The availability of these proteins is necessary for the activation of signal transduction during dermatophyte infection. The degradation of keratin and other fibrous proteins releases high amounts of cysteine, proline, serine or lysine, and the metabolism of these amino acids leads to the secretion of ammonia, which raises the extracellular pH from acidic to alkaline values. Transcriptomic analyses suggest that during the first 48 h after infection9, the highest transcriptional activity of genes responsible for the synthesis of proteases and adhesins occurs, which allows colonization of the host tissue10. Analysis of changes in dermatophyte gene expression profiles under defined growth conditions can improve our knowledge of the mechanisms associated with the pathogenicity of dermatophytes and of the other biological properties of this group of pathogens. Information gathered during such study may be useful in the search for new therapeutic and prophylactic strategies. Quantitative reverse transcription PCR (RT-qPCR) is a powerful technique used to quantify the mRNA levels of different genes of interest under various experimental conditions. However, different experimental and technical variations can lead to incorrect data analysis. Therefore, it is necessary to establish a set of optimal reference genes before conducting target gene expression analysis. Due to the limited knowledge regarding reference genes useful for RT-qPCR analysis in dermatophytes10 and the particularly insufficient information on the complete genome sequence of T. interdigitale, eight reference genes, including adp-rf (ADP ribosylation factor), β-act (β-actin), ef1-α (elongation factor 1-alpha), gapdh (glyceraldehyde 3-phosphate dehydrogenase), psm1 (mitotic cohesion complex subunit Psm1), sdha (succinate dehydrogenase complex flavoprotein subunit A), rpl2 (ribosomal protein L2) and ubc (ubiquitin) (Table 1) were ultimately selected and evaluated in a T. interdigitale strain subjected to 13 different environmental conditions (Table 2). The selected candidate reference genes were chosen from among internal controls used in some species of fungi, including dermatophytes10,11, and in other eukaryotic organisms9,10,12,13,14,15. The expression stability of each candidate reference gene was calculated by the following algorithms: the geNorm module of qbase + (Biogazelle)16, NormFinder17, and BestKeeper18. The online comprehensive tool RefFinder was ultimately used to compare and rank the candidate reference genes.
Results
Amplification efficiency and specificity of eight candidate reference genes
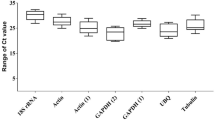
The specificity of the primer sets was validated based on the identification of a single band of the expected size on 8% polyacrylamide gels and a single homogenous peak in melting curve analysis (Table 1, Supplementary Fig. S1A,B). The PCR efficiencies (E%) ranged from 99–110%, with correlation coefficient (R2) values varying from 0.996 to 0.999 (Table 1). The expression profiles of the eight reference gene candidates (Table 1) were analysed under control and experimental conditions by calculating the mean raw Ct value from three independent repetitions (Supplementary Table S1). As shown in Fig. 1, the Ct values of the eight candidate housekeeping genes ranged from 15.20 to 26.32 across all experimental conditions.
Expression levels of eight reference genes in T. interdigitale. The mean Ct values for all experimental conditions for each candidate reference gene are shown as box plot representations. Each box indicates the 25th and 75th percentiles. The line across the box represents the median, and the whisker caps show the maximum and minimum values.
Expression stability analyses
The GeNorm algorithm, which is a module of qbase + (Biogazelle), was used to evaluate the candidate reference genes based on their expression stability values (M-values) and pairwise variations (Vn/Vn+1). Psm1 (M-value = 0.483), adp-rf (M-value = 0.502) and sdha (M-value = 0.520) were the most stable reference genes under all experimental conditions (Fig. 2). In contrast, the β-act gene had the highest M-value, with the lowest expression stability (M-value = 1.132) in all analysed samples (Fig. 2). The pairwise variation (Vn/Vn+1) results indicated that five reference genes (psm1, adp-rf, sdha, ubc and rpl2) should be used for reliable normalization (V5/6 = 0.136) (Fig. 3). The most stable reference genes (in order) among all chosen candidates for T. interdigitale under each experimental condition were as follows (Table 3): ef1-α, rpl2, sdha, adp-rf, ubc, psm1, gapdh, and β-act for control conditions (MM-Cove); adp-rf, ubc, gapdh, psm1, ef1-α, sdha, rpl2, and β-act for glucose supplementation; ef1-α, ubc, sdha, psm1, rpl2, β-act, adp-rf, and gapdh for keratin supplementation; rpl2, gapdh, ef1 α-α, ubc, β-act, psm1, sdha, and adp-rf for keratin and soy protein supplementation; sdha, psm1, rpl2, adp-rf, ubc, β-act, ef1-α, and gapdh for elastin supplementation; ef1-α, psm1, adp-rf, rpl2, β-act, sdha, gapdh, and ubc for collagen supplementation; rpl2, psm1, adp-rf, β-act, ef1-α, ubc, gapdh, and sdha for colloidal chitin supplementation; adp-rf, ef1-α, β-act, sdha, psm1, ubc, rpl2, and gapdh for low-Pi MM; and β-act, adp-rf, psm1, ef1-α, gapdh, ubc, rpl2, and sdha for low-Pi YEM. Furthermore, pairwise variation (Vn/Vn+1) calculation with a V-value < 0.15 showed that only two internal controls were sufficient for normalizing gene expression under all experimental conditions (Fig. 3).
According to NormFinder17, across all experimental conditions, psm1 had the lowest stability value (SV = 0.080) (Fig. 2). Psm1 and rpl2 constituted the best combination of internal control genes with SV = 0.061 under all experimental conditions. Psm1 was found to be the most stably expressed gene in the presence of colloidal chitin (SV = 0.032) (Fig. 4G), and rpl2 was the most stable gene under control conditions (SV = 0.100) (Fig. 4A). β-act was the most stable gene in the medium supplemented with keratin and soy protein (SV = 0.138) (Fig. 4D) and under low-Pi conditions in MM (SV = 0.045) (Fig. 4I). In the case of collagen supplementation and under the low-Pi condition in YEM, NormFinder calculations revealed that adp-rf had the lowest stability values, with SV = 0.259 (Fig. 4E) and SV = 0.028 (Fig. 4H), respectively. Ef1-α, ubc and gapdh were the most stably expressed genes in the presence of glucose (SV = 0.096) (Fig. 4B), keratin (SV = 0.010) (Fig. 4C), and elastin (SV = 0.010) (Fig. 4F).
Assessment of the expression variation of the candidate reference genes using the BestKeeper algorithm18 revealed that seven of the genes had standard deviation values defined as acceptable [0.5 < SD[±Ct] ≤ 1.00], while gapdh had an unacceptable standard deviation, as it was higher than 1.0 (SD = 1.32) (Table 3). Analyses showed also significant expression correlations with the BestKeeper index, which is the geometric mean of the Ct values of the analysed genes (correlation coefficient r = 0.698–0.901), except for the β-act gene (r = 0.145). The expression of all genes correlated with the BI with p values < 0.001, except the β-act gene (p-value = 0.543). The overall order of the most stable genes based on BestKeeper was psm1, adp-rf, ef1-α, sdha, ubc and rpl2 (Fig. 2, Table 4).
In the final step, RefFinder, a free online tool for the identification of stable reference genes that integrates all methods applied in the present study, was used to generate a final ranking of the eight reference genes according to their geomean ranking values. As shown in Fig. 2, psm1 and adp-rf were ranked as the best reference genes for measuring target gene expression levels under the chosen conditions.
Stability and validation of adp-rf and psm1 as reference genes
To confirm adp-rf and psm1 as the most stable reference genes, their expression was compared in the T. interdigitale CBS 124408 reference strain and two clinical isolates: T. interdigitale 12/2010 and T. interdigitale 45/10. These three strains of T. interdigitale were incubated at 28 °C for 48 h in control medium and in medium supplemented with keratin. The obtained Ct values (Fig. 5) for the adp-rf and psm1 genes were not significantly different under both conditions (ppsm1 = 0.93; padp-rf = 0.89, ANOVA) (Fig. 5), which confirmed that these reference genes can be used for accurate expression level evaluation in various T. interdigitale strains. To confirm the reliability of adp-rf and psm1 as reference genes for RT-qPCR normalization, the expression of Tri m4 was examined19. Tri m4 is known as an aminopeptidase gene whose expression increases in the presence of keratin and elastin, which suggests that the product of this gene may play an important role as a virulence factor19. The validation was performed using templates from the T. interdigitale 45/10 strain incubated at 28 °C for 48 h in control medium (MM-Cove) and in medium supplemented with keratin or elastin. Three different sets of reference genes were analysed: set A included the most stable reference genes (adp-rf and psm1), set B included the least stable reference genes (β-act and gapdh), and set C included all eight candidate reference genes. The relative expression of the target gene was determined using the 2−ΔΔCt method20. As shown in Fig. 6, an increase in Tri m4 transcript levels in T. interdigitale growing in the presence of keratin or elastin in relation to control conditions was noticed only when the adp-rf and psm1 reference genes (set A) previously selected by all four algorithms were used.
Stability of adp-rf and psm1 reference gene expression in three T. interdigitale strains cultivated in control medium (MM-Cove) and MM-Cove supplemented with keratin. The gene expression levels are presented as the average Ct values. The adp-rf (p = 0.93, ANOVA) and psm1 (p = 0.89, ANOVA) gene expression levels were not significantly different across the analysed culture conditions. The error bars indicate the standard error.
Relative quantification of Tri m4 gene expression in control, keratin and elastin protein samples using different reference gene variants: (A) (p = 0.55, ANOVA), which included the two most stable reference genes, adp-rf and psm1; (B) (p = 0.46, ANOVA), which included the two least stable reference genes, β-act and gapdh; and (C) (p = 0.36, ANOVA), which included all candidate reference genes. The error bars indicate the standard error.
Discussion
Quantitative reverse transcription PCR (RT-qPCR) is an efficient method for analysing target gene expression but requires comprehensive normalization with properly selected reference genes. The internal controls should have relatively stable expression levels in different types of cells or tissues, and their expression should be constant under various growth conditions21. Many reports on target gene expression analysis have used a single housekeeping gene for RT-qPCR normalization, such as 18S rRNA, gapdh, β-act, β-tub or ef1-α22; these genes regulate basic cellular functions. Such an approach has a strong tradition and history of use since the introduction of reverse-transcription-based assays. However, since 2009, according to the MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments21, no single reference gene should be used to quantify target gene expression under different conditions, and candidates for reference genes should be carefully selected for each study to comply with these guidelines. Numerous studies have shown that the expression of the above housekeeping genes can vary between individual tissues or experimental conditions such that no universal reference gene can be used in all situations18,23,24,25,26,27. Moreover, a reference gene with stable expression in one organism may not be suitable for normalization of gene expression in another organism, even a closely related organism. Additionally, genes such as 18S rRNA and 28S rRNA, despite their stability, are often expressed at very high levels and thus should not be used as internal controls28,29. To avoid biased normalization, following the MIQE guidelines21, the use of multiple candidate reference genes is highly recommended to obtain reliable RT-qPCR results. Based on the limited literature reports regarding the analysis of gene expression in dermatophytes10,11, supplemented with information on reference genes used in studies on other eukaryotes9,12,13, 12 genes, including adp-rf (ADP ribosylation factor), β-act (β-actin), β-tub (β-tubulin), ef1-α (elongation factor 1-alpha), gapdh (glyceraldehyde 3-phosphate dehydrogenase), mbp1 (multiubiquitin chain-binding protein 1), fis1 (mitochondrial fission 1 protein), psm1 (mitotic cohesion complex subunit Psm1), rGTPa (rho GTPase activating-protein 5), rpl2 (ribosomal protein L2), sdha (succinate dehydrogenase complex flavoprotein subunit A) and ubc (ubiquitin) (Table 1 and Supplementary Table S2), were selected as putative candidates based on a BLAST search of available T. interdigitale genomic sequences. Despite several attempts at primer modification, only 8 candidate amplification products were obtained, and these genes were used in further studies. Unfortunately, only three genomes of this species are currently available in the databases, of which two are at a scaffold level, while the third is at a contig level. An in-depth analysis of these genomes revealed the presence of regions of predicted sequences, indicating that the full nucleotide sequences have yet to be established (https://www.ncbi.nlm.nih.gov/genome/genomes/44693). It can therefore be assumed that both these problems and others related to the difficulties in correctly determining the taxonomic affiliation of many strains of T. interdigitale, as described in the literature30 (which may affect the correct genome assembly of this species), were responsible for the unsuccessful attempts to develop a larger number of correct primers, making it impossible to test a greater number of putative reference genes.
In this study, the geNorm, NormFinder, BestKeeper and RefFinder algorithms were used to evaluate the selected candidate reference genes as internal controls for analysis of target gene expression in T. interdigitale growing under different environmental stimuli, such as supplementation with various carbon sources, low Pi, and different pH values10. Some of these circumstances have been suggested to promote adhesion to the host tissue and are essential for the expression of specific genes associated with adaptation and interactions between T. interdigitale and its host. Our study is the first report on the identification and validation of reference genes for T. interdigitale that indicates psm1 and adp-rf as the most stable genes among the analysed candidates (Fig. 2).
Mitotic cohesion complex ATPase subunit (psm1) is involved in mitotic cohesion loading/unloading and is required for the cohesion of sister chromatids after DNA replication. In addition, ADP-ribosylation factor (adp-rf) is a ubiquitous GTP-binding protein essential for mitotic growth. These two candidates were found in the present study to be reliable internal controls for accurate expression level analysis of target genes of T. interdigitale growing under adhesion-inducing conditions.
Llanos et al.31 showed that according to geNorm analysis, psm1, ubcB (ubiquitin carrier protein) and sac7 (Rho GTPase activator) were ranked as the most stably expressed reference genes in the fungus Talaromyces versatilis grown under various conditions, such as in the presence of different carbon sources; under different temperatures and pH levels; and under salt stress and carbon/nitrogen starvation. However, in our previous report on the validation of reference genes for the dermatophyte Microsporum canis11, psm1 was classified in the group of unstable reference genes. To date, there have been only two studies on the validation of psm1 as a stably expressed reference gene in RT-qPCR analysis.
The ADP ribosylation factor gene (adp-rf) was found to be the best reference gene for analyses of target transcript levels in an exotic invasive insect, Leptinotarsa decemlineata32; the melon Cucumis melo L.33; the cereal wheats Triticum spp.12, the Pacific oyster, Crassostrea gigas34; the monkey Macaca fascicularis35; and the desert willow shrub, Salix psammophila36. Again, our previous study11 demonstrated that the expression stability of adp-rf was low in M. canis. Furthermore, the β-act gene, which encodes a cytoskeletal protein involved in many cellular processes, and the gapdh gene, which encodes an enzyme of the glycolytic pathway, are often used as reliable reference genes in expression analyses37; however, these genes were in the group of the least stable reference genes in the present study. On the other hand, in the search for reliable reference genes for RT-qPCR analysis of target gene expression in M. canis, the β-act gene was classified as one of the three most stable genes11. The present and the previous results11 of our team confirmed that the stability of housekeeping gene expression should be verified for each condition and each particular species (Table 3, Fig. 4), which again highlights the fact that there is no ideal and universal internal control gene for RT-qPCR analysis.
To validate the reference genes selected by the four algorithms, the genes were used as reference genes for the measurement of the relative expression of Tri m4, a gene that encodes aminopeptidase and is known to be upregulated in the presence of keratin and elastin as inducers19. Elevated Tri m4 expression was detected in T. interdigitale growing under inducing conditions only when the two internal controls psm1 and adp-rf, which were selected as the most stable internal controls by the four algorithms, were used in combination (Fig. 6). Upregulation was detected neither for the least stable pair of reference genes (set B) nor for the whole set of eight candidate genes (set C). These results clearly confirmed that the chosen best pair of internal control genes can be preferentially used for RT-qPCR normalization in the case of T. interdigitale grown under the described experimental conditions.
Conclusion
The present study was the first attempt to identify and validate T. interdigitale internal control genes. The psm1 and adp-rf genes were found to be the most stable reference genes appropriate for gene expression analysis in T. interdigitale. The use of these genes as internal controls may further improve the robustness of RT-qPCR for T. interdigitale grown under adhesion-inducing conditions.
Materials and Methods
Reference gene selection and primer design
Twelve putative candidate reference genes (adp-rf, β-act, β-tub, ef1-α, fis1, gapdh, mbp1, psm1, sdha, rpl2, rGTPa and ubc) (Table 1 and Supplementary Table S2) were chosen in the present study based on the NCBI database (http://www.ncbi.nlm.nih.gov) and our previous study11. Primers were designed and theoretically evaluated using Primer 3 software38. PCR products within the 80–150 bp range were obtained only in the case of 8 candidates (adp-rf, β-act, ef1-α, gapdh, psm1, sdha, rpl2 and ubc), and these genes were analysed by PCR in a Gradient Thermal Cycler T1000 (BioRad) (Table 1, Supplementary Fig. S1B).
Fungal material and growth conditions
The Trichophyton interdigitale 45/10 strain, isolated from tinea pedis of a 42-year-old man, was used in all RT-qPCR analyses. Trichophyton interdigitale CBS 124408 (a reference strain from the CBS-KNAW Collection, Utrecht, The Netherlands) and Trichophyton interdigitale 12/2010, a clinical isolate from the onychomycosis case of a 61-year-old man, were used in the evaluation step of the reference genes. The clinical strains were chosen from the collection maintained in the Department of Microbial Genetics, Faculty of Biology and Environmental Protection, University of Łódź, Poland. PCR-RFLP analysis of the ITS1-5.8S-ITS2 region followed by sequencing was performed for standard mycological identification39. Germinated conidia of the T. interdigitale strain (approximately 107 cells/ml)40 were incubated separately in minimal liquid medium (MM-Cove)41 under 7 different conditions (Table 2), in low-Pi MM, and in YEM (yeast extract medium) under 3 different conditions42 (Table 2).
RNA extraction, cDNA synthesis and quantitative reverse transcription PCR
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. RNA integrity was verified by electrophoretic and spectrophotometric (NanoPhotometerPearl Version 1.0, IMPLEN) analyses, according to the MIQE guidelines21 for RT-qPCR. RNA samples with A260/A280 ratios between 1.9 and 2.1 were used for further analysis. First-strand cDNA was synthesized using 2 μg of total RNA (DNA-free), RevertAid Transcriptase (Thermo Scientific) and random hexamer primers (Thermo Scientific) following the manufacturer’s protocol. The qRT-PCR reactions were conducted on a RotorGene Q System (Qiagen) based on a method described previously11 using SsoAdvanced Universal SYBR® Green Supermix (2X) (Bio-Rad). The reactions were subjected to an initial step of 95 °C for 1 min followed by 40 cycles at 95 °C for 20 s, 60.5 °C for 20 s, at 72 °C for 15 s. Melting curve analysis was performed by heating the amplicon from 72 °C to 95 °C.
Data analysis
The expression stability of the candidate reference genes in T. interdigitale was analysed using four bioinformatic tools: geNorm16, NormFinder17, BestKeeper18 and RefFinder (http://leonxie.esy.es/RefFinder/). The geNorm tool was used to calculate the gene expression stability according to the M-value, which is defined as the average pairwise variation with all other tested candidate reference genes. The algorithm recommends selecting genes with M-values below 1.0 to ensure the choice of the most stably expressed internal control genes11. Moreover, Vandesompele et al.16 suggested that an M-value lower than 0.5 indicates very good stability of expression. GeNorm also suggests that the best combination of reference genes based on pairwise variations (Vn/Vn+1) between two sequential normalization factors (NFn and NFn+1) has a V-value < 0.15. NormFinder is a VBA tool for Microsoft Excel used to calculate stability values (SVs) by combining intra- and inter-group variations in reference gene expression17. Lower SV values correspond to lower variations and, hence, higher stability of the reference genes. BestKeeper ranks the candidate reference genes according to their correlation coefficients (r values) for correlation with the BestKeeper Index (BI), which is the geometric mean of the Ct values of the candidate reference genes determined by calculating the standard deviation (SD) and coefficient of variance (CV)18. The online tool RefFinder was used to measure the geometric mean of the attributed weights for the overall final ranking. Box-and-whisker plots were drawn and one-way ANOVA was performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, California, USA).
References
Summerbell, R. Trichophyton, Microsporum, Epidermophyton, and Agents of Superficial Mycoses. In Manual of Clinical Microbiology (eds Versalovic, J., Carroll, K., Funke, G., Jorgensen, J. & Landry, M. W. D.) 1919–1942 (ASM Press, Washington, DC, 2011).
Szepietowski, J. C., Reich, A., Garlowska, E., Kulig, M. & Baran, E. Factors Influencing Coexistence of Toenail Onychomycosis With Tinea Pedis and Other Dermatomycoses A Survey of 2761 Patients. Arch Dermatol 142, 1279–1284 (2006).
Ilkit, M., Tanir, F., Hazar, S., Gümüşay, T. & Akbaba, M. Epidemiology of tinea pedis and toenail tinea unguium in worshippers in the mosques in Adana, Turkey. J. Dermatol. 32, 698–704 (2005).
Perea, S. et al. Prevalence and risk factors of tinea unguium and tinea pedis in the general population in Spain. J. Clin. Microbiol. 38, 3226–3230 (2000).
Triviño-Duran, L. et al. Prevalence of tinea capitis and tinea pedis in Barcelona Schoolchildren. Pediatr. Infect. Dis. J. 24, 137–141 (2005).
Drakensjö, I. T. & Chryssanthou, E. Epidemiology of dermatophyte infections in Stockholm, Sweden: A retrospective study from 2005-2009. Med. Mycol. 49, 484–488 (2011).
Ilkit, M. & Durdu, M. Tinea pedis: The etiology and global epidemiology of a common fungal infection. Critical Reviews in Microbiology 41, 374–388 (2015).
Leng, W., Liu, T., Wang, J., Li, R. & Jin, Q. Expression dynamics of secreted protease genes in Trichophyton rubrum induced by key host’s proteinaceous components. Med. Mycol. 47, 759–765 (2009).
Peres, N. Ta et al. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. . BMC Microbiol. 10, 39 (2010).
Jacob, T. R. et al. Rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 50, 368–377 (2012).
Ciesielska, A. & Stączek, P. Selection and validation of reference genes for qRT-PCR analysis of gene expression in Microsporum canis growing under different adhesion-inducing conditions. Sci. Rep. 8, 1197 (2018).
Giménez, M. J., Pistón, F. & Atienza, S. G. Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233, 163–173 (2011).
Dheda, K. et al. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37, 112–119 (2004).
Rocha, D. J. P., Santos, C. S. & Pacheco, L. G. C. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. https://doi.org/10.1007/s10482-015-0524-1 (2015).
Carvalho, D. M. et al. Reference genes for RT-qPCR studies in Corynebacterium pseudotuberculosis identified through analysis of RNA-seq data. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbio. https://doi.org/10.1007/s10482-014-0231-3 (2014).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 34–1 (2002).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Kaufman, G., Berdicevsky, I., Woodfolk, J. A. & Horwitz, B. A. Markers for host-induced gene expression in Trichophyton dermatophytosis. Infect. Immun. 73, 6584–6590 (2005).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative. Methods 25, 402–8 (2001).
Bustin, S. A. et al. The MIQE guidelines:Minimum Information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Huggett, J., Dheda, K., Bustin, S. & Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes and Immunity 6, 279–284 (2005).
Thellin, O. et al. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 75, 291–295 (1999).
Czechowski, T. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. PLANT Physiol, https://doi.org/10.1104/pp.105.063743 (2005).
Jain, M., Nijhawan, A., Tyagi, A. K. & Khurana, J. P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun, https://doi.org/10.1016/j.bbrc.2006.04.140 (2006).
Huis, R., Hawkins, S. & Neutelings, G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol, https://doi.org/10.1186/1471-2229-10-71 (2010).
Manoli, A., Sturaro, A., Trevisan, S., Quaggiotti, S. & Nonis, A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol, https://doi.org/10.1016/j.jplph.2012.01.019 (2012).
Tao, Y. et al. Identification of novel and robust internal control genes from Volvariella volvacea that are suitable for RT-qPCR in filamentous fungi. Sci. Rep, https://doi.org/10.1038/srep29236 (2016).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. Journal of Applied Genetics 54, 391–406 (2013).
Zhan, P. et al. Phylogeny of dermatophytes with genomic character evaluation of clinically distinct Trichophyton rubrum and T. violaceum. Stud. Mycol, https://doi.org/10.1016/j.simyco.2018.02.004 (2018).
Llanos, A., François, J. M. & Parrou, J. L. Tracking the best reference genes for RT-qPCR data normalization in filamentous fungi. BMC Genomics 16 (2015).
Shi, X.-Q. et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 6, 93 (2013).
Kong, Q. et al. Assessment of Suitable Reference Genes for Quantitative Gene Expression Studies in Melon Fruits. Front. Plant Sci. 7, 1–10 (2016).
Huan, P., Wang, H. & Liu, B. Assessment of housekeeping genes as internal references in quantitative expression analysis during early development of oyster. Genes Genet. Syst. 91, 257–265 (2016).
Park, S. J. et al. Selection of new appropriate reference genes for RT-qPCR analysis via transcriptome sequencing of cynomolgus monkeys (Macaca fascicularis). PLoS One 8, e60758 (2013).
Li, J. et al. Selection of Reliable Reference Genes for Gene Expression Analysis under Abiotic Stresses in the Desert Biomass Willow, Salix psammophila. Front. Plant Sci. 7 (2016).
de Jonge, H. et al. Evidence based selection of housekeeping genes. PLoS One 2 (2007).
Koressaar, T. & Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291 (2007).
Dobrowolska, A., Sta̧czek, P., Kaszuba, A. & Kozłowska, M. PCR-RFLP analysis of the dermatophytes isolated from patients in Central Poland [3]. Journal of Dermatological Science 42, 71–74 (2006).
Dobrowolska, A. & Staczek, P. Development of transformation system for Trichophyton rubrum by electroporation of germinated conidia. Current Genetics 55, 537–542 (2009).
Cove, D. J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta - Enzymol. Biol. Oxid. 113, 51–56 (1966).
Trevisan, G. L. et al. Transcription of Aspergillus nidulans pacC is modulated by alternative RNA splicing of palB. FEBS Lett. 585, 3442–3445 (2011).
Acknowledgements
This work was supported by grant 2014/13/B/NZ7/02307 and by grant 2016/23/D/NZ7/03964 from the National Science Centre, Poland. The experiments were partly performed using equipment from the Laboratory of Microscopic Imaging and Specialized Biological Techniques, Faculty of Biology and Environmental Protection, University of Łódź, Poland. We thank Dr. Marek Gadzalski for help with candidate reference gene selection and primer design.
Author information
Authors and Affiliations
Contributions
A.C. conceived and conducted the experiments, analysed the data, interpreted the results, and wrote the manuscript. B.O. conducted the experiments. P.S. conceived and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ciesielska, A., Oleksak, B. & Stączek, P. Reference genes for accurate evaluation of expression levels in Trichophyton interdigitale grown under different carbon sources, pH levels and phosphate levels. Sci Rep 9, 5566 (2019). https://doi.org/10.1038/s41598-019-42065-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-42065-5
This article is cited by
-
Dispersal of Aphanoascus keratinophilus by the rook Corvus frugilegus during breeding in East Poland
Scientific Reports (2022)
-
Metabolomic analysis of Trichophyton rubrum and Microsporum canis during keratin degradation
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.