Abstract
Lytic bacteriophages are valuable therapeutic agents against bacterial infections. There is continual effort to obtain new phages to increase the effectivity of phage preparations against emerging phage-resistant strains. Here we described the genomic diversity of spontaneous host-range mutants of kayvirus 812. Five mutant phages were isolated as rare plaques on phage-resistant Staphylococcus aureus strains. The host range of phage 812-derived mutants was 42% higher than the wild type, determined on a set of 186 methicillin-resistant S. aureus strains representing the globally circulating human and livestock-associated clones. Comparative genomics revealed that single-nucleotide polymorphisms from the parental phage 812 population were fixed in next-step mutants, mostly in genes for tail and baseplate components, and the acquired point mutations led to diverse receptor binding proteins in the phage mutants. Numerous genome changes associated with rearrangements between direct repeat motifs or intron loss were found. Alterations occurred in host-takeover and terminal genomic regions or the endolysin gene of mutants that exhibited the highest lytic activity, which implied various mechanisms of overcoming bacterial resistance. The genomic data revealed that Kayvirus spontaneous mutants are free from undesirable genes and their lytic properties proved their suitability for rapidly updating phage therapeutics.
Similar content being viewed by others
Introduction
Phage therapy is an alternative to antibiotics used against Staphylococcus aureus infections, especially for the treatment of chronic infections caused by multidrug-resistant strains. Virulent bacteriophages of the genus Kayvirus from the family Myoviridae are the most promising due to their high lytic activity, therefore some of them are now commercialized1,2,3,4. The emergence of phage-resistant bacterial strains requires innovation in phage preparations, which is usually achieved by isolating new phages from the environment5,6,7 or time-consuming procedures of phage adaptation such as phage training8. Upgraded preparations necessitate the safety assessments required by pharmaceutical authorities9,10. Phages adapted to different hosts exhibit distinct phenotypes and their specialization can be manifested as both improved infection of the host used for evolution and decreased infection of the other host types11. Phage and host co-evolution, driven by the resistance of bacterial hosts, results in intraspecies heterogeneity in phage genomes via emerging recombination events and single-nucleotide polymorphisms (SNPs) whose frequency and distribution vary depending on the phage and growth conditions12,13. The genes for host recognition, attachment, and infection are often affected by SNPs or major re-arrangements11,14,15. The fixation of these genome changes by natural selection enables the phages to recognize new hosts16 and avoid anti-phage systems17 such as the type II R-M system Sau3AI in kayviruses18.
Due to their polyvalence, kayviruses are valuable agents which can be used to treat infections caused by a broad range of S. aureus and even non-S. aureus staphylococcal species1,3,7. Bacteriophage K is the best studied member of the genus Kayvirus18,19. The unit genomes of kayviruses are about 140 kb in size with more than 200 genes, as was reviewed by Łobocka et al.20. Early expressed host-takeover genes are located in identical left and right long terminal repeats (L-LTR and R-LTR, respectively), in which R-LTR forms genomic redundancy21,22,23. The majority of Kayvirus genomes contain group I introns, often encoding a homing endonuclease, which are located in the genes for DNA metabolism, morphogenesis and lysis24,25,26.
In our previous studies, we isolated almost twenty mutants of kayvirus 812, which, when used in cocktails, extended the amount of susceptible S. aureus strains of different origin1,27,28. Phage 812 was later studied at the proteomic and structural level29,30, however detailed genomic characterization has not been performed yet. Here, spontaneous mutants of phage 812 exhibiting a broad host range on recently circulating methicillin-resistant Staphylococcus aureus (MRSA) strains were isolated. With respect to interest in using the mutants in phage therapy, we focused on deep sequence analysis of their genomes, characterization of genomic variations, and exclusion of potential risks for phage therapy as required by pharmaceuticals authorities31.
Results
The host range of phage 812 and its mutants
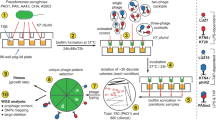
Mutants of phage 812 exhibiting a broader host range were isolated as rare plaques (with frequencies 10−6–10−9) on various S. aureus strains resistant to the parental phage (Fig. 1). The mutant phage 812a was a starting phage for several subsequent multi-step mutants isolated on different strains (Fig. 1). As the phage 812a-derived mutants did not exhibit a satisfactory lytic effect on livestock-associated MRSA (LA-MRSA) strains (Supplementary Table S1), a new lineage of mutants represented by phage 812h1 was selected from wild-type phage 812 on phage-resistant LA-MRSA strains (Fig. 1).
Diagram showing the isolation procedure of spontaneous host-range mutants of phage 812 represented by boxes. All phage mutants were isolated from a single plaque on selection S. aureus strains shown next to the corresponding mutants. Phage mutants were then propagated on S. aureus strains that are colour coded according to the phage mutant. The sequence type (ST) and the staphylococcal protein A (spa)-type of each strain are given in parentheses.
To evaluate the efficiency of the phage mutants, their lytic ability was tested on a set of well-characterized MRSA isolates (n = 186) and compared with type phage K and kayviruses isolated from commercial therapeutic preparations Stafal (Bohemia Pharmaceuticals, Czech Republic), PyoPhage (Eliava Biopreparations, Georgia) and StaphPhage (Microgen, Russia). The tested S. aureus strains belonged to 18 different sequence types (ST) and 45 staphylococcal protein A gene (spa)-types chosen to represent the major global MRSA clones (Supplementary Table S1). The susceptibility to phages varied among the strains of the same ST and even of the same spa-type. Of the tested strains, 59.1% (n = 110) were susceptible to all the tested bacteriophages. In contrast, 4.8% (n = 9) of isolates, mostly of ST45, were completely resistant to all the tested phages (Fig. 2, Supplementary Table S1). Overall, the number of tested strains lysed by wild-type phage 812 or at least one of its mutants (n = 177) was higher than phages from commercial preparations Stafal (Bohemia Pharmaceuticals, Czech Republic), PyoPhage (Eliava Biopreparations, Georgia), and StaphPhage (Microgen, Russia) (n = 165) (Supplementary Table S1). The mutant 812h1 exhibited the broadest host range, lysing almost 90% (n = 166) of the strains in total (Fig. 2). Nevertheless, 20 strains remained resistant to this mutant, but 11 of them were susceptible to some other phage 812-derived mutant, mostly to 812K1/420 (Supplementary Table S1).
Lytic activity of phages K, 812 and its mutants, and kayviruses from commercial preparations Stafal (Bohemia Pharmaceuticals, Czech Republic), PyoPhage (Eliava Biopreparations, Georgia) and StaphPhage (Microgen, Russia) determined on a set of 186 methicillin-resistant S. aureus strains. The number of strains susceptible or resistant to a given phage is shown in each column. The susceptibility patterns of strains are listed in Supplementary Table S1. Susceptibility of strains to at least one of the 812-derived mutants and at least one of the commercial preparations is also shown.
Genomic analysis of phage 812 and its mutants
The phage 812 unit genome, 141,906 bp in size with a GC content of 30.4%, starts with a 8,484-bp left long terminal repeat (L-LTR) and ends with a tandem repeat with a 35–bp-long repeat unit sequence TATTAYTACTACTAAGTACCTTTGTTATGTACTAC (Figs 3 and 4). The copy number of the repeat unit is variable, ranging from four to more than twenty complete copies in the phage 812 population and down to two copies in the mutant 812K1/420 (Supplementary Fig. S1). The right LTR (R-LTR), identical to L-LTR, comes after the unit genome and forms the terminal redundancy (Fig. 4). In total, 218 coding sequences (CDS), 63 putative promoters, and 30 putative intrinsic terminators were predicted in the unit genome (Figs 3 and 5). Three genes for tRNA (tRNA-Met-CAT, tRNA-Phe-GAA, and tRNA-Asp-GTC) and one gene for pseudo tRNA with an unknown function were predicted in the phage 812 genome. At least five introns were detected, of which three encode putative endonucleases, as in phage K23,32. The genomes of phage 812 and phage K exhibit 98.5% identity; although phage 812 contains two additional non-coding introns in the tail tube protein gene and the adjacent gene for putative endonuclease, which were also localized in phage 812h1 (Fig. 3) and in four previously described kayviruses, i.e. A3R, 676Z, Fi200W, and P4W20.
Phage 812 unit genome (GenBank accession number MH844528) and its comparison with phage K (NC_005880), phage mutant 812h1 (MH844529), phage mutant 812a (KJ206560), and 812a-derived mutant 812K1/420 (KJ206563). The major differences are labeled by red numbers: (1) host-takeover region showing low similarity between 812 and 812h1; (2) orf23 and orf24 of 812h1 that were not found in phage 812; (3) orf73 encoding endolysin (LysK) that contains a partial deletion in 812a-derived mutant 812K1/420; (4) orf103 for a tail tube protein that does not contain non-coding introns in 812a-derived mutants, and orf104 for a putative intron-encoded nuclease which is missing in phage K and 812a-derived mutants; (5) orf119 for tail fibre complex that contains a partial deletion in 812K1/420; (6) orf173 for putative membrane-associated protein which is split into two open reading frames in 812h1 due to a partial deletion; (7) orf190 and orf191 deleted in all 812a-derived mutants, which led to fusion of orf189 for regulatory protein and orf192 for putative membrane protein; and (8) terminal part of the genome which is highly different from 812h1. Mutations and fixed polymorphisms with impact on protein sequences in compared phages are shown. The genome comparison is based on tblastx with an identity cut-off of 95%.
Coverage differences of genomic regions achieved by mapping of sequencing reads to unit genomes of phages 812 and 812h1. Long terminal repeats (LTRs) were delimited according to excessively increased coverage at the left end of the genomes due to the alignment of reads of both the LTRs. The increased coverage at the right end of the phage 812 genome is caused by a variable number of 35-bp tandem repeats. The genome annotations are illustrated on three feature lines: the top line shows coding sequences; the middle line indicates the location of LTR, tRNA genes, introns, and the direct repeats (DR) serving as recombination sites in the endolysin gene of phages 812F1 and 812K1/420, and 2-kb deletion/insertion polymorphism in phage 812h1; and the bottom lines show the positions of all polymorphisms.
Sequence logos of phage 812 promoters. (A) Bacterial promoters in long terminal repeats contain typical −35 and −10 sequences. (B) Putative early promoters recognised by host sigma factor. (C) Putative late promoters recognised by phage sigma factor. The late promoters drive the transcription of genes for homing endonuclease in long terminal repeat and in the polymerase gene intron, and genes for packaging, the structure of the head, tail and baseplate, and lysis. No conserved −35 sequence was found in phage late promoters.
In total, 176 variations were identified in the sequencing data of the wild-type phage 812 genome (Supplementary Table S2): 171 single-nucleotide polymorphisms (SNPs) and 5 deletion/insertion polymorphisms (DIPs), often localized in host range-associated genes (Fig. 4, Supplementary Table S2). A lot of minor SNPs and DIPs with a frequency in the genome < 50% of wild-type phage 812 were found to predominate in the mutant 812a and all its derivatives 812F1, 812K1, and 812K1/420 (Table 1). Such polymorphisms are called “fixed” in the text below and some of them affected encoded proteins (Fig. 3).
In addition, a few gene product alterations specific for each of the multi-step mutants were found. Phage 812K1 differs from the others by an 11-aa intrinsic deletion in an encoded ligase and 78-aa deletion along with the substitution of 4 aa in an encoded primase. Both the phages 812F1 and 812K1/420 encode truncated endolysin (a 211-aa deletion) due to a 1509-bp deletion in the endolysin gene as reported previously26. The gene for the tail fibre protein complex (orf119) of the phage 812K1/420 contains a premature stop codon due to a 77-bp deletion that divides this gene into 2 ORFs. Another premature stop codon caused by a nonsense mutation divides the gene for ribonuclease H of phage 812K1/420 into two ORFs without changing the reading frame.
Genomic analysis of phage 812h1
The phage 812h1 was selected in two steps using two distinct strains (Fig. 1). The unit genome of phage 812h1 is 142,150 bp long with an 8,432-bp-long L-LTR. The unit genome completely lacks the tandem repeat locus typical for phage 812, 812a and its subsequent mutants, as well as for phage K (Fig. 4 and Supplementary Fig. S1). In total, 219 CDS, 62 putative promoters, 31 putative intrinsic terminators, 3 tRNA genes and 1 pseudo tRNA gene were predicted in the unit genome. 255 SNPs and 5 DIPs were identified mostly in the genes for structural proteins and replication, as in phage 812 (Supplementary Tables S2 and S3). On the other hand, many SNPs of phage 812h1 are located in the genes for terminal repeat-encoded proteins (Fig. 4, Supplementary Table S3). One of the most apparent variabilities is the duplication of the gene for terminal repeat-encoded protein (orf7) located in the host-takeover region. The orf7 duplication is bordered by the nucleotide motif TAGGAT, which is present in more than 90 copies in all Kayvirus genomes. The next apparent DIP is for a 2-kb-long sequence including orf195 – orf200 of unknown function, bordered by a unique direct repeat GGAGGTTAATTWATGAA (Fig. 4). The phage 812h1 genome differed from wild-type phage 812 and all mutants derived from phage 812a in multiple loci (Fig. 3, Supplementary Table S4).
Two different types of genes encoding receptor binding proteins (RBPs) corresponding to phage 812 orf123 and orf125 determine the host range of kayviruses through sequence variability and by the adsorption mechanism that may vary even in closely related phages29,33,34. These genes exhibit a high variability among Kayvirus genomes (Fig. 6), which could help the phages to adapt to a new host strain. The phages 812 and 812h1 differ in both the RBPs (Supplementary Table S4) and by SNPs content in both RBP-encoding genes (Supplementary Tables S2 and S3). The 812a-derived mutants only fixed SNPs of the parental phage 812 in the orf123 homologue (Table 1).
Unrooted phylogenetic trees showing similarity of two Kayvirus receptor binding proteins homologous to (A) protein encoded by phage 812 orf123 and (B) protein encoded by phage 812 orf125. The phylogenetic trees were reconstructed using the maximum likelihood method (aLRT test) performed on the Phylogeny.fr platform. GenBank accession numbers of the compared phage genomes are listed next to the trees. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site.
Safety assessment
Because of the extensive presence of variable genetic elements and virulence or resistance genes in the genome of S. aureus strains used for the selection and propagation of the phage 812-derived mutants, it was necessary to check the phage genomes for the possible acquisition of genes with potential safety concerns. None of the isolated phage mutants carried genes for antibiotic resistance, bacterial virulence factors, or mobile elements-derived sequences. Similarly, no acquisition of the genes for lysogeny was found in the analysed phage genomes.
To exclude the possibility of generalized transduction, the frequency of packaging of the genes mecA, tetK and blaZ (encoding resistance to methicillin, tetracycline, and penicillin, respectively) into phage particles was analysed by relative quantification (Supplementary Table S5). Kayviruses K, and 812K1/420 as a representative of 812-derived phages, and PyoPhage was propagated on well-characterized MRSA strains: the monolysogenic strain COL with plasmid pT181 (tetK) and double lysogenic strain 08/019 of a USA300 clone with penicillinase plasmid pUSA-Houmr-like (blaZ). The restriction-deficient prophage-less methicillin-susceptible S. aureus (MSSA) strain RN4220 (pT181 and pUSA-Houmr-like) was used to prove the absence of spontaneously induced prophages from the COL and USA300 strains. Compared to simultaneously tested transducing siphovirus 80α, the resistance gene packaging by the studied kayviruses was 1000-fold lower (Supplementary Table S5).
Discussion
As shown in this study, the isolation of host-range spontaneous mutants is another and effective way to obtain improved therapeutic phages against almost every S. aureus strain. The strains exhibited distinctive phage susceptibility patterns (Supplementary Table S1), which indicates the action of several anti-phage defence mechanisms. This indicates that some of the mechanisms are shared among the strains of different genotypes, as was demonstrated e.g. for strains susceptible only to phage mutants with a truncated endolysin gene26. Since there is no apparent association between the strain genotype and the phage susceptibility, it can be assumed that anti-phage defence mechanisms are mainly encoded by the accessory genome. To overcome various defence mechanisms, it seems advantageous to use phage cocktails or apply the phages sequentially during the phage therapy.
Compared to the parental phage, all the isolated mutants exhibited point mutations and/or larger genomic rearrangements, some of which were reported before27. Deep sequencing analysis of wild-type phage 812 and the most effective mutant 812h1 showed that numerous genomic polymorphisms emerged during the phage propagation. In agreement with the recent evolutionary study13, most of the SNPs accumulated in genes with an assumed effect on phage fitness.
Because of the different history of isolation and various genotypes of selection and propagation strains, the conditions for SNP accumulation in the analysed phages were different. The number of SNPs in both the wild-type phage 812 and recently isolated phage mutant 812h1 was higher than reported in Pseudomonas phages13. The high prevalence of polymorphisms indicates a high mutation rate that enables the diversification of the phage populations. In the genomes of 812a-derived phages obtained by 454-sequencing technology, a deep sequence analysis was not possible, so only the SNP/DIP fixation was evaluated. The procedure for isolating a phage mutant effective against to-date resistant strains were based on natural selection on the phage-resistant strain and the random picking up of a rare single plaque which may lead to genetic drift. Therefore, the understanding of evolutionary processes in natural Kayvirus populations requires further study, including the deep sequence analysis of isolates with a unified history.
Sequences of 812h1 distinct from the wild-type phage 812 were found in the genomes of related phages ISP, vB_SauM-fRuSau02, Fi200W, MSA6 or G12,3,20,32. Some of the sequences were also found in wild-type phage 812 sequencing data, but with very low coverage. This suggests that a large pool of phage sequences is maintained in minor genome variants persisting in Kayvirus populations.
Numerous direct repeat motifs found across the genomes of kayviruses enable genomic rearrangements, therefore in the phage progeny, the polymorphic loci can be detected in different combinations, which plays an important role in phage diversification. Propagation on strains with different genotypes can cause fixation of the particular polymorphisms beneficial for phage fitness, as was reported in streptococcal phages able to avoid host defence mechanisms17. Another repeat-associated change in phage 812-derived mutants is of the copy number of tandem repeats in the variable right terminus as a characteristic feature of Kayvirus genomes, which probably plays a significant role in replication and packaging35.
Compared to phage 812, host-range mutant 812h1 exhibited differences in both the genes for RBPs, while in the 812a-derived mutants they were only in one of them (orf123). As recently described for bacteriophage SA012, RBP encoded by a gene homologous to orf125 of phage 812 seems to play an essential role in binding onto the backbone of wall teichoic acids (WTA); the additional binding of RBP encoded by a gene homologous to orf123 of phage 812 to N-acetyl-α-D-glucosamine (α-GlcNAc) of WTA is required for the effective infection of some phages34. In kayviruses, the homologs of the α-GlcNAc-binding RBPs exhibit higher variability than WTA-backbone-binding RBPs, which indicates that they determine the host specificity.
Phages suitable for therapy should not contain genes for the lysogenic life cycle, such as genes for integrases and those for virulence or drug resistance factors, which could be horizontally transferred into the genome of the bacterial host8,9,31. A recent comparison of the complete genomes of myoviruses, including 14 kayviruses, proved that they are all strictly lytic and do not carry antibiotic resistance or bacterial virulence genes36. Except for the non-spontaneous T4 mutants37,38, the potential of bacterial gene transduction has not been reported for the strictly virulent phages, some of which even break down the bacterial DNA to generate the building blocks required for the synthesis of progeny DNA39. The genomes of these phages feature specific gene products involved in nucleotide metabolism, such as ribonucleotide reductase subunits. It has been shown that the genes for Nrd are host-like auxiliary metabolic genes promoting phage replication40. Importantly, the above safety requirements hold for phage 812 and all its host-range mutants described in this paper. Therefore, a shorter assessment time frame, similar to that of the influenza vaccine, should be considered with regard to the data which indicate that bacteriophage strain replacement or addition has no impact on the safety of the preparation10.
The results of our study help to clarify the molecular basis of differences in phage lytic properties. These findings also contribute to a better understanding of Kayvirus genomics and to establishing the rules for updating phage preparations. During commercial phage production, confirmation of the phage identity including its genome consistency is required from the master seed lots to the final products9. Since the genomic sequences of host-range phage mutants are almost identical, sequencing PCR amplicons of the most variable regions could distinguish the phages in commercial products from different companies. Alternatively, phage mutants can be distinguished using a multiplex PCR assay designed to target sequences specific for individual phage mutants (Supplementary Fig. S2). Nevertheless, it is necessary to determine the level of acceptable genome variability between the phage master seed lots and the final products. Our results show that the emergence of genomic alterations in therapeutic phages is a naturally occurring process beneficial for rapidly gaining new host-range mutants. Importantly, we showed that the phage mutants have no undesirable gene acquisitions with impact on the safety of phage therapeutics.
Materials and Methods
Bacterial strains
186 MRSA strains of various origin (Supplementary Table 1) were obtained from the National Reference Laboratory for Antibiotics and National Reference Laboratory for Staphylococci (National Institute of Public Health, Prague, Czech Republic), University Hospital Brno (Czech Republic), and farm livestock and the environment from different locations in the Czech Republic, deposited in the collection of the Bacteriology Department (Veterinary Research Institute, Brno, Czech Republic). Propagating S. aureus strains CCM 4028 and CCM 8428 were provided by the Czech Collection of Microorganisms (Masaryk University, Brno, Czech Republic). Strain RN4220 was obtained from Prof. F. Götz (University of Tübingen, Germany). Strain COL was kindly provided by Prof. Hermínia de Lencastre (Universidade Nova de Lisboa, Portugal and The Rockefeller University, New York, NY). Other propagating and selection strains were isolated in this or earlier studies1 and are genotypically characterized below. Propagation strain CAPM 6630 for phage 812h1 was deposited in the Collection of Animal Pathogenic Microorganisms (Veterinary Research Institute, Brno, Czech Republic).
Genotyping of S. aureus strains
All strains were genotypically characterized using multi-locus sequence typing (MLST) according to Enright et al.41 and spa-typing according to the Ridom StaphType standard protocol (www.ridom.org). The software package Ridom StaphType v2.2.1 (Ridom, Germany) was used for spa-type assignment. Clonal complex analysis was performed with eBURST v342. All strains were tested for the mecA gene as described previously43. The prophage content in propagating and selection strains was investigated by multiplex PCR assay44.
Phages, selection and propagation
Bacteriophage K was obtained from Dr. Christiane Wolz (University of Tübingen, Germany). Phage 812 was described previously1. Phages designated Stafal, Pyophage, and StaphPhage were isolated from the commercial therapeutic preparations Stafal (Bohemia Pharmaceuticals, Czech Republic), PyoPhage (Eliava Biopreparations, Georgia), and Staphylococal bacteriophage liquid (Microgen, Russia), respectively, and propagated on strain CCM 8428 as described previously45. Phages 812 and 812h1 were deposited in the Czech Collection of Microorganisms under accession number CCM 7911 and in the Collection of Animal Pathogenic Microorganisms under accession number CAPM V-689, respectively. Phage 812K1/420 was patented for treating human MRSA infections46.
Phage 812 was propagated using stock lysate. After the seeding of the parental phage with a strain resistant to this phage (selection strain) into 0.7% meat-peptone soft agar overlaid on 1.5% meat peptone agar (MPA) plates, individual rare plaques of the mutant phages were picked into 0.1 ml of a liquid medium, meat-peptone broth (MPB), prepared from 13 g of nutrient broth (Oxoid, CM0001), 3 g of yeast extract (Oxoid, LP0021), and 5 g of peptone (Oxoid, LP0037) dissolved in distilled water to a volume of 1000 ml (pH 7.4). Suspensions were purified through a 0.45 µm OlimPeak nylon filter (Teknokroma, Barcelona, Spain) to remove the remaining bacterial cells. Phage mutant 812a was derived from the wild-type phage 812 by selection on S. aureus strain NCTC 8511 (ST254, t118) laboratory lysogenized by phage ϕ5347. The mutant 812a served as a parental phage to produce the mutants 812K1 and 812F1 isolated on S. aureus strains FN34 (ST5, t002) and FN4 (ST1021, t11517), respectively1. Phage mutant 812K1/420 was derived from phage 812K1 by selection on human MRSA strain NRL Atb 420 (ST5, t179). Phage 812h1 was selected on MRSA strain 1098 (ST8, t064) from mutant 812h which was selected from phage 812 on MSSA strain 347 (ST5, t002).
Large-scale phage propagation on S. aureus strains was performed in 250 ml MPB with aeration. The phages 812, 812a, 812F1, and 812K1 were propagated on strain CCM 4028 (ST30, t021). Phage 812K1/420 was propagated on prophage-less strain CCM 8428 (ST30, t021). Phage 812h1 was propagated on prophage-less MSSA strain CAPM 6630 (ST6, t304). To prove the genetic stability of the obtained phage mutants and to exclude the possibility of phage DNA modification, the phage lytic activity was tested on the selection strain after passaging on the propagation strain, and the efficiency of plating (EOP) was determined. EOP was calculated by dividing the phage titer obtained on selection strains by the phage titer on the propagating strain1.
Phage susceptibility testing
S. aureus strains were grown in MPB to the logarithmic phase at 37 °C. Meat peptone agar plates (1,5% agar) with 2 mM CaCl2 were flooded with the broth culture, drained, and dried. The phage lysates of titer of 109 plaque forming units (PFU) per ml were diluted 10−2 (i.e. 107 PFU/ml) and 10−4 (i.e. 105 PFU/ml) and applied in triplicate for both the dilutions by spotting 10 µl aliquots onto soft agar lawns inoculated with tested S. aureus strain. Plates were incubated overnight at 37 °C. Then the results were evaluated (Supplementary Fig. S3). If single plaques appeared, the strain was considered susceptible (Supplementary Fig. S3A). If no plaques appeared in any dilution, the strain was considered phage-resistant (Supplementary Fig. S3B).
Phage genome sequencing and analysis
Phenol/chloroform extraction of DNA was performed according to Sambrook et al.48. The procedure was modified so that Phase Lock Gel tubes (5 PRIME, Hamburg, Germany) were used for separating the aqueous and organic phases. The complete nucleotide sequences (30 to 50-fold coverage) of bacteriophages 812a, 812K1, 812F1, and 812K1/420 were determined using the GS Junior system (Roche, 454 Life Science, USA) according to a previous study49. The genomes of phages 812 and 812h1 were determined using the MiSEQ system (Illumina, USA) according to Botka et al.50. The reads were assembled using the A5-miseq pipeline51. The genomes were covered, on average, 2272-fold in phage 812 and 3076-fold in phage 812h1. Sequence gaps were closed using PCR and primer walking. Amplified PCR products were purified using a QIAquick PCR Purification kit (Qiagen, Netherlands) and sequenced by Eurofins MWG Operon (Germany). The same approach was used for PCR assays to verify extensive DIPs of phage 812h1 and to analyse tandem repeat regions. Primers and polymerase chain reaction conditions for differentiation of the phage 812-derived mutants are described in Supplementary Table S6.
The GenBank (NCBI) accession numbers of the phage genomes are as follows: updated genome of phage 812 (MH844528), and genomes of phages 812a (KJ206560), 812K1 (KJ206561), 812F1 (KJ206562), 812K1/420 (KJ206563), and 812h1 (MH844529).
Variations in the genomes obtained with the MiSEQ system were detected using CLC Genomic Workbench v3.6.5 (Qiagen Bioinformatics, Denmark) by analysing the set of pair-end reads aligned to the genome sequence under the following conditions: coverage of reads ≥90% and identity ≥90%, ignoring non-specific matches. To exclude sequencing errors, only the SNPs and DIPs with at least 10% frequency and, at the same time, a minimum of 30-fold coverage in low-coverage parts were considered. The quality conditions were as follows: quality of central base and average quality of surrounding bases ≥Q30. DNA samples were verified by multiplex PCR assay (Supplementary Table S6) to exclude cross-contamination.
Taking into account that the redundant regions exhibit multiplied coverage compared to the rest of the genomes35, the long terminal repeats were predicted based on a significant change in the MiSEQ read number of the alignments. Open reading frames (ORFs) were identified using GeneMarkS, optimized for phage sequences (http://exon.gatech.edu) and inspected manually. Genomes were annotated using RAST52, Phobius (http://phobius.sbc.su.se), and BLAST (http://blast.ncbi.nlm.nih.gov/). The EMBOSS tools Needle and Stretcher (https://www.ebi.ac.uk/) were used for pairwise sequence comparisons. The tRNA genes were identified using tRNAscan-SE53 and RAST. To predict promoters specific for the host and phage sigma factors, 200-bp sequences upstream of start codons were investigated using GLAM2 v.5.0.2 from MEME Suite54 and BPROM55, with only the promoters that reached an overall score (LDF) above 5 being considered. Only one promoter per transcription unit was taken into account. ARNold56 was applied to Rho-independent terminator identification. Only the predicted terminators with a dG value of less than −7.0 kcal/mol and a functional position were taken into consideration. The genome maps were constructed using GCView57. The ARDB58 and VFDB59 databases were used to search for antibiotic resistance and virulence factor genes. Parameters for ARDB searching were as follows: blastp program, complete database of resistance genes, 40% identity, and an E-value of 0.0001. Parameters for VFDB searching were as follows: tblastn program, using whole phage sequences against DNA sequences from the VFDB datasets A and B. Phylogenetic trees were constructed using the maximum likelihood method (aLRT test) performed in the on-line tool Phylogeny.fr60. Genomic sequences of related phages ISP (FR852584), fRuSau02 (MF398190), JD007 (NC_019726), Staph1N (JX080300), A3R (JX080301), 676Z (JX080302), Fi200W (JX080303), P4W (JX080305), K (KF766114), SA012 (NC_023573), G1 (AY954969), IME-SA1 (KP687431), IME-SA2 (KP687432), GH15 (NC_019448), JA1 (MF405094), IPLA-RODI (NC_028765), S25-4 (NC_022918), SA3 (MF001365), SA5 (JX875065), Sb-1 (HQ163896), Team1 (NC_025417), A5W (EU418428), P108 (NC_025426), and qdsa002 (KY779849)2,3,4,6,20,23,32,33,34,61,62,63,64,65,66,67 were retrieved from the NCBI Nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide).
qPCR quantification of bacterial genes blaZ, tetK and mecA inside phage particles
Phages K, 812K1/420, PyoPhage and 80α were propagated on MRSA strains COL (pT181) and USA300 08/019 (pUSA-Houmr-like)68, and the strain RN4220 with plasmids pT181 and pUSA-Houmr-like prepared in this study. Extraction of phage DNA including DNase treatment was performed as described previously69. A LightCycler 480 Instrument II (Roche) was used for qPCR performed according to Mašlaňová et al.69. Reactions were carried out in triplicates in optical 96-well reaction plates. Each reaction mixture (25 μl) contained 12.5 μl LightCycler 480 SYBR Green I Master (Roche), 300 µM of each primer (Supplementary Table S5) and 25 ng of template DNA in a volume of 2.5 µl. Genes encoding distinctive siphoviral and myoviral structural proteins44 were used as a reference.
Data Availability
All data generated or analysed during this study are included in this published article (and its Supplementary files), or the relevant depository/collection/database is indicated in Materials and Methods. The large datasets (qPCR, raw sequence data) generated during and/or analysed during the current study are available from the corresponding author on request.
References
Pantůček, R. et al. The polyvalent staphylococcal phage phi 812: its host-range mutants and related phages. Virology 246, 241–252, https://doi.org/10.1006/viro.1998.9203 (1998).
Vandersteegen, K. et al. Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS One 6, e24418, https://doi.org/10.1371/journal.pone.0024418 (2011).
Leskinen, K. et al. Characterization of vB_SauM-fRuSau02, a Twort-like bacteriophage isolated from a therapeutic phage cocktail. Viruses 9, 258, https://doi.org/10.3390/v9090258 (2017).
Kvachadze, L. et al. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb Biotechnol 4, 643–650, https://doi.org/10.1111/j.1751-7915.2011.00259.x (2011).
Synnott, A. J. et al. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol 75, 4483–4490, https://doi.org/10.1128/AEM.02641-08 (2009).
Cui, Z. et al. Characterization and complete genome of the virulent Myoviridae phage JD007 active against a variety of Staphylococcus aureus isolates from different hospitals in Shanghai, China. Virol J 14, 26, https://doi.org/10.1186/s12985-017-0701-0 (2017).
Abatángelo, V. et al. Broad-range lytic bacteriophages that kill Staphylococcus aureus local field strains. PLoS One 12, e0181671, https://doi.org/10.1371/journal.pone.0181671 (2017).
Rohde, C. et al. Expert opinion on three phage therapy related topics: bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 10, 178, https://doi.org/10.3390/v10040178 (2018).
Pirnay, J.-P. et al. Quality and safety requirements for sustainable phage therapy products. Pharm Res 32, 2173–2179, https://doi.org/10.1007/s11095-014-1617-7 (2015).
Pelfrene, E., Willebrand, E., Cavaleiro Sanches, A., Sebris, Z. & Cavaleri, M. Bacteriophage therapy: a regulatory perspective. J Antimicrob Chemother 71, 2071–2074, https://doi.org/10.1093/jac/dkw083 (2016).
Enav, H., Kirzner, S., Lindell, D., Mandel-Gutfreund, Y. & Béjà, O. Adaptation to sub-optimal hosts is a driver of viral diversification in the ocean. Nat Commun 9, 4698, https://doi.org/10.1038/s41467-018-07164-3 (2018).
Brüssow, H. Population Genomics of Bacteriophages in Population Genomics: Microorganisms (eds Polz, M. F. & Rajora, O. P.) 297–334, https://doi.org/10.1007/13836_2018_16 (Springer, 2019).
Magill, D. J. et al. Localised genetic heterogeneity provides a novel mode of evolution in dsDNA phages. Sci Rep 7, 13731, https://doi.org/10.1038/s41598-017-14285-0 (2017).
Essoh, C. et al. Investigation of a large collection of Pseudomonas aeruginosa bacteriophages collected from a single environmental source in Abidjan, Côte d’Ivoire. PLoS One 10, e0130548, https://doi.org/10.1371/journal.pone.0130548 (2015).
Moreno Switt, A. I. et al. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genomics 14, 481, https://doi.org/10.1186/1471-2164-14-481 (2013).
Meyer, J. R. et al. Repeatability and contingency in the evolution of a key innovation in phage Lambda. Science 335, 428–432, https://doi.org/10.1126/science.1214449 (2012).
Paez-Espino, D. et al. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio 6, e00262–15, https://doi.org/10.1128/mBio.00262-15 (2015).
O’Flaherty, S. et al. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G + C content. J Bacteriol 186, 2862–2871, https://doi.org/10.1128/JB.186.9.2862-2871.2004 (2004).
O’Flaherty, S. et al. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl Environ Microbiol 71, 1836–1842, https://doi.org/10.1128/AEM.71.4.1836-1842.2005 (2005).
Łobocka, M. et al. Genomics of staphylococcal Twort-like phages-potential therapeutics of the post-antibiotic era. Adv Virus Res 83, 143–216, https://doi.org/10.1016/B978-0-12-394438-2.00005-0 (2012).
Stewart, C. R., Yip, T. K. S., Myles, B. & Laughlin, L. Roles of genes 38, 39, and 40 in shutoff of host biosyntheses during infection of Bacillus subtilis by bacteriophage SPO1. Virology 392, 271–274, https://doi.org/10.1016/j.virol.2009.06.046 (2009).
Stewart, C. R., Deery, W. J., Egan, E. S. K., Myles, B. & Petti, A. A. The product of SPO1 gene 56 inhibits host cell division during infection of Bacillus subtilis by bacteriophage SPO1. Virology 447, 249–253, https://doi.org/10.1016/j.virol.2013.09.005 (2013).
Gill, J. J. Revised genome sequence of Staphylococcus aureus bacteriophage K. Genome Announc 2, e01173–13, https://doi.org/10.1128/genomeA.01173-13 (2014).
Lavigne, R. & Vandersteegen, K. Group I introns in Staphylococcus bacteriophages. Future Virol 8, 997–1005, https://doi.org/10.2217/fvl.13.84 (2013).
Gu, J. et al. Genomic characterization of lytic Staphylococcus aureus phage GH15: providing new clues to intron shift in phages. J Gen Virol 94, 906–915, https://doi.org/10.1099/vir.0.049197-0 (2013).
Benešík, M. et al. Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity. Virus Genes 54, 130–139, https://doi.org/10.1007/s11262-017-1507-2 (2018).
Kašpárek, P., Pantůček, R., Kahánková, J., Růžičková, V. & Doškař, J. Genome rearrangements in host-range mutants of the polyvalent staphylococcal bacteriophage 812. Folia Microbiol 52, 331–338, https://doi.org/10.1007/BF02932087 (2007).
Rosypal, S., Rosypalová, A., Doškař, J., Pakrová, E. & Genovová, S. The use of the polyvalent phage 812 and its host-range mutants to the differentiation of Staphylococcus aureus from other staphylococci. Scripta Fac Sci Nat Univ Purk Brun 16, 317–336 (1986).
Nováček, J. et al. Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc Natl Acad Sci USA 113, 9351–9356, https://doi.org/10.1073/pnas.1605883113 (2016).
Eyer, L. et al. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics 7, 64–72, https://doi.org/10.1002/pmic.200600280 (2007).
Sybesma, W., Pirnay, J.-P. & Expert round table on acceptance and re-implementation of bacteriophage therapy. Silk route to the acceptance and re-implementation of bacteriophage therapy. Biotechnol J. 11, 595–600, https://doi.org/10.1002/biot.201600023 (2016).
Kwan, T., Liu, J., DuBow, M., Gros, P. & Pelletier, J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA 102, 5174–5179, https://doi.org/10.1073/pnas.0501140102 (2005).
Takeuchi, I. et al. The presence of two receptor-binding proteins contributes to the wide host range of staphylococcal Twort-like phages. Appl Environ Microbiol 82, 5763–5774, https://doi.org/10.1128/AEM.01385-16 (2016).
Azam, A. H., Hoshiga, F., Takeuchi, I., Miyanaga, K. & Tanji, Y. Analysis of phage resistance in Staphylococcus aureus SA003 reveals different binding mechanisms for the closely related Twort-like phages ɸSA012 and ɸSA039. Appl Microbiol Biotechnol 102, 8963–8977, https://doi.org/10.1007/s00253-018-9269-x (2018).
Zhang, X. et al. Conserved termini and adjacent variable region of Twortlikevirus Staphylococcus phages. Virol Sin 30, 433–440, https://doi.org/10.1007/s12250-015-3643-y (2015).
Cui, Z. et al. Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci Rep 7, 41259, https://doi.org/10.1038/srep41259 (2017).
Wilson, G. G., Young, K. K. Y., Edlin, G. J. & Konigsberg, W. High-frequency generalised transduction by bacteriophage T4. Nature 280, 80–82, https://doi.org/10.1038/280080a0 (1979).
Young, K. K., Edlin, G. J. & Wilson, G. G. Genetic analysis of bacteriophage T4 transducing bacteriophages. J Virol 41, 345–347 (1982).
Carlton, R. M., Noordman, W. H., Biswas, B., de Meester, E. D. & Loessner, M. J. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul Toxicol Pharmacol 43, 301–312, https://doi.org/10.1016/j.yrtph.2005.08.005 (2005).
Dwivedi, B., Xue, B., Lundin, D., Edwards, R. A. & Breitbart, M. A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biol 13, 33, https://doi.org/10.1186/1471-2148-13-33 (2013).
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J. & Spratt, B. G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38, 1008–1015 (2000).
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P. & Spratt, B. G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186, 1518–1530, https://doi.org/10.1128/JB.186.5.1518-1530.2004 (2004).
Milheiriço, C., Oliveira, D. C. & de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother 51, 3374–3377, https://doi.org/10.1128/AAC.00275-07 (2007).
Pantůček, R. et al. Identification of bacteriophage types and their carriage in Staphylococcus aureus. Arch Virol 149, 1689–1703, https://doi.org/10.1007/s00705-004-0335-6 (2004).
Štveráková, D. et al. Rapid identification of intact staphylococcal bacteriophages using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Viruses 10, 176, https://doi.org/10.3390/v10040176 (2018).
Moša, M., Boštík, J., Pantůček, R. & Doškař, J. Medicament in the form of anti-Staphylococcus phage lysate, process of its preparation and use. CZ201200668-A3. Patent Application (2012).
Rosypal, S. & Rosypalová, A. A spontaneus mutant of polyvalent phage 812 capable of growth in Staphylococcus aureus NCTC 8511 carrying prophage 53. Folia Fac Sci Nat Univ Purk Brun 11, 37–47 (1970).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual. 2nd ed., (Cold Spring Harbor Laboratory, 1989).
Botka, T. et al. Complete genome analysis of two new bacteriophages isolated from impetigo strains of Staphylococcus aureus. Virus Genes 51, 122–131, https://doi.org/10.1007/s11262-015-1223-8 (2015).
Botka, T. et al. Two highly divergent lineages of exfoliative toxin B-encoding plasmids revealed in impetigo strains of Staphylococcus aureus. Int J Med Microbiol 307, 291–296, https://doi.org/10.1016/j.ijmm.2017.05.005 (2017).
Coil, D., Jospin, G. & Darling, A. E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31, 587–589, https://doi.org/10.1093/bioinformatics/btu661 (2015).
Brettin, T. et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5, 8365, https://doi.org/10.1038/srep08365 (2015).
Lowe, T. M. & Chan, P. P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44, W54–W57, https://doi.org/10.1093/nar/gkw413 (2016).
Frith, M. C., Saunders, N. F. W., Kobe, B. & Bailey, T. L. Discovering Sequence Motifs with Arbitrary Insertions and Deletions. PLoS Computational Biology 4, e1000071, https://doi.org/10.1371/journal.pcbi.1000071 (2008).
Solovyev, V. & Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences in Metagenomics and its applications in agriculture, biomedicine and environmental studies (ed. Li, R. W.) 61–78 (Nova Science Publishers, 2011).
Lesnik, E. A. et al. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res 29, 3583–3594, https://doi.org/10.1093/nar/29.17.3583 (2001).
Grant, J. R. & Stothard, P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36, W181–W184, https://doi.org/10.1093/nar/gkn179 (2008).
Liu, B. & Pop, M. ARDB-Antibiotic Resistance Genes Database. Nucleic Acids Res 37, D443–D447, https://doi.org/10.1093/nar/gkn656 (2009).
Chen, L., Zheng, D., Liu, B., Yang, J. & Jin, Q. VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res 44, D694–D697, https://doi.org/10.1093/nar/gkv1239 (2016).
Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–W469, https://doi.org/10.1093/nar/gkn180 (2008).
Gu, J. et al. Complete genome sequence of Staphylococcus aureus bacteriophage GH15. J Virol 86, 8914–8915, https://doi.org/10.1128/JVI.01313-12 (2012).
Ajuebor, J. et al. Comparison of Staphylococcus phage K with close phage relatives commonly employed in phage therapeutics. Antibiotics (Basel). 7, 37, https://doi.org/10.3390/antibiotics7020037 (2018).
Gutiérrez, D. et al. Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species staphylococcal biofilms. Appl Environ Microbiol 81, 3336–3348, https://doi.org/10.1128/AEM.03560-14 (2015).
Takemura-Uchiyama, I. et al. Genomic and phylogenetic traits of Staphylococcus phages S25-3 and S25-4 (family Myoviridae, genus Twort-like viruses). Ann Microbiol 64, 1453–1456, https://doi.org/10.1007/s13213-013-0762-2 (2014).
Kim, S. G. et al. Complete genome sequence of Staphylococcus aureus bacteriophage pSa-3. Genome Announc 5, e00182–17, https://doi.org/10.1128/genomeA.00182-17 (2017).
Kirby, A. E. Synergistic action of gentamicin and bacteriophage in a continuous culture population of Staphylococcus aureus. PLoS One 7, e51017, https://doi.org/10.1371/journal.pone.0051017 (2012).
El Haddad, L. et al. Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS One 9, e102600, https://doi.org/10.1371/journal.pone.0102600 (2014).
Varga, M. et al. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol Lett 332, 146–152, https://doi.org/10.1111/j.1574-6968.2012.02589.x (2012).
Mašlaňová, I. et al. Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies. Environ Microbiol Rep 5, 66–73, https://doi.org/10.1111/j.1758-2229.2012.00378.x (2013).
Acknowledgements
The authors would like to thank their colleagues from the participating hospitals for providing the S. aureus strains. Thanks are also due to Ms. Eva Kodytková and Dr. Marta Fridrichová (National Institute of Public Health, Prague), Dr. Petra Vídeňská (Veterinary Research Institute, Brno), and Dr. Jiří Jarkovský (Institute of Biostatistics and Analyses at the Masaryk University, Brno) for valuable help. This work was supported by grants from the Czech Science Foundation (18-13064S), Ministry of Agriculture of the Czech Republic (QJ1510216), Ministry of Health of the Czech Republic (16-29916A) and project AdmireVet CZ.1.05/2.1.00/01.0006-ED0006/01/01 from the Czech Ministry of Education, Youth and Sports of the Czech Republic.
Author information
Authors and Affiliations
Contributions
R.P., T.B., I.M., J.D. and V.R. conceived the experiments; T.B., I.M., M.B., P.P., P.H., I.K., M.F. and V.J. conducted the experiments; T.B., R.P., I.M., M.B., M.V., H.Ž. and R.K. analysed the data; T.B., R.P., J.D., V.R. and I.M. wrote the main manuscript; T.B. and R.P. prepared tables and figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Botka, T., Pantůček, R., Mašlaňová, I. et al. Lytic and genomic properties of spontaneous host-range Kayvirus mutants prove their suitability for upgrading phage therapeutics against staphylococci. Sci Rep 9, 5475 (2019). https://doi.org/10.1038/s41598-019-41868-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41868-w
This article is cited by
-
Phage activity against Staphylococcus aureus is impaired in plasma and synovial fluid
Scientific Reports (2023)
-
Phage therapy of wound-associated infections
Folia Microbiologica (2022)
-
Microbiota’s role in health and diseases
Environmental Science and Pollution Research (2021)
-
Engineered Bacteriophage Therapeutics: Rationale, Challenges and Future
BioDrugs (2021)
-
A virophage cross-species infection through mutant selection represses giant virus propagation, promoting host cell survival
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.