Abstract
Insertional mutagenesis is a powerful means of identifying cancer drivers in animal models. We used the Sleeping Beauty (SB) transposon/transposase system to identify activated oncogenes in hematologic cancers in wild-type mice and mice that express a stabilized cyclin E protein (termed cyclin ET74AT393A). Cyclin E governs cell division and is misregulated in human cancers. Cyclin ET74AT393A mice develop ineffective erythropoiesis that resembles early-stage human myelodysplastic syndrome, and we sought to identify oncogenes that might cooperate with cyclin E hyperactivity in leukemogenesis. SB activation in hematopoietic precursors caused T-cell leukemia/lymphomas (T-ALL) and pure red blood cell erythroleukemias (EL). Analysis of >12,000 SB integration sites revealed markedly different oncogene activations in EL and T-ALL: Notch1 and Ikaros were most common in T-ALL, whereas ETS transcription factors (Erg and Ets1) were targeted in most ELs. Cyclin E status did not impact leukemogenesis or oncogene activations. Whereas most SB insertions were lost during culture of EL cell lines, Erg insertions were retained, indicating Erg’s key role in these neoplasms. Surprisingly, cyclin ET74AT393A conferred growth factor independence and altered Erg-dependent differentiation in EL cell lines. These studies provide new molecular insights into erythroid leukemia and suggest potential therapeutic targets for human leukemia.
Similar content being viewed by others
Introduction
Insertional mutagenesis is a powerful means of identifying the molecular drivers of cancer initiation and progression in animal models. Sleeping Beauty (SB) is a transposon/transposase insertional mutagenesis system that is designed to either overexpress nearby genes or inactivate genes, depending on the transposon’s integration site and orientation1,2. By combining conditional expression of the SB transposase with the T2Onc transposon in various genetic backgrounds, SB screens have been used extensively to identify\cancer genes and how they cooperate with one another in wild type and cancer-sensitizing backgrounds, and across many cancer types3,4,5,6.
In this study, we employed SB to identify oncogenes that might promote multi-step carcinogenesis in a mouse model engineered to express a stabilized version of the cyclin E protein. Cyclin E, in conjunction with its catalytic partner CDK2, has crucial roles in cell division, and cyclin E-CDK2 deregulation causes genome instability and contributes to cancer development and progression7. One important means of cyclin E regulation is phosphorylation-dependent degradation by the SCFFbw7 ubiquitin ligase8,9,10,11,12. To study the physiologic consequences of abnormal cyclin E degradation, we previously created a knock-in mouse model that ablated two cyclin E phosphorylation sites (T74 and T393) that trigger its degradation13,14. The cyclin ET74AT393A mutation caused increased cyclin E abundance and epithelial cell hyperproliferation. However, these mice did not spontaneously develop epithelial dysplasia or tumors, suggesting that compensatory mechanisms maintain tissue architecture and suppressed tumorigenesis. Cyclin ET74AT393A expression also caused ineffective erythropoiesis with marked expansion of immature erythroid precursors in the spleen and bone marrow, impaired erythroid differentiation, and mild anemia. These features resemble the early stages of human refractory anemia/myelodysplastic syndrome (MDS).
Because MDS can evolve to leukemia in humans, we speculated that cyclin ET74AT393A mice may provide a sensitized background to identify genetic events that cooperate with abnormal cyclin E regulation to promote leukemia. We thus used interferon-inducible Mx-Cre to activate the SB transposase in hematopoietic precursors to identify genes that might cooperate with abnormal cyclin E regulation to promote leukemia. The stabilized cyclin E allele neither predisposed mice to hematologic cancers nor altered gene activations by SB. Strikingly however, Mx-Cre-induced SB activation caused highly penetrant hematologic cancers within 8–13 weeks after Cre induction. To control for biases in transposon integrations that frequently occur proximal to the T2Onc array15,16, we used two different T2/Onc2 strains that contained the transposon array on different chromosomes. The most common malignancies were immature T-cell leukemia/lymphomas (T-ALL) and pure red blood cell erythroleukemias (EL), and there was a non-significant trend towards more EL in the cyclin ET74AT393A mice.
To identify activated oncogenes in these neoplasms, we determined the transposon insertion sites in all ELs and T-ALLs. Transposon insertions that are shared by multiple independent tumors, termed common insertion sites (CIS), often occur in the vicinity of cancer-associated genes, which provides the selective pressure for these shared insertions. We identified CIS using two different statistical methods and found that the CIS profile of ELs and T-ALLs differed markedly. Whereas Notch and Ikaros insertions were most common in T-ALL, ETS family transcription factors (Erg and Ets1) were the most commonly activated genes in ELs and were activated in the almost all of these tumors.
While T-ALL is common in SB screens performed in blood cells, EL has not, and we thus examined EL in more detail by developing 5 transplantable EL cell lines. Transposon analyses indicated that the vast majority of CISs found in primary ELs were lost during culture of EL cell lines, suggesting that they were not required for their proliferation and maintenance in vitro. However, all EL lines retained their Erg insertions and overexpressed ERG protein, further supporting the key role of Erg activation in EL. Insertions near the Bach2 transcription factor were also retained in several cell lines, and one EL line also retained a Flt3 insertion and exhibited FLT3-dependence. Finally, although cyclin ET74AT393A expression did not impact leukemogenesis or CIS involvement, we found two phenotypic differences between EL cell lines derived from WT and cyclin ET74AT393A mice. First, while WT EL lines remained dependent on exogenous growth factors (GFs), the cyclin E mutant cell lines did not and proliferated in the absence of GFs. Next, Erg knockdown resulted in erythroid differentiation of the WT but not the cyclin ET74AT380A cell lines. These findings suggest that properly regulated cyclin E activity influences both growth factor requirements and differentiation in EL cells.
Results
Development and Characterization of SB-induced hematologic cancers
We performed a SB insertional mutagenesis screen in WT and cyclin ET74AT393A mice. Mice homozygous for the cyclin ET74AT393A mutation and Rosa26SB11 LSL SB transposase and containing one of two different T2/Onc2 arrays were crossed with mice homozygous for cyclin ET74AT393A and expressing the Mx1-Cre transgene, which generated the experimental cohort (cyclin ET74AT393A; Rosa26SBLSL; T2/Onc2+; Mx1-Cre1) (Fig. 1a). The matched control cohort was wild-type cyclin E; Rosa26SBLSL; T2/Onc2+; Mx1-Cre1. Parallel crosses were made using two separate T2/Onc2 lines with transposon arrays on different chromosomes (strain 6113- chromosome 1, strain 6070 -chromosome 4)17,18 Mice (7–10 weeks old) were injected with poly I:C to activate Cre recombinase expression and subsequent transposase expression. Mx1-Cre induction and SB expression was monitored by loss of GFP expression in peripheral blood leukocytes from a subset of study mice, which demonstrated 71–86% loss of GFP-expressing cells following induction (Table S1).
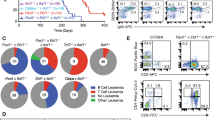
Breeding scheme, survival, and tumor spectrum of Mx-1 sleeping beauty mice. (a) Breeding scheme for transposon mutagenesis in cyclin ET74AT393A and wild-type mice. Cyclin ET74AT393A or wild-type mice homozygous for both a T2/Onc transposon array and Cre recombinase-inducible SB transposase allele (T2/Onc2/T2/Onc2; RosaSBLSL/RosaSBLSL) were mated with cyclin ET74AT393A mice heterozygous for Mx-1 Cre to generate cyclin ET74AT393A or wild-type mice with T2/Onc transposon array and Mx-1 Cre recombinase. (b) Kaplan Myer survival curve of SB-Cyclin ET74AT393A and SB-wild-type mice following induction of Mx-1 Cre and transpose expression by poly I/C injection. Time indicates intervention due to illness related to hematologic malignancy. Induction of malignancy was completely penetrant in both backgrounds with a median survival of 11.9 weeks post-injection for wild-type and 12.9 weeks for cyclin ET74AT393A mice (p = 0.08). (c) Spectrum of hematologic neoplasms that arise in Mx-1 Sleeping beauty mice. Diagnosis was made by a combination of histology and flow cytometry (see text).
SB insertional mutagenesis in immature hematopoietic cells resulted in progressive serious illness, including weight loss, poor grooming, and hunched posture, leading to euthanasia within 3 months of Mx-Cre induction. Necropsy showed hematologic malignancies in all animals, including lymphoid, myeloid, erythroid and megakaryocytic tumors (Fig. 1c, Table S2). Surprisingly, rather than accelerating tumorigenesis, the cyclin ET74AT393A allele imparted a non-significant trend towards longer survival (11.6 weeks post injection for wild-type and 12.9 weeks for cyclin ET74T393A mice, p = 0.08) (Fig. 1b). The most frequent neoplasm was immature T-cell lymphoblastic lymphoma with leukemia (T-ALL), which usually presented with a massively enlarged thymus. T-ALL typically involved the spleen and bone marrow and in some instances, disseminated to the liver, kidneys and lungs. Leukemia was also noted with leukocytosis and circulating lymphoblasts. Histologic sections of affected organs revealed homogeneous tumors composed of tightly packed lymphoblasts with a high mitotic rate and scattered apoptotic cells admixed with phagocytic histiocytes (Fig. S1). Immunophenotyping by flow cytometry showed expression of CD3 and CD8 with frequent variable co-expression of CD4, findings consistent with immature T-cell lymphoblastic leukemia (Fig. 2f,g, Fig. S1, Table S2).
Morphology and immunophenotype of hematopoietic neoplasms in Mx-1 sleeping beauty mice. (a,b) Histologic sections (H&E) of erythroleukemia involving spleen (A: 20X objective) and liver (B: 40X objective). (c) Bone marrow cytospin preparations (Wright Giemsa) of erythroleukemia with block-like clumped chromatin and dark blue basophilic cytoplasm characteristic of erythroid precursor cells (63X objective). (d) Peripheral blood smear (Wright Giemsa) with circulating erythroblasts (40X objective). (e) Histologic section (H&E) of erythroleukemia with prominent megakaryocytic differentiation (large atypical cells) involving the liver (20X objective). (f) Representative immunophenotype of immature T-cell leukemia in blue (top row) and erythroleukemia in red (bottom row). (g) Summary of T-cell leukemia and erythroleukemia immunophenotype.
Many (40%, n = 100) mice developed EL with intermixed megakaryocytic differentiation, which caused prominent splenomegaly and involved the bone marrow and sometimes the liver (Figs 1c, 2a,b,e and Table S2). The mice with EL exhibited peripheral involvement with leukocytosis consisting of a prominent population of circulating erythroblasts with dark basophilic cytoplasm (Fig. 2c,d). Multiparameter flow cytometry studies revealed immature erythroblasts (as shown by CD45/side scatter) with dim CD45 expression, variable CD117 (c-Kit) expression, bright CD71 (transferrin receptor) expression, and occasional Ter119 (glycophorin associated protein) expression (Fig. 2f,g, Table S2). ELs varied somewhat with respect to their degree of erythroid differentiation, with variable expression of CD117 (immature immunophenotype) and Ter119 (mature immunophenotype) (Fig. S2). The immunophenotype of the neoplastic population was identical in samples isolated from peripheral blood, spleen and bone marrow (Fig. S3). Some ELs showed megakaryocytic differentiation with scattered enlarged cells with abundant cytoplasm and multilobulated nuclei, morphologically consistent with megakaryocytes (Fig. 2e). Additional flow cytometry studies showed variable expression of CD41, consistent with megakaryocytic differentiation in these cases (Fig. S4). Overall, these tumors displayed many features resembling those of pure red blood cell EL in humans (see Discussion)19. Some mice (7%) contained mixed tumors of both lymphoid and erythroid lineages identified by parallel flow cytometry studies (Table S2). The spectrum of hematologic malignancies was similar in the cyclin E-T74AT393A and wild-type mice, with a small trend toward increased EL in the mutant cyclin E mice (Fig. 1c).
Common Insertion Site (CIS) Identification in T-ALL and EL
To identify CIS, we isolated DNA from 40 ELs (spleens) and 53 T-ALLs (thymus or lymph node), amplified the transposon integration sites via ligation-mediated PCR, and identified the transposon integration sites by next generation sequencing. We mapped 5002 EL insertion sites and 7012 T-ALL sites (Table S3). Each tumor contained between 50 and 150 unique insertions, presumably representing both driver and passenger insertions. We identified CIS by using two different statistical methods that differ in the parameters used to identify CIS and their associated genes: (1) gene-centric common insertion site analysis (gCIS), and (2) TAPDANCE20,21.
gCIS identified 74 total CIS in T-ALLs (Table S4). The chromosome that contains the transposon concatemer (the “donor chromosome”) is disproportionately targeted by transposon insertions (“local hopping”) in SB screens. As expected, many CIS were mapped to the donor chromosome. Although these insertions could represent true genetic drivers of cancer, we initially filtered these CIS from subsequent analyses, resulting in 25 T-ALL CIS (Table 1). This analysis identified many established drivers of T-ALL, including Notch1, Ikzf1, Rasgrp1, and Akt26. The presence of the cyclin E mutation did not impact the spectrum of CIS observed. Since T-ALLs have been well studied in insertional mutagenesis studies, we focused on the ELs for the remainder of our study4,22,23,24.
gCIS identified 36 CIS genes in ELs prior to filtering out the donor chromosome CIS (Table S5), and 13 CIS after donor chromosome filtering (Table 2). Many of the EL CIS thus fell within the donor chromosomes. We observed a very high insertion rate in the ETS family of transcription factors (TFs), with insertions in Erg and Ets1 in 65% and 44% of ELs, respectively. In fact, only 4 of the 40 ELs lacked insertions targeting either Erg or Ets1 (Table S4). Interestingly, a previous SB insertional mutagenesis study performed in Jak2 mutant mice also identified ETS transcription factors as CIS in EL, and Erg has been implicated in human EL (see discussion), highlighting the importance of ETS TFs in EL25. Many CIS occurred near genes implicated in hematologic cancers and hematopoietic development (Erg, Ets1, Epo, Il2rb, Flt3, Kras, Stat5b, Fli1). While CIS within donor chromosomes were filtered from our analyses, some of the CIS found within donor chromosomes have been reported in other transposon-mediated insertion studies targeting hematologic neoplasms, and thus may represent physiologically significant gene alterations rather than local hopping. Examples of these types of CIS (on chromosome 4) that are likely to have contributed to EL in our study include Csf3r (35% of ELs), Bach2 (41% of ELs) and Cdkn2a/Arf (5% of ELs). Because we found each of these CIS in both T2Onc strains, these insertions were not restricted to potential local hops. TAPDANCE, an alternate statistical method that eliminates donor chromosome insertions, was also used to annotate CIS and gene associations in T-ALL and EL. The most frequent CIS found by TAPDANCE in T-ALL included Notch1, Ikzf1, Rasgrp1, Akt2, Runx1, FoxP1, whereas the most frequent CIS present in EL were Erg and Ets1, followed by Epo, Gata1, Pik3ca and Fli1 (Tables S6 and S7).
CIS evolution in EL cell lines
We used marrow transplantation to demonstrate that ELs were truly neoplastic and capable of self-replication. Five primary ELs were transplanted into sub-lethally irradiated mice, and recipient mice developed signs of progressive leukemia with a latency of 10–20 days post-injection. Analysis of peripheral blood revealed similar features to the parental tumors, including circulating erythroblasts with the same immunophenotypes as the original tumor, albeit more homogeneously (Fig. 3a). Necropsy of the transplanted mice demonstrated splenomegaly, similar to that seen in the original model. Stable EL cultures were established from five of these transplanted mice using murine erythroblast cell culture media containing c-Kit ligand and erythropoietin. No major immunophenotypic or morphologic changes occurred during the prolonged cell culture (Fig. 3a, Table S2).
Immunophenotypic change and loss of insertion sites of erythroleukemia following transplantation and cell culture models. (a) Flow cytometry results from primary leukemia (top), transplanted leukemia (middle), and cell culture (bottom). The abnormal erythroblasts are CD45 dim/low side scatter/high CD71/variable CD117/variable Ter119 (red). (b) Loss of insertion sites following cell culture. Number of total insertion sites and common insertion site (CIS) identified in primary tumor (blue and green) and cell lines (red and purple) in five separate tumors and derived cell lines. (c) Quantification of inserts and CIS in tumor-derived cell lines.
We used these long-term EL cultures to further identify putative driver insertions that promote EL. Since the SB transposase is constitutively active in these cell lines, we predicted that serial passage might result in the loss of irrelevant or passenger transposon insertions that were not required for EL proliferation or maintenance in culture, and conversely, that insertions required for tumor maintenance would be preserved. All five EL primary cultures were cultured for 12 weeks in the presence of growth factors and then analyzed for transposon insertions as described above (Table S3). Long-term cell culture dramatically altered the number and identity of the insertions. On average, 113 unique insertions (ranging between 57–143) were identified from the original ELs. Following long-term culture, the number of insertions decreased to an average of 24 unique insertions (ranging from 13–39) per cell line, and only a few of these (1–4) had been previously identified as CIS (Fig. 3b,c). The loss of insertions may result from continued transposon hopping and tumor evolution in culture, or may alternatively reflect clonal selection from an initially heterogeneous population of cells in primary ELs. In all cases, just a few previously identified CIS were maintained during long-term culture (Fig. 3b,c and Table S8). In one cell line, we detected an Erg insertion that had not been found in the primary tumor, which may reflect either a small subclonal population present in the original tumor that was not detected in the initial sequencing, or alternatively, clonal evolution in vitro. Of the previously identified CIS, only Erg insertions were retained in all the cell lines tested, suggesting that persistent Erg deregulation is required for EL proliferation and/or maintenance in culture. Interestingly, Bach2, a chromosome 4 insertion site that was found in both T2Onc strains, was identified as a CIS in 3/5 cell lines, suggesting that this hematopoietic TF may have an important role in EL24,26.
Effects of Erg insertions and cyclin ET74AT393A expression on EL differentiation and growth factor requirements
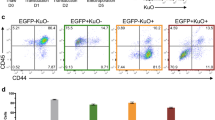
Erg was the most common EL CIS in our study, with 65% of tumors containing at least one Erg insertion. These integrations were found in the same transcriptional orientation as the ERG gene and upstream of exons 3 and 4, suggesting that the insertions led to overexpression of truncated Erg mRNAs and proteins (Fig. 4a). Erg overexpression drives murine erythroid-megakaryocytic leukemia and has been associated with various human leukemias and poor outcomes27,28,29. We used shRNA to reduce ERG protein expression to study the role of persistent Erg deregulation in long-term EL cultures. Primary EL cell lines expressed variable levels of ERG protein, and Erg shRNA only partially reduced ERG abundance (Fig. 4b). The inability to fully silence Erg expression may reflect the essential role of Erg deregulation in these cells. Partial Erg knockdown induced immunophenotypic differentiation in the cell lines derived from the WT background, with decreased CD117 (cKit) expression and increased Ter119 (glycophorin B), consistent with erythroid maturation (Fig. 4b,c). These results are consistent with prior studies that have shown that ERG expression in human and murine leukemias inhibits differentiation30,31,32. Surprisingly, unlike WT EL cell lines, EL lines derived from cyclin ET74AT393A mice did not exhibit immunophenotypic markers of differentiation after partial ERG knockdown (Fig. 4c). Even more unexpectedly, both cell lines derived from the cyclin E mutant mice proliferated without growth factor supplementation, while the three cell lines derived from wild-type mice required erythropoietin and c-Kit supplementation (Fig. 4d). These results suggest that inappropriate cyclin E regulation confers growth factor independence. This is reminiscent of observations with the related cell cycle protein, cyclin D2, which promotes growth factor independence in B-cell lines33. Thus, despite the similar disease spectrum in both strains, cyclin ET74AT393A expression overcame normal growth factor dependency, indicating that cyclin E may be a key mediator of mitogenic signaling in EL cells.
Erg over-expression inhibits erythroid differentiation. (a) Transposon insertion sites within Erg are predicted to promote the expression of a truncated transcript containing the full ETS domain. All of the insertions are in the same transcriptional orientation as the Erg gene. (b) Immunoblot showing partial knockdown of ERG with mErg-specific shRNA in EL cell lines. (c) ERG knockdown in erythroleukemia cell lines induces immunophenotypic maturation (decreased CD117 and increased Ter119 expression) in wt EL cell lines (4230 and 4460) but not in cyclin ET74AT393A EL cell lines (3945 and 4489). (d) Growth factor dependence of the erythroleukemia-derived cell lines. Two cell lines (3945 and 4489) from cyclin ET74AT393A mice are growth factor independent, and three cell lines (4230, 4456, 4460) from wt mice are growth factor dependent (EPO and c-Kit ligand).
Flt3 (FMS-like tyrosine kinase 3) is a receptor tyrosine kinase that is mutated in AML and confers a poor clinical prognosis34. We identified three Flt3 insertions in ELs, all of which were located downstream of exon 9 (Fig. 5a). These insertions are predicted to result in overexpression of a truncated FLT3 protein containing only the tyrosine kinase domain without the receptor domain. One of the tumors selected for culture (4489) had the Flt3 insertion and retained this insertion after serial passage. Western blot analysis shows that this cell line over-expressed a truncated ~65 kDa form of FLT3 protein without apparent expression of endogenous full length Flt3 (Fig. 5b). To test if the Flt3 insertion is required for proliferation of this EL line, we treated all five of the tumor-derived cell lines with three different pharmacologic inhibitors that target the FLT3 kinase and have been used in clinical trials of patients with FLT3 mutant AML (Lestaurtinib, PKC412, and Sorafenib). Compared with the other cell lines, line 4489 (with the Flt3 insertion) was exquisitely sensitive to all three inhibitors, showing that this EL cell line remained dependent on FLT3 activity for proliferation (Fig. 5c–e).
(a) Transposon insertion site within the gene encoding Flt3. All of the insertions are in the same orientation of the gene and are predicted to result in overexpression of a truncated protein containing the tyrosine kinase domain of Flt3. (b) Immunoblot showing over-expression of truncated FLT3 protein (65 kDa) in the 4489 cell line that contained an insertion in the Flt3 gene. Endogenous FLT3 was not detected (113 KDa). (c–e) Over-expression of the truncated FLT3 in 4489 imparts increased sensitivity to a panel of FLT3 tyrosine kinase inhibitors including Lestaurtinib (c), PKC412 (d), and Sorafenib (e).
Discussion
Based on our previous work showing that mice with a stabilized cyclin E protein develop proliferative and differentiation anomalies in hematopoietic cells, we undertook this study to use SB to identify putative cyclin E-cooperating oncogenes. In light of the phenotypic consequences of cyclin ET74AT393A expression in mice, as well as the roles for cyclin E in human cancer and murine cancer models7,14,35,36,37, it was surprising to find that the cyclin ET74AT393A allele did not alter the incidence, latency, or spectrum of hematologic cancers, or the gene activations observed. The reasons why cyclin ET74AT393A failed to contribute to multistep tumorigenesis in this model remain unclear. The T74AT393A mutation only stabilizes cyclin E in catalytically active complexes, and because many cyclin E-CDK2 complexes are inactive, this mutation has a greater impact on cyclin E-CDK2 activity than cyclin E abundance. In contrast, cyclin E amplifications in human tumors elevate cyclin E abundance, which may or may not increase cyclin E activity (due to compensatory mechanisms that suppress CDK2 activity). Because cyclin E has non-catalytic activities (e.g. replication origin licensing), one possibility is that mutations that differentially impact cyclin E abundance versus activity may lead to different biologic outcomes38. However, we do not have evidence to directly support this speculation from this study.
While cyclin ET74AT393A expression did not alter carcinogenesis, it did impact growth factor requirements and differentiation of EL cell lines. Cyclin E-CDK2 activation has previously been proposed to be a crucial feature of the G1 restriction point, which is defined at the point in the G1 phase of the cell cycle after which cells no longer require extracellular mitogens for S-phase entry39,40. Our findings that cyclin ET74AT393A rendered two EL cell lines growth factor independent supports this notion, although these studies involved a limited number of cell lines. Of note, our attempts to convert WT-EL cell lines to GF independence through ectopic cyclin ET74AT393A expression were unsuccessful because ectopic cyclin ET74AT393A expression was toxic.
SB activation by Mx-Cre caused two distinct hematologic neoplasms, T-ALL and EL, that differed morphologically, immunophenotypically, and with respect to the identified CIS associated with each disease. While T-ALL has been extensively described in SB models22, EL is much less common25. In humans, acute erythroleukemia (EL) is a rare and relatively poorly characterized form of acute myeloid leukemia19. Until recently, the diagnostic criteria for EL encompassed a diverse set of hematopoietic malignancies that share some degree of erythroid hyperplasia, ranging from an expanded abnormal myeloid blast population with erythroid hyperplasia to a pure erythroid leukemia with erythroid blasts encompassing greater than 80% of the marrow cellularity (2008 WHO; see below). The prior diagnostic criteria for acute erythroleukemia thus included both erythroleukemia (erythroid/myeloid) and pure erythroid leukemia, which occurs less commonly. Because erythroleukemia (erythroid/myeloid) frequently arises from pre-existing MDS (and shares many clinical features of MDS), the WHO recently modified the diagnostic criteria for EL: the more common erythroleukemia (erythroid/myeloid) is now classified as MDS or AML based on the percentage of myeloid blast population, irrespective of the erythroid component41,42. Therefore, many cases that had been previously diagnosed as EL are now reclassified as low grade MDS. Pure erythroid leukemia (PEL) remains a distinct diagnostic entity per WHO criteria and is characterized by malignant immature erythroid cells that represent greater than 80% of the marrow. Because of its rare occurrence and previously imprecise diagnostic criteria, our understanding of PEL remains limited. A few small-scale clinical studies on PEL have demonstrated that the leukemic cells usually have complex chromosomal abnormalities, frequent p53 mutations, and extremely poor overall survival43,44.
The murine EL that developed in this study most resembled human PEL, with an expanded erythroid blast population (>80% of the marrow cellularity) associated with peripheral cytopenia, circulating erythroid blasts and frequent extramedullary involvement of the liver and spleen. We identified many insertion sites in ELs that target genes involved in myeloproliferative disorders, growth factor signaling, and signal transduction. In accordance with previous work on murine and human EL27,28,29, our studies support a crucial role for ETS transcription factors in the development and maintenance of EL. Most of the ELs that we studied (90%) had inserts in one of the ETS transcription factors (ERG, ETS, or FLI1) and it is tempting to speculate that the few ELs without these insertions may have activated these pathways through other mechanisms. However, we do not have direct evidence to support this speculation. Strikingly, ERG-associated SB insertions were retained in each of the five EL cell lines that we derived from primary ELs, while the vast majority of other CIS found in the primary tumors were not (Fig. 3c). This supports the idea that EL cell lines depend upon continued ERG deregulation, which is consistent with the differentiation caused by partial ERG knockdown (Fig. 4c). Our studies also suggest that Erg may be a good therapeutic target for several forms of AML, including PEL.
The other CIS that was retained in multiple EL cell lines was Bach2 (B-lymphoid transcription factor, BTB and CNC Homology 1 Basic Leucine Zipper Transcription Factor 2), a transcriptional repressor that regulates antibody class switching and has been implicated in B-cell neoplasm45,46. Bach1 and Bach2 are highly regulated and homologous proteins with key functions in iron hemostasis, erythropoiesis, megakaryopoiesis, and autoimmunity47. Our finding that Bach2 insertions were retained in EL cell lines suggests a biologic selection for persistent Bach2 deregulation and perhaps a role for Bach2 in EL development and/or maintenance. During erythropoiesis, elevated levels of free heme bind to and inhibit Bach1, thereby activating the expression of Beta-globin and other genes involved in erythroid differentiation48. Although Bach2 functions as a transcriptional repressor during B-cell maturation and has been implicated in B-cell neoplasms, other studies have shown the Bach2 is also regulated by heme binding and that overexpression of Bach2 in hematopoietic stem cells promotes erythroid commitment, suggesting it may also play an early role in directing erythroid lineage fate decisions49.
Finally, pure erythroid leukemia is an aggressive poorly characterized form of AML. Now that the WHO has redefined PEL as a distinct form of AML, new clinical studies are needed to identify the molecular pathways that promote PEL. The functional studies presented here provide candidate genes and pathways that should serve as a resource for future studies of human PEL and help define altered pathways that promote this rare, but highly aggressive, leukemia.
Materials and Methods
Mice
All mice used in these studies were previously described. Homozygous cyclin ET74AT393A mice were developed and maintained as described13. The Cre inducible RosaSBase-LSL transposase and the T2/Onc2 transposon (Lines 6113 and 6070) mice were obtained from N. Copeland17,18. Mx1-Cre transgenic mice were obtained from Jackson Laboratories (Bar Harbor, ME). The breeding schemes to generate study mice are shown in Fig. 1a. For Mx1-Cre induction, mice received intraperitoneal injections of polyinosinic: polycytidylic acid (pIpC) (10 μg/g body weight) every other day for a total of five injections. For transplant studies, erythroleukemia cells were expanded in vitro prior to injection. Recipient C57/Bl6 mice were sublethally irradiated (700 Gy) 24 hours prior to transplantation and infused with 1 × 106 cultured erythroleukemia cells. Each cell line was transplanted into five recipient mice. Mice were monitored daily using institutional standard procedures and euthanized when indicated. Animal studies and all animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and were carried out at the Fred Hutchinson Cancer Research Center (FHCRC). All methods were performed in accordance with the guidelines and regulation established by FHCRC IACUC, Institutional Review, and Institutional Biosafety Committees.
Histopathological and Hematological Analysis
Mice were euthanized via carbon dioxide inhalation and necropsies were performed. All tissues were harvested and fixed in 10% formalin for 5–7 days and stored in 70% ethanol until submitted for pathological evaluation. Sternums for bone marrow analysis were decalcified in Formical-4 (Decal Chemical Corp) for 24 hrs. after fixation in formalin. After fixation, all samples were submitted to the Experimental Histopathology Core Facility at FHCRC where tissues were processed, embedded in paraffin, cut into slides, and stained with H&E according to standard techniques. Tissues were reviewed by a board-certified hematopathologist (KRL) and a board-certified veterinary pathologist (SEK). Peripheral blood was harvested and analyzed via Siemens Advia 2120i with multispecies software and a manual differential analysis. Lymphomas and leukemias were classified using the Bethesda recommendations for mice50,51.
Flow cytometry
Primary splenocytes and thymocytes were recovered by crushing excised spleen or thymic tissue between glass microscope slides, washing with PBS + 2% FBS, and passing through an 80 µm mesh filter (Sefar, Nitex 03-80/37). Cells were washed once in PBS + 2% FBS and were resuspended in PBS + 2% FBS and kept on ice until analysis. For analysis of bone marrow, marrow was extracted from femurs using a mortar and pestle and processed as above. For analysis of peripheral blood, red blood cells were lysed (155 mM NH4CL, 12 mM NaHCO3, 0.13 mM EDTA) at room temperature and washed twice in PBS + 2% FBS. Flow cytometry analysis included a 4-color T-cell panel (CD8a-PE, CD4-APC, CD19-APC-Cy7and CD45r/B220-PerCP (BD Biosciences)) and a 6-color erythroid/myeloid panel (CD45-APC-Cy7, CD71-FITC, CD117-PE-Cy7, Ter119-APC, Ly-6G/Ly-6C (Gr1)-PerCP-Cy5.5, and B220-FITC (BD Biosciences) and CD3e Alexa Fluor 488 (BioLegend). Cells were also stained with DAPI to determine cell viability. Flow cytometry was performed on a Canto 2 (Becton Dickinson) or a custom designed LSR2 (Becton Dickinsin, Franklin Lakes, NJ) and analyzed using either FlowJo Software (FlowJo, Ashland, OR) or Woodlist, a noncommercial software program developed in our clinical laboratory52.
Ligation-mediated PCR to amplify transposon junction sequences
Genomic DNA was extracted from frozen tissue using the Puregene Core Kit A (Qiagen) according to manufacturer’s instructions. Five μg of genomic DNA in a volume of 100 μl of ddH2O was sheared to 300 bp lengths using the Covaris E-series sonicator according to the manufacturer’s protocol. One μg of DNA (20 μl) was end-repaired in a reaction containing T4 DNA polymerase and polynucleotide kinase and incubated at 20°C for 30 minutes. Then 1 μl of 0.5 M EDTA was added and enzymes were heat-inactivated at 75°C for 20 minutes. Blunt/Alu adaptors were ligated to ends of repaired DNA and amplification of transposon junctions was conducted using a ligation-mediated PCR approach as previously described20,22. Individual sample library quality was analyzed by running an aliquot via agarose gel electrophoresis. Remaining PCR products were purified, pooled (20–25 ng per sample), and submitted for direct sequencing on an Illumina HiSeq 2000. Sequencing reads were analyzed using gene-centric common insertion site analysis (gCIS) or TAPDANCE20,21.
Cell lines
Primary tumor cell lines were derived by plating splenocytes in IMDM media containing 20% FBS, 4.5 × 10−5 M monothioglycerol, 2 U/ml erythropoietin (Epoetin alpha; Amgen Biotechnology), and 50 ng/ml mouse stem cell factor. Cell lines were then maintained in culture as indicated in the text. Lentivirus infection with Erg-specific shRNA, and retroviral infection with cyclin ET74AT393A and wildtype cyclin E were performed as previously described53.
Western Blotting
Western blotting was performed as described11. Anti-ERG antibody (CM421AC) was from Biocare Medical (Pacheco, CA) and anti-FLT3 antibody (8F2) was from Cell Signaling (Danvers, MA).
Cytotoxicity Assays
Cell lines were incubated with the indicated concentrations of drugs and incubated for 72 hours. Viability was measured using a resazurin-based assay and % viability was normalized to the untreated control.
References
Largaespada, D. A. Transposon-mediated mutagenesis of somatic cells in the mouse for cancer gene identification. Methods 49, 282–286, https://doi.org/10.1016/j.ymeth.2009.07.002 (2009).
Dupuy, A. J. Transposon-based screens for cancer gene discovery in mouse models. Semin Cancer Biol 20, 261–268, https://doi.org/10.1016/j.semcancer.2010.05.003 (2010).
Abbott, K. L. et al. The Candidate Cancer Gene Database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res 43, D844–848, https://doi.org/10.1093/nar/gku770 (2015).
Copeland, N. G. & Jenkins, N. A. Harnessing transposons for cancer gene discovery. Nat Rev Cancer 10, 696–706, https://doi.org/10.1038/nrc2916 (2010).
Moriarity, B. S. & Largaespada, D. A. Sleeping Beauty transposon insertional mutagenesis based mouse models for cancer gene discovery. Curr Opin Genet Dev 30, 66–72, https://doi.org/10.1016/j.gde.2015.04.007 (2015).
Newberg, J. Y., Mann, K. M., Mann, M. B., Jenkins, N. A. & Copeland, N. G. SBCDDB: Sleeping Beauty Cancer Driver Database for gene discovery in mouse models of human cancers. Nucleic Acids Res 46, D1011–D1017, https://doi.org/10.1093/nar/gkx956 (2018).
Hwang, H. C. & Clurman, B. E. Cyclin E in normal and neoplastic cell cycles. Oncogene 24, 2776–2786 (2005).
Koepp, D. M. et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 (2001).
Strohmaier, H. et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413, 316–322 (2001).
Moberg, K. H., Bell, D. W., Wahrer, D. C., Haber, D. A. & Hariharan, I. K. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413, 311–316 (2001).
Welcker, M. et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Molecular cell 12, 381–392 (2003).
Davis, R. J., Welcker, M. & Clurman, B. E. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26, 455–464, https://doi.org/10.1016/j.ccell.2014.09.013 (2014).
Minella, A. C. et al. Cyclin E phosphorylation regulates cell proliferation in hematopoietic and epithelial lineages in vivo. Genes & development 22, 1677–1689 (2008).
Loeb, K. et al. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell 8, 35–47 (2005).
Carlson, C. M. et al. Transposon mutagenesis of the mouse germline. Genetics 165, 243–256 (2003).
Keng, V. W. et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nature methods 2, 763–769, https://doi.org/10.1038/nmeth795 (2005).
Dupuy, A. J., Akagi, K., Largaespada, D. A., Copeland, N. G. & Jenkins, N. A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436, 221–226, https://doi.org/10.1038/nature03691 (2005).
Dupuy, A. J. et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 69, 8150–8156, https://doi.org/10.1158/0008-5472.CAN-09-1135 (2009).
Boddu, P. et al. Erythroleukemia-historical perspectives and recent advances in diagnosis and management. Blood Rev 32, 96–105, https://doi.org/10.1016/j.blre.2017.09.002 (2018).
Brett, B. T. et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PloS one 6, e24668, https://doi.org/10.1371/journal.pone.0024668 (2011).
Sarver, A. L., Erdman, J., Starr, T., Largaespada, D. A. & Silverstein, K. A. TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics 13, 154, https://doi.org/10.1186/1471-2105-13-154 (2012).
Berquam-Vrieze, K. E. et al. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood 118, 4646–4656, https://doi.org/10.1182/blood-2011-03-343947 (2011).
Collier, L. S. et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res 69, 8429–8437, https://doi.org/10.1158/0008-5472.CAN-09-1760 (2009).
Wotton, S. et al. Proviral insertion indicates a dominant oncogenic role for Runx1/AML-1 in T-cell lymphoma. Cancer Res 62, 7181–7185 (2002).
Tang, J. Z. et al. Transposon mutagenesis reveals cooperation of ETS family transcription factors with signaling pathways in erythro-megakaryocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America 110, 6091–6096, https://doi.org/10.1073/pnas.1304234110 (2013).
Liu, J. et al. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol Biol 10, 2, https://doi.org/10.1186/1471-2199-10-2 (2009).
Carmichael, C. L. et al. Hematopoietic overexpression of the transcription factor Erg induces lymphoid and erythro-megakaryocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America 109, 15437–15442, https://doi.org/10.1073/pnas.1213454109 (2012).
Martens, J. H. Acute myeloid leukemia: a central role for the ETS factor ERG. Int J Biochem Cell Biol 43, 1413–1416, https://doi.org/10.1016/j.biocel.2011.05.014 (2011).
Pigazzi, M. et al. Presence of high-ERG expression is an independent unfavorable prognostic marker in MLL-rearranged childhood myeloid leukemia. Blood 119, 1086–1087; author reply 1087–1088, https://doi.org/10.1182/blood-2011-10-385815 (2012).
Xie, Y. et al. Reduced Erg Dosage Impairs Survival of Hematopoietic Stem and Progenitor Cells. Stem Cells 35, 1773–1785, https://doi.org/10.1002/stem.2627 (2017).
Tsuzuki, S., Taguchi, O. & Seto, M. Promotion and maintenance of leukemia by ERG. Blood 117, 3858–3868, https://doi.org/10.1182/blood-2010-11-320515 (2011).
Knudsen, K. J. et al. ERG promotes the maintenance of hematopoietic stem cells by restricting their differentiation. Genes & development 29, 1915–1929, https://doi.org/10.1101/gad.268409.115 (2015).
Parada, Y. et al. BCR-ABL and interleukin 3 promote haematopoietic cell proliferation and survival through modulation of cyclin D2 and p27Kip1 expression. The Journal of biological chemistry 276, 23572–23580, https://doi.org/10.1074/jbc.M101885200 (2001).
Grunwald, M. R. & Levis, M. J. FLT3 Tyrosine Kinase Inhibition as a Paradigm for Targeted Drug Development in Acute Myeloid Leukemia. Seminars in hematology 52, 193–199, https://doi.org/10.1053/j.seminhematol.2015.03.004 (2015).
Bortner, D. M. & Rosenberg, M. P. Induction of mammary gland hyperplasia and carcinomas in transgenic mice expressing human cyclin E. Molecular and cellular biology 17, 453–459 (1997).
Ma, Y. et al. Transgenic cyclin E triggers dysplasia and multiple pulmonary adenocarcinomas. Proceedings of the National Academy of Sciences of the United States of America 104, 4089–4094, https://doi.org/10.1073/pnas.0606537104 (2007).
Smith, A. P. et al. Deregulated cyclin E promotes p53 loss of heterozygosity and tumorigenesis in the mouse mammary gland. Oncogene 25, 7245–7259, https://doi.org/10.1038/sj.onc.1209713 (2006).
Geng, Y. et al. Kinase-independent function of cyclin E. Molecular cell 25, 127–139 (2007).
Sherr, C. J. Cancer cell cycles. Science 274, 1672–1677 (1996).
Ohtsubo, M. & Roberts, J. M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science 259, 1908–1912 (1993).
Arber, D. A. et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405, https://doi.org/10.1182/blood-2016-03-643544 (2016).
Sabattini, E., Bacci, F., Sagramoso, C. & Pileri, S. A. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica 102, 83–87 (2010).
Wang, W., Wang, S. A., Medeiros, L. J. & Khoury, J. D. Pure erythroid leukemia. Am J Hematol 92, 292–296, https://doi.org/10.1002/ajh.24626 (2017).
Montalban-Bravo, G. et al. More than 1 TP53 abnormality is a dominant characteristic of pure erythroid leukemia. Blood 129, 2584–2587, https://doi.org/10.1182/blood-2016-11-749903 (2017).
Muto, A. et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 429, 566–571, https://doi.org/10.1038/nature02596 (2004).
Ge, Z. et al. Ikaros regulation of the BCL6/BACH2 axis and its clinical relevance in acute lymphoblastic leukemia. Oncotarget 8, 8022–8034, https://doi.org/10.18632/oncotarget.14038 (2017).
Igarashi, K. & Watanabe-Matsui, M. Wearing red for signaling: the heme-bach axis in heme metabolism, oxidative stress response and iron immunology. Tohoku J Exp Med 232, 229–253 (2014).
Zhou, Y., Wu, H., Zhao, M., Chang, C. & Lu, Q. The Bach Family of Transcription Factors: A Comprehensive Review. Clin Rev Allergy Immunol 50, 345–356, https://doi.org/10.1007/s12016-016-8538-7 (2016).
Kato, H. et al. Infection perturbs Bach2- and Bach1-dependent erythroid lineage ‘choice’ to cause anemia. Nat Immunol 19, 1059–1070, https://doi.org/10.1038/s41590-018-0202-3 (2018).
Kogan, S. C. et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 100, 238–245 (2002).
Morse, H. C. 3rd et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 100, 246–258 (2002).
Cherian, S. et al. A novel flow cytometric assay for detection of residual disease in patients with B-lymphoblastic leukemia/lymphoma post anti-CD19 therapy. Cytometry B Clin Cytom 94, 112–120, https://doi.org/10.1002/cyto.b.21482 (2018).
Minella, A. C., Grim, J. E., Welcker, M. & Clurman, B. E. p53 and SCF(Fbw7) cooperatively restrain cyclin E-associated genome instability. Oncogene (2007).
Acknowledgements
This work was supported by the following grants: NIH R01 CA193808 (B.E.C.), NIH 1R21CA229922 (A.J.D), and American Cancer Society 125006-RSG-13-183-01-CSM (J.E.G). This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA015704 and a pilot grant from Core Center of Excellence in Hematology P30 DK 56465. B.E.C is supported by the Rosput Reynolds Endowed Chair.
Author information
Authors and Affiliations
Contributions
K.L. co-wrote the main manuscript, contributed to preparing all figures and tables, and performed experiments for each section of the manuscript. He also provided hematopathology interpretations for all flow and pathologic specimens. B.H. contributed to the main text and performed many of the mouse and sequencing experiments. B.F. assisted with the mouse colony, crosses, and genotyping. N.O. provided technical assistance for Figures 4 and 5 and flow cytometry. S.K.- provided expert veterinary pathology services. J.G.- contributed to the development of the mouse strategies and crosses and edited the manuscript. L.D. assisted with DNA sequencing and analyses. A.S. provided statistical guidance for the analyses of insertion sites with TAPDANCE. A.D. provided expertise and oversight in all aspects of Sleeping Beauty strategy and analyses, assisted in analyses of transposon insertions through gCIS, coordinated sequencing strategies, and edited the manuscript. B.C. oversaw the entire project and co-wrote the manuscript and co-prepared all figures and tables.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Loeb, K.R., Hughes, B.T., Fissel, B.M. et al. Insertional mutagenesis using the Sleeping Beauty transposon system identifies drivers of erythroleukemia in mice. Sci Rep 9, 5488 (2019). https://doi.org/10.1038/s41598-019-41805-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41805-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.