Abstract
Papillary thyroid microcarcinoma accounts for a large proportion of papillary thyroid carcinoma, especially among new cases. Many PTMC patients have regional lymph node metastasis, with some experiencing recurrence and even death. However, the risk factors and mechanism by which PTMC relates to these factors are unknown. In this study, differentially expressed genes were identified with microarray from The Cancer Genome Atlas, followed by analysis using the Kyoto Encyclopedia of Genes and Genomes. Immunohistochemistry, immunofluorescence, western blot and Oil Red O staining were carried out to evaluate expression levels and functional alterations. Mesenteric Estrogen Dependent Adipogenesis expression was observed in almost all cases of papillary thyroid microcarcinomas, and the location of expression was associated with histological subtype. High expression was correlated with metastasis and poor disease-free survival. Furthermore, the enrichment analysis indicated that Mesenteric Estrogen Dependent Adipogenesis expression may be associated with metabolic reprogramming to influence metastasis and prognosis. These findings contribute to a better understanding of how Mesenteric Estrogen Dependent Adipogenesis affects metastasis and the prognosis of papillary thyroid microcarcinoma patients and suggest that Mesenteric Estrogen Dependent Adipogenesis expression may be a novel prognostic marker in these patients.
Similar content being viewed by others
Introduction
Papillary thyroid microcarcinoma (PTMC) is defined as a papillary thyroid carcinoma (PTC) not more than 1 cm, accountings for the majority of new cases of PTC. PTC is the most common malignancy of the endocrine system, with a rapid increase incidence in recent years1,2,3,4,5,6. Clinically, most of PTMCs are distinct indolent and follow a rather benign course with good prognosis, while a small portion of cases are characterized by aggressive disease. The clinical management of PTMC is thus a source of significant debate.
In clinical practice, PTMC is usually detected by ultrasound as a thyroid nodule with distinct ultrasonic characteristics. However, ultrasound examination is not ideal for the detection of the lymph node metastasis, which may influence the surgical margins and TNM staging directly. Previous studies showed that approximately 29.0–58.2% of PTMC patients had regional lymph node metastasis (LNM), some of which could not be detected before surgery, with some patients experiencing lymph node recurrence7,8,9. PTMC can be diagnosed by means of fine needle aspiration(FNA), combined with detection of genetic alterations of genes, such as BRAF, KRASand NRAS, but no single molecular aberration has been proven to be useful in directing PTMC treatment to affect clinical outcomes at present10,11,12,13,14,15,16.
According to the most recent guidelines, active surveillance may be considered as a substitute for immediate surgery in PTMC patients without clinically evident metastases or local invasion17. However, these guidelines also emphasize that additional studies are needed to identify the molecular abnormalities that may distinguish the small number of PTMC patients destined to develop clinically significant disease progression18. In short, clarification of the biomarkers and hallmarks associated with poor outcome is urgently needed to further guide PTMC treatment.
To identify the gene expression changes related to the aggressive characteristics of PTMC and its progression, we downloaded the microarray data from TCGA (The Cancer Genome Atlas: https://cancergenome.nih.gov/) and the corresponding clinical data. We found that high expression of Mesenteric Estrogen Dependent Adipogenesis (MEDAG) may be associated with LNM and poor prognosis (p < 0.05, fold change >2). MEDAG, also known as Meda4, is located at 13q12.3 and acts as a regulator of the transcription factor peroxisome proliferator activated receptor gamma (PPARG). Previous investigations suggest that MEDAG overexpression promotes adipogenesis and enhances glucose uptake19, and this gene is also significantly differentially expressed in healing gingiva20.
Materials and Methods
Collection of TCGA public data
RNAseq datasets and clinical data were downloaded from the TCGA database. Gene expression data from 492 PTC samples were downloaded from cBioPortal for Cancer Genomics (https://www.cbioportal.org). A total of 76 patients were included in this study and were each divided into experimental group and control group according to aggressive characteristics (extrathyroid invasion (ETE) or LNM)or poor outcome (recurrence or lethal). RNAseq dataset comparisons between the two groups were performed to find differentially expressed genesusing Student’s t-test. Then Venn diagrams were generated to identify the differentially expressed genes in both the aggressive characteristics group and poor outcome group.
Patients and Thyroid tumors
In total, 67 patient specimens were gathered from December 2014 to December 2015 at the First Hospital Affiliated to ZhengZhou University and diagnosed in the pathology department; BRAFv600 mutation status was detected by qPCR near the time of collection. All surgically resected specimens were fixed in 4% paraformaldehyde. Among the 67 PTMC patients, 57 had undergone total thyroidectomy, and 10 patients had undergone lobectomy with no evidence of LNM during surgery. Total thyroidectomy and simultaneous neck lymph node dissection was performed in 35 patients with suspicious neck LNM on ultrasonic or surgical exploration. Postoperative radioactive iodine (RAI) remnant ablation therapy was conducted after (T4) withdrawal in patients who showed regional lymph node involvement and extrathyroidal invasion. Higher dose RAI therapy was administered when distant metastasis was detected in whole-body scanning post-therapy. There were 29 patients who received RAI therapy. Stages were determined according to the seventh edition of the tumor node metastasis staging system recommended by the American Joint Committee on Cancer. Clinicopathologic parameters such as age at diagnosis, sex, tumor size, extrathyroidal invasion, and presence of lymph node or distant metastasis were obtained from patients’ medical records. The follow-up time ranged from 1 to 32 months with a median follow-up time of 15 months. Then, an additional 14 PTMC and 4 nodular goiter tissue samples were collected from February to April in 2017 and stored under liquid nitrogen for western blot (WB) and Oil Red O (ORO) staining. Mesenteric specimens were collected from 3 patients (female, age> 60 years) who underwent unrelated colon operations as positive controls for immunohistochemistry (IHC) and ORO. Permission to use tumor or normal tissues and paraffin slides was granted by all the patients, in accordance with regulations of the Hospital Human Ethics Committee.
Immunohistochemistry and immunofluorescence staining procedure
IHC staining method was used to detect MEDAG expression in different pathological types of thyroid tissues in 3–µm-thick paraffin sections. Briefly, the slides were first dewaxed in dimethyl benzene, then rehydrated through a gradient of 100%, 95%, 75% and 50% ethanol in water, with antigen retrieval performed in 0.01% sodium citrate solution at 98 °C for 20 min. The slides were treated with 3% H2O2 in methanol to eliminate endogenous peroxidase activity and blocked with normal goat serum for 40 min at room temperature to prevent nonspecific binding; then, the primary antibody was added and incubated at 4 °C overnight. The next morning, the slides were incubated at 37 °C for 40 min, then washed with PBS and incubated with the secondary antibody (PV-6000 polymer detection system, ZsBio Ltd., China) at 37 °C for 2 h. A color substrate solution from the DAB Detection Kit was applied for 3 minutes, followed by rinsing with tap water for 15 min and then counterstaining with hematoxylin for 15 seconds. Finally, the slides were dehydrated in an ethanol gradient and sealed with neutral gum. Negative controls were included by omitting the primary antibody. Visceral fat depot specimens were used as positive controls. The subcellular localization of the staining and the percentage of stained cells were noted. Immunostaining was blinded and semiquantitatively evaluated in terms of staining intensity (0, negative/−; 1, weak/+; 2, intermediate/++; 3, strong/+++) and percentage of stained cells (0, 0–5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4, >75%). An IHC score was calculated by multiplying the proportion of positive cells by the intensity of the staining, with 12 being the maximum score. The cellular localization was also evaluated as cytoplasmic and/or nuclear. All the sections were assessed by two experienced pathologists. In addition, 7 cases of PTMC with inconsistent IHC results were reviewed again by the two pathologists to obtain the final pathological diagnosis. All stained sections were observed under an optical microscope by two experienced pathologists. Immunofluorescence (IF) was used only for special category stain.
For IF procedure, fluorescein-conjugated secondary antibody was substituted for the PV-6001 polymer detection system, with incubation at 37 °C for 2 h, and the slides were counterstained with DAPI (Beyotime, China) for 60 s.
Western blot analysis
Protein expression of MEDAG was evaluated by WB. Total protein was extracted from the paracancer, nodular goiter, and tumor tissues using radioimmunoprecipitation (RIPA) lysis buffer with 1% PMSF (Solarbio, China). The protein concentrations were then determined by measuring the absorbance at 280 nm (A280) (NanoDrop 2000C spectrophotometer, ThermoFisher scientific, US). Equal amounts of proteins were loaded, separated using 12% SDS-PAGE, and transferred to a nitrocellulose membrane (Bio-Rad, USA), which was then blocked for 2 h at room temperature in 5% bovine serum albumin. Subsequently, blots were washed and incubated overnight at 4 °C in Tris-buffered saline containing 0.05% Tween 20 (TBST) with a 1:1000 dilution of the first antibody. Membranes were washed three times with TBST and incubated with secondary antibody at a 1:2000 dilution for 1.5 h at room temperature and then washed three times with TBST. Protein expression was detected by an enhanced chemiluminescence (ECL) method and imaged with a FluorChem Systems instrument (Proteinsimple, USA).
The following antibodies were used in IHC, IF and WB. Primary antibodies: rabbit anti-MEDAG (Signalway Antibody LLC., USA.), mouse anti-β Tubulin (ZsBio Ltd., China), mouse anti-PPARG (Proteintech Group, China). Secondary antibody: fluorescein(FITC)- conjugated goat anti-rabbit IgG (ZsBio Ltd., Beijing, China), TRITC- conjugated goat anti- mouse IgG (ZsBio Ltd., China), goat anti-rabbit IgG HRP conjugated (Signalway Antibody LLC., USA.), HRP-conjugated affinipure goat anti-rabbit IgG (Proteintech Group, China).
Oil Red O staining procedure
Oil Red O (ORO) was performed conventionally. Briefly, frozen PTC and para-cancer tissues were cryosectioned at 10 μm thickness on a freezing microtome (CRYOSTAR NX50, Thermo, China) and stained with Oil Red O(Servicebio, China) for 10 min. After rinsing in distilled water, sections were counterstained for 5 min in hematoxylin. The percentage of ORO-stained cells was determined (0, 0–5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4, >75%).
Differentially expressed gene analysis
Enrichment analysis was carried out by KEGG Mapper21 (https://www.genome.jp/kegg/mapper.html).
Statistical analysis
With SPSS 21.0, the statistical significance of categorical data was determined by χ2 tests or Fisher’s exact test. The Wilcoxon rank sum test was used to compare two sets of data. Survival was analyzed by the Kaplan–Meier method and compared by the log-rank test. The odds ratio (OR) and the confidence interval (CI) for the relationship between MEDAG expression and disease free survival (DFS) were calculated. The survival analysis was based on disease-free survival. Survival time was calculated as the time from the beginning of diagnosis or operation to recurrence or last follow-up. P values of <0.05 or less were deemed to be statistically significant.
Ethics approval and consent to participate
All protocols performed in studies involving human participants were reviewed and approved by the Scientific Research and Clinical Laboratory Ethics Committee of the First Affiliated Hospital of Zhengzhou University, China. Informed consent was obtained from all the patients.
Results
Bioinformatics analysis of gene expression data and clinical data from TCGA
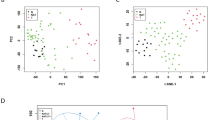
MEDAG was identified as a differentially expressed gene with biological significance and was significantly high in lethal outcome groups (P < 0.05, fold change >2), the LNM group (P < 0.05, fold change >2) and in the recurrence group(P < 0.05, fold change >1.5). Meanwhile, the differentially expressed genes showed almost no intersection between the poor outcome (recurrence/lethal) group and the aggressive (invasion and metastasis) group (Fig. 1).
Differentially expressed gene screen. (A) Heatmap: A subset of genes whose expression was significantly associated with PTMC lymph node metastasis. (B) Venn diagram: Only two genes were overexpressed in both the lymph node metastasis group and the lethal outcome group (P < 0.05, fold change >2). MEDAG was one of these genes and exhibited a fold change >1.5 in the recurrence outcome group. The other instances of expression of this gene were so low as to be of no biological significance. (C) MEDAG mRNA Seq expression between the LNM-positive and LNM-negative groups (#P = 0.029).
Association between MEDAG expression and clinical pathological characteristics in patients with PTMC
Of the 67 patients, 48 (63%) were female and 19 (37%) were male. Their average age at diagnosis was 45.3 years (range, 26–74 years). ETE of the tumor was found in 6 patients (8%). Initial lymph node metastases were noted in 12 patients (30%). Multicentric tumors were detected in 21 patients (28%). Histologically, 63 (94%) tumors were of the classical type, 3 (4%) were follicular variant of papillary thyroid carcinoma (FVPTC), and 1 (1%) was tall cell variant (TCV). There were 61 patients with stage I/II disease (91%) according to the American Joint Committee on Cancer staging criteria. High MEDAG expression in patients with PTMC was positively correlated with LNM (P = 0.003) and multicenter tumors (P = 0.049). However, no obvious relationship was found with age, gender, non-classical status, extrathyroid invasion, distant metastasis, tumor stage or BRAFv600 mutation (P > 0.05) (Table 1).
Expression of MEDAG in PTMC
MEDAG staining was absent in the vast majority of para-cancer tissues (64/67, 95.5%) (Fig. 2A). MEDAG expression was detected in 65 out of 67 tumor samples (97%) (Fig. 2B–H). MEDAG staining was either cytoplasmic, nuclear or both. The staining intensity varied from negative to high (Fig. 2C–H). MEDAG expression in nodular goiter and normal tissue was essentially negative as detected by IHC (Fig. 2I, 2J) and WB analysis (Fig. 2L) compared with PTMC at any TNM stage. The expression of MEDAG was greater in LNM samples than in LNM-negative samples (P = 0.042), and there was a distinct difference in expression between PTC and normal tissue (P = 0.001) (Fig. 2L,M).
Immunohistochemical and western blot analysis of MEDAG in PTC, para-cancer and nodular goiter tissues: Paraffin-embedded sections were stained using an anti-MEDAG monoclonal antibody as described in the Methods section. Benign tissue sections and negative control samples showing no detectable immunohistochemical staining (A,B). Negative(−) or faint(+) cytoplasmic staining was observed in PTC without LNM (C,D). Moderate(++) to strong(+++) cytoplasmic staining were visible in LNM cases (E,F). Nuclear and/or cytoplasmic staining were observed in FVPTC (G,H). Scores for LNM-positive sections are 2.71 (±0.62), compared with LNM-negative 2.20 (±0.98). #P < 0.05, Mann-Whitney Test (K). Immunohistochemical staining of MEDAG in nodular goiter (I). Immunohistochemical staining of MEDAG PTC and para-cancer tissues (J). Ratio of total MEDAG signal to β-tubulin signal in the WB. The data are presented as the mean values ± S.E.M. One-way ANOVA: *P < 0.05; **P, ***P < 0.01 (M). Representative images of WB analysis of benign thyroid tissue, nodular goiter and different stages of PTMC (D). Original magnification (A–G), (I): ×400; (H,J): ×100. (L) The β-Tubulin blots were exposed for 4 s and MEDAG blots exposed for 3 min.
Expression of MEDAG in different histological subtypes
In 62 classical PTC cases, staining was nearly always cytoplasmic (98%) (Figs 2C–F and 3A–D), and only 1 of 62 classical PTCs exhibited both cytoplasmic and nuclear staining (Fig. 2G), which was indicative of the BRAFv600 mutation. MEDAG staining was different in 4 FVPTC cases, in which the BRAFv600 mutation was not detected. Both cytoplasmic and nuclear staining were observed in three cases, and one case exhibited only nuclear staining (Figs 2H and 3E–H). Nuclear or both cytoplasmic and nuclear staining were associated with the FVPTC histopathological subtype (P = 0.025). In tall cell variant PTC, MEDAG staining was observed in the cytoplasm but was limited to the margin (Fig. 3I–L).
Representative photomicrographs show immunostaining of MEDAG in different subtypes of PTC. Strong cytoplasmic staining of MEDAG in the classical PTMC with a detectable BRAFv600 mutation (A), (C) and (D). Strong nuclear staining of MEDAG in the follicular variant of PTMC without a detectable BRAFv600 mutation (E), (G) and (H). Staining of MEDAG was limited to the margin in tall cell variants PTMC. The black arrow indicates the high expression in the cytoplasm (I), (K) and (L) compared to PPARG (red fluorescent in L). Nuclear staining by DAPI (blue fluorescent in B, F and J). MEDAG (green fluorescent in C, D, G, H, K and L). Original magnifications: E × 100; A–D, F–L × 400.
The representative photomicrographs show immunostaining of MEDAG, PPARG and ORO staining of normal thyroid and thyroid carcinomas. In normal thyroid samples, MEDAG staining was negative (A), and in some cases, PPARG immunostaining was negative (B), but ORO staining was positive (C). Lipid droplets were rarely found in PTMC (F), even though MEDAG and PPARG were highly expressed (D,E). There was a statistically significant association between the MEDAG over expression and PPARG expression (G). The ORO staining scores were different between paracancer and PTMC samples (P = 0.00009) (H). The potential for metabolic reprogramming was observed in PTMC (I). Yellow triangles show the lipid droplets in the cytoplasm. *Fisher’s Exact Test, **Wilcoxon test. Original magnifications: B × 100; A,C–F × 400.
Association between MEDAG expression and adipogenesis in PTMC
Previous in vivo and in vitro studies have confirmed the function of MEDAG in adipogenesis. We detected lipid droplets by Oil Red O (ORO) staining in PTMC compared with mesentery samples. MEDAG expression was also detected at the same time. However, few lipid droplets were detected in PTMC, even when MEDAG was highly expressed (Fig. 4D,F), compared with mesentery and normal thyroid tissue (P = 0.00009) (Fig. 4A,C,H). Additionally, there was a statistically significant association between the MEDAG over expression and PPARG expression in PTMC (P = 0.023) (Fig. 4D,E,G). The high expression of MEDAG in PTMC with adipogenesis may indicate that the downstream process of adipogenesis was disrupted or hindered.
Pathway and differential expression analysis of PTMC metabolism in adipogenesis
To explore metabolic reprogramming and its role in PTMC, we performed the the categorization of annotations of the differentially expressed genes (P < 0.05, fold change >2) in KEGG metabolic pathways, and included fatty acid metabolism, lipid metabolism, sterol biosynthesis, glycolysis and the citrate cycle (TCA cycle). The main metabolic pathways in which differentially expressed genes are enriched are highlighted by colored lines on the KEGG metabolism pathway diagram (Fig. 5). Genes with downregulated expression were enriched in fatty acid biosynthesis, triacylglycerol biosynthesis, acylglycerol degradation, and ceramide biosynthesis pathways, etc. in the LNM group(Fig. 5A). In the poor outcome group, the genes with upregulated differential expression were involved in fatty acid biosynthesis, ketone body biosynthesis, triacylglycerol biosynthesis, acylglycerol degradation, ceramide biosynthesis, sphingosine biosynthesis, and sterol biosynthesis pathways, etc. (Fig. 5B). In the lethal outcome group, the genes with upregulated expression were enriched in triacylglycerol biosynthesis, acylglycerol degradation, phosphatidylcholine biosynthesis, glycosphingolipid biosynthesis, bile acid biosynthesis, and steroid hormone biosynthesis pathways. Genes with downregulated expression were enriched in carbon fixation, glycolysis, and citrate cycle pathways (TCA cycle) (Fig. 5C).
The metabolic pathways of differentially expressed genes (P < 0.05, fold change >2). Pathways enriched in up-regulated genes are indicated by red lines, and pathways enriched for down-regulated genes are indicated by blue lines. (A) Differential expression states for genes enriched in the LNM group. (B) Differential expression states for genes enriched in the poor outcome group. (C) Differential expression states for genes enriched in the lethal outcome group21.
Survival analysis
To investigate the prognostic value of MEDAG for PTMC, we evaluated the association between MEDAG expression and survival duration using a Kaplan–Meier analysis based on data from both the TCGA database (Fig. 6A,B) and on our cohort (Fig. 6C). In our study, the median duration of follow-up was 9.6 months (range, 1–32 months). Locoregional recurrence was noted in patients (20.9%), while 1 (1.5%) patient developed metastases; no patients deaths were reported. The log-rank test showed that the DFS was different between the group of PTMC patients with high MEDAG expression and the group with low MEDAG expression (P = 0.0495). In short, the MEDAG over-expression group had a shorter DFS duration. The result in our cohort was similar to that from the TCGA database. In our cohort, MEDAG over-expression was associated with poorer DFS (OR = 1.350, 95% CI 1.153–1.581, P = 0.039), and TNM stage was associated with poorer DFS (OR = 4.352, 95% CI 1.090–17.378, P = 0.028) as well. Other variables, including age ≥45 (OR = 0.670, 95% CI 0.204–2.197, P = 0.507), male sex (OR = 2.308, 95% CI 0.675–7.892, P = 0.176), ETE (OR = 2.042, 95% CI 0.334–12.488, P = 0.432), LNM (OR = 2.308, 95% CI 0.675–7.892, P = 0.176), multifocality (OR = 1.900, 95% CI 0.563–6.407, P = 0.296), and BRAF mutation (OR = 0.780, 95% CI 0.075–8.130, P = 0.835), were not associated with poorer DFS.
High expression of MEDAG was associated with poor overall survival and poor disease-free survival. Kaplan-Meier curves for overall survival (A) and disease-free survival (B) in PTMC patients from the TCGA database. (C) In our cohort, disease-free status is different, as estimated by the Kaplan–Meier method and compared by the log-rank test (P < 0.05).
Discussion
In this study, we initially investigated the potential prognostic biomarkers of PTMC. To identify the molecular aberrations associated with progression and clinical characteristics in PTMCs, we downloaded gene expression data and clinical data from TCGA, and examined the differentially expressed genes. We then used IHC to verify the results in our cohort and confirm that overexpression of MEDAG was related to LNM and poor prognosis in PTMC. Finally, the effects on Oil Red O staining and pathway analysis of differential expression genes indicated that the biological process of adipose tissue synthesis mediated by MEDAG involves metabolic reprogramming in PTMC. In summary, our results imply that MEDAG could be a potential biomarker of PTMC. Our findings may be useful in directing PTMC treatment to affect clinical outcomes, especially in screening low risk PTMC patients with active surveillance to avoid immediate surgery.
This study showed for the first time that MEDAG expression plays a role in cancer. Our result show that MEDAG localized to the cytoplasm in most classical PTMCs, but few lipid droplets were found in PTMC compared with para-cancer or adipose tissue, even when MEDAG was highly expressed in PTMC. However, previous investigations indicate that MEDAG plays an important role in carbohydrate and lipid metabolism in the cytoplasm and acts as a regulator of the transcription factor PPARG. To explain this phenomenon, we hypothesize that PPARG could be depressed and down-regulated by the MAPK signaling pathway as previously reported22,23,24. This signaling pathway is usually activated by RAS mutation, BRAF mutation, or RET/PTC rearrangement in PTC25,26. This indicated that the biological process of adipose tissue synthesis mediated by MEDAG involves metabolic reprogramming27,28,29 (Fig. 4K). Furthermore, since MEDAG is also significantly differentially expressed in healing gingiva20, we believe the overexpression in PTMC could be in line with the hypothesis–“Tumors: wounds that do not heal”30.
In practice, PTMC is more likely to be considered as a clinical subset of PTC with an indolent course, rather than a histological subtype. No animal model or cell lines have been reported as suitable for PTMC research. For example, TERT promoter mutation, a prognostic genetic marker for thyroid cancer31,32,33, is universal in almost all types of thyroid cancer cell lines34, but is a rare genetic event in PTMC35,36. To accurately reflect the lipid metabolism characteristics and mechanisms underlying the distinct prognoses of invasive PTMC, in which MEDAG may be involved, differentially expressed gene analysis was carried out by KEGG Mapper. As shown in Fig. 5, the metabolic features in the LNM group may lead to cytomembrane flexibility to overcome hurdles for cell motility and initiation of ER stress. The former is thought to be necessary for metastasis37,38, and the latter may inhibit PTMC cells growth39,40,41. However, the lethal prognosis group seemed to easily tolerate ER stress, and the poor prognosis group exhibited characteristics of lipid accumulation, which was also reported in the cribriform variant of PTC42. Additionally, we were surprised by the results of the Venn diagram, which shows that the greatest difference overexpressed and underexpressed genes (data not shown) was between the poor outcome group and aggressive clinical characteristics group. This suggests that the poor prognosis group may be a special subset in PTMC in which tumor progression and metastasis may be distinct processes38.
The localization of MEDAG to the nucleus was also observed in FVPTC, especially in BRAFv600 mutant-negative samples in our study. FVPTC is considered to have fewer BRAFv600 mutations but more PAX/PPARG rearrangements, which can also be combined with other genetic mutations. In PTC with PAX/PPARG rearrangement, abnormal hyperactivation of the PAX8 promoter leads to increased expression of the fusion protein43. Therefore, we hypothesized that MEDAG, which acts as a positive transcription factor, may be restricted within PTC’s nuclei with PAX/PPARG rearrangement.
In our study, we observed a trend towards decreased expression and localization of MEDAG in TCV. Although it is unknown whether the cause of this tendency is protein or mRNA degradation, it may be beneficial to TCV cancer cells during cellar stress. Similar to anaplastic thyroid carcinoma (ATC), TCV exhibits a higher proportion of TP53 mutation than does classical PTC, and both of them have poorer prognosis44,45. This phenomenon may be in line with the metabolic feature of the poor prognosis group and the “second hit” hypothesis46. Additionally, we suspect that there is a potential mechanism by which the reducing in MEDAG would free the tumor from inertia in non-PTMC.
In our resaerch, we investigated the potential prognostic biomarkers of PTMC and their underlying mechanisms using clinical samples and by bioinformatics methods, as no animal model or cell lines have been reported suitable for PTMC research. Thus, additional rationally designed experiments need to be carried out in vivo and in vitro. And we also observed that the K-M curve is not sufficient with regards to DFS (Fig. 6A,6C). This partly rely on differences in the definition of DFS, expression platform or that follow-up time is not long enough for this clinically indolent tumor. As classical PTC accounts for the majority of PTC in both the TCGA database and our cohort, the biomarker analysis was almost entirely based on classical PTMC. So, we speculate that various histological subtypes of PTC should be taken into account, when the MEDAG expression will be evaluated in PTMC FNA samples.
Conclusion
In summary, our results are useful for diagnosing PTMC and providing information about prognosis and the likelihood of recurrence after surgery. MEDAG expression could be a novel prognostic marker for PTMC patients. Additional studies are needed in non-PTMC cases as well as in vivo and in vitro studies.
Data Availability
Microarray data and clinical data referred to in this article can be found in TCGA. Please contact author for all other data requests.
References
Olaleye, O. et al. Increasing incidence of differentiated thyroid cancer in South East England: 1987–2006. European archives of oto-rhino-laryngology: official journal of the European Federation of Oto-Rhino-Laryngological Societies 268, 899–906, https://doi.org/10.1007/s00405-010-1416-7 (2011).
Chen, A. Y., Jemal, A. & Ward, E. M. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer 115, 3801–3807, https://doi.org/10.1002/cncr.24416 (2009).
Li, N., Du, X. L., Reitzel, L. R., Xu, L. & Sturgis, E. M. Impact of enhanced detection on the increase in thyroid cancer incidence in the United States: review of incidence trends by socioeconomic status within the surveillance, epidemiology, and end results registry, 1980–2008. Thyroid: official journal of the American Thyroid Association 23, 103–110, https://doi.org/10.1089/thy.2012.0392 (2013).
Tuttle, R. M. et al. Thyroid carcinoma, version 2.2014. Journal of the National Comprehensive Cancer Network: JNCCN 12, 1671–1680; quiz 1680 (2014).
Liu, C. et al. Reevaluating the prognostic significance of male gender for papillary thyroid carcinoma and microcarcinoma: a SEER database analysis. Scientific reports 7, 11412, https://doi.org/10.1038/s41598-017-11788-8 (2017).
Rahib, L. et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research 74, 2913–2921, https://doi.org/10.1158/0008-5472.CAN-14-0155 (2014).
Xu, J. M. et al. A Risk Model for Predicting Central Lymph Node Metastasis of Papillary Thyroid Microcarcinoma Including Conventional Ultrasound and Acoustic Radiation Force Impulse Elastography. Medicine 95, e2558, https://doi.org/10.1097/MD.0000000000002558 (2016).
Li, M. et al. Risk factors for predicting central lymph node metastasis in papillary thyroid microcarcinoma (CN0): a study of 273 resections. European review for medical and pharmacological sciences 21, 3801–3807 (2017).
Lin, D. Z. et al. Risk prediction and clinical model building for lymph node metastasis in papillary thyroid microcarcinoma. OncoTargets and therapy 9, 5307–5316, https://doi.org/10.2147/OTT.S107913 (2016).
Cancer Genome Atlas Research, N. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690, https://doi.org/10.1016/j.cell.2014.09.050 (2014).
Vuong, H. G. et al. A meta-analysis of prognostic roles of molecular markers in papillary thyroid carcinoma. Endocrine connections 6, R8–R17, https://doi.org/10.1530/EC-17-0010 (2017).
Liu, C., Chen, T. & Liu, Z. Associations between BRAF (V600E) and prognostic factors and poor outcomes in papillary thyroid carcinoma: a meta-analysis. World journal of surgical oncology 14, 241, https://doi.org/10.1186/s12957-016-0979-1 (2016).
Vuong, H. G., Altibi, A. M. A., Duong, U. N. P. & Hassell, L. Prognostic implication of BRAF and TERT promoter mutation combination in papillary thyroid carcinoma-A meta-analysis. Clinical endocrinology 87, 411–417, https://doi.org/10.1111/cen.13413 (2017).
Chen, Y. et al. BRAF (V600E) Is Correlated with Recurrence of Papillary Thyroid Microcarcinoma: A Systematic Review, Multi-Institutional Primary Data Analysis, and Meta-Analysis. Thyroid: official journal of the American Thyroid Association 26, 248–255, https://doi.org/10.1089/thy.2015.0391 (2016).
McKelvie, P. A. et al. The prognostic significance of the BRAF V600E mutation in papillary thyroid carcinoma detected by mutation-specific immunohistochemistry. Pathology 45, 637–644, https://doi.org/10.1097/PAT.0000000000000008 (2013).
Gouveia, C. et al. Lack of association of BRAF mutation with negative prognostic indicators in papillary thyroid carcinoma: the University of California, San Francisco, experience. JAMA otolaryngology–head & neck surgery 139, 1164–1170, https://doi.org/10.1001/jamaoto.2013.4501 (2013).
Ito, Y. & Miyauchi, A. Nonoperative management of low-risk differentiated thyroid carcinoma. Current opinion in oncology 27, 15–20, https://doi.org/10.1097/CCO.0000000000000143 (2015).
Haugen, B. R. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: What is new and what has changed? Cancer 123, 372–381, https://doi.org/10.1002/cncr.30360 (2017).
Zhang, H., Chen, X. & Sairam, M. R. Novel genes of visceral adiposity: identification of mouse and human mesenteric estrogen-dependent adipose (MEDA)-4 gene and its adipogenic function. Endocrinology 153, 2665–2676, https://doi.org/10.1210/en.2011-2008 (2012).
Wang, Y. & Tatakis, D. N. Human gingiva transcriptome during wound healing. Journal of clinical periodontology 44, 394–402, https://doi.org/10.1111/jcpe.12669 (2017).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research 45, D353–D361, https://doi.org/10.1093/nar/gkw1092 (2017).
Camp, H. S., Tafuri, S. R. & Leff, T. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma1 and negatively regulates its transcriptional activity. Endocrinology 140, 392–397, https://doi.org/10.1210/endo.140.1.6457 (1999).
Buffet, C. et al. DUSP5 and DUSP6, two ERK specific phosphatases, are markers of a higher MAPK signaling activation in BRAF mutated thyroid cancers. PloS one 12, e0184861, https://doi.org/10.1371/journal.pone.0184861 (2017).
Gertz, R. J., Nikiforov, Y., Rehrauer, W., McDaniel, L. & Lloyd, R. V. Mutation in BRAF and Other Members of the MAPK Pathway in Papillary Thyroid Carcinoma in the Pediatric Population. Archives of pathology & laboratory medicine 140, 134–139, https://doi.org/10.5858/arpa.2014-0612-OA (2016).
Zou, M. et al. Concomitant RAS, RET/PTC, or BRAF mutations in advanced stage of papillary thyroid carcinoma. Thyroid: official journal of the American Thyroid Association 24, 1256–1266, https://doi.org/10.1089/thy.2013.0610 (2014).
Cordioli, M. I. et al. Fusion Oncogenes Are the Main Genetic Events Found in Sporadic Papillary Thyroid Carcinomas from Children. Thyroid: official journal of the American Thyroid Association 27, 182–188, https://doi.org/10.1089/thy.2016.0387 (2017).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674, https://doi.org/10.1016/j.cell.2011.02.013 (2011).
Parrales, A. & Iwakuma, T. p53 as a Regulator of Lipid Metabolism in Cancer. International journal of molecular sciences 17, https://doi.org/10.3390/ijms17122074 (2016).
Swierczynski, J., Hebanowska, A. & Sledzinski, T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World journal of gastroenterology 20, 2279–2303, https://doi.org/10.3748/wjg.v20.i9.2279 (2014).
Dvorak, H. F. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. The New England journal of medicine 315, 1650–1659, https://doi.org/10.1056/NEJM198612253152606 (1986).
Lee, S. E. et al. Prognostic Significance of TERT Promoter Mutations in Papillary Thyroid Carcinomas in a BRAF (V600E) Mutation-Prevalent Population. Thyroid: official journal of the American Thyroid Association 26, 901–910, https://doi.org/10.1089/thy.2015.0488 (2016).
Moon, S. et al. Effects of Coexistent BRAF (V600E) and TERT Promoter Mutations on Poor Clinical Outcomes in Papillary Thyroid Cancer: A Meta-Analysis. Thyroid: official journal of the American Thyroid Association 27, 651–660, https://doi.org/10.1089/thy.2016.0350 (2017).
Insilla, A. C. et al. TERT promoter mutations and their correlation with BRAF and RAS mutations in a consecutive cohort of 145 thyroid cancer cases. Oncology letters 15, 2763–2770, https://doi.org/10.3892/ol.2017.7675 (2018).
Schweppe, R. E. et al. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. The Journal of clinical endocrinology and metabolism 93, 4331–4341, https://doi.org/10.1210/jc.2008-1102 (2008).
Yabuta, T. et al. TERT Promoter Mutations Were Not Found in Papillary Thyroid Microcarcinomas That Showed Disease Progression on Active Surveillance. Thyroid: official journal of the American Thyroid Association 27, 1206–1207, https://doi.org/10.1089/thy.2016.0645 (2017).
de Biase, D. et al. TERT Promoter Mutations in Papillary Thyroid Microcarcinomas. Thyroid: official journal of the American Thyroid Association 25, 1013–1019, https://doi.org/10.1089/thy.2015.0101 (2015).
Yamaguchi, H. & Oikawa, T. Membrane lipids in invadopodia and podosomes: key structures for cancer invasion and metastasis. Oncotarget 1, 320–328, https://doi.org/10.18632/oncotarget.100907 (2010).
Li, J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nature communications 1, 34, https://doi.org/10.1038/ncomms1033 (2010).
Colgan, S. M., Hashimi, A. A. & Austin, R. C. Endoplasmic reticulum stress and lipid dysregulation. Expert reviews in molecular medicine 13, e4, https://doi.org/10.1017/S1462399410001742 (2011).
Han, J. & Kaufman, R. J. The role of ER stress in lipid metabolism and lipotoxicity. Journal of lipid research 57, 1329–1338, https://doi.org/10.1194/jlr.R067595 (2016).
Leignadier, J., Dalenc, F., Poirot, M. & Silvente-Poirot, S. Improving the efficacy of hormone therapy in breast cancer: The role of cholesterol metabolism in SERM-mediated autophagy, cell differentiation and death. Biochemical pharmacology 144, 18–28, https://doi.org/10.1016/j.bcp.2017.06.120 (2017).
Takada, N. et al. Cytoplasmic Lipid Accumulation Characteristic of the Cribriform Variant of Papillary Thyroid Carcinoma. Pathobiology: journal of immunopathology, molecular and cellular biology 84, 251–257, https://doi.org/10.1159/000477910 (2017).
Armstrong, M. J. et al. PAX8/PPARgamma rearrangement in thyroid nodules predicts follicular-pattern carcinomas, in particular the encapsulated follicular variant of papillary carcinoma. Thyroid: official journal of the American Thyroid Association 24, 1369–1374, https://doi.org/10.1089/thy.2014.0067 (2014).
Morita, N., Ikeda, Y. & Takami, H. Clinical significance of p53 protein expression in papillary thyroid carcinoma. World journal of surgery 32, 2617–2622, https://doi.org/10.1007/s00268-008-9756-9 (2008).
Ruter, A., Dreifus, J., Jones, M., Nishiyama, R. & Lennquist, S. Overexpression of p53 in tall cell variants of papillary thyroid carcinoma. Surgery 120, 1046–1050 (1996).
Smallridge, R. C., Marlow, L. A. & Copland, J. A. Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocrine-related cancer 16, 17–44, https://doi.org/10.1677/ERC-08-0154 (2009).
Acknowledgements
We thank the department of Biochemical and Molecular Biology, Basic Medical Science, Zhengzhou University for Advanced Microscopy and FluorChem Systems. We also thank Li Hao-hao, Gastrointestinal Surgery Department of The First Affiliated Hospital of Zhengzhou University, for the collection of mesenteric specimens. This work was supported by grants from the Foundation of Henan Educational Committee (18A320008).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Y.S., X.G.Q. Performed theexperiments: Y.S., L.J.F., X.G.Q. Analyzed the data: Y.S., H.T.L., X.G.Q. Contributed reagents/materials/analysis tools: Y.S., L.J.F., H.T.L., X.G.Q. Wrote the manuscript: Y.S., L.J.F., H.T.L., X.G.Q. Clinical evaluation of subjects and procurement of tissue samples: Y.S., H.T.L., X.G.Q. Revised the manuscript: X.G.Q.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, Y., Fu, Lj., Li, Ht. et al. Evaluation of MEDAG gene expression in papillary thyroid microcarcinoma: associations with histological features, regional lymph node metastasis and prognosis. Sci Rep 9, 5800 (2019). https://doi.org/10.1038/s41598-019-41701-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41701-4
This article is cited by
-
Papillary thyroid microcarcinoma with lung metastases: a case report and review of the literature
Thyroid Research (2021)
-
MEDAG enhances breast cancer progression and reduces epirubicin sensitivity through the AKT/AMPK/mTOR pathway
Cell Death & Disease (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.