Abstract
Rev-erbα is a ligand-dependent nuclear receptor and a key repressor of the molecular clock transcription network. Accumulating evidence indicate that the circadian clock machinery governs diverse biological processes in skeletal muscle, including muscle growth, repair and mass maintenance. The physiological function of Rev-erbα in myogenic regulation remains largely unknown. Here we show that Rev-erbα exerts cell-autonomous inhibitory effects on proliferation and differentiation of myogenic precursor cells, and these actions concertedly inhibit muscle regeneration in vivo. Mechanistic studies reveal Rev-erbα direct transcriptional control of two major myogenic mechanisms, proliferative pathway and the Wnt signaling cascade. Consistent with this finding, primary myoblasts lacking Rev-erbα display significantly enhanced proliferative growth and myogenic progression. Furthermore, pharmacological activation of Rev-erbα activity attenuates, whereas its inhibition by an antagonist promotes these processes. Notably, upon muscle injury, the loss-of-function of Rev-erbα in vivo augmented satellite cell proliferative expansion and regenerative progression during regeneration. Collectively, our study identifies Rev-erbα as a novel inhibitory regulator of myogenic progenitor cell properties that suppresses postnatal myogenesis. Pharmacological interventions to dampen Rev-erbα activity may have potential utilities to enhance regenerative capacity in muscle diseases.
Similar content being viewed by others
Introduction
Rev-erbα is a member of the nuclear receptor superfamily (Subfamily 1 group D member 1, NR1D1), and a key circadian clock repressor that inhibits core clock activator Brain and Muscle Arnt-line 1 (Bmal1) transcription1. Rev-erbα is highly expressed in metabolic tissues, with known functions in conferring circadian clock integration to glucose, lipoprotein and bile acid metabolism2,3. Interestingly, although the Rev-erbα metabolic functions have been extensively studied, its role in skeletal muscle growth and repair processes is largely unknown.
Accumulating evidence indicate that the circadian clock is critical for maintenance of skeletal muscle mass and growth4. The circadian clock circuit, composed of an inter-locking molecular network of transcriptional and translational regulators5,6, participates in diverse aspects of skeletal muscle functions ranging from oxidative metabolism, sarcomeric structure, contractile performance to muscle mass maintenance7,8,9,10,11. A remarkable ~17% of all genes examined in skeletal muscle exhibit circadian oscillations12, with many involved in proliferative or growth processes.
Bmal1 is the essential transcription regulator of the clock transcriptional feedback circuit. Bmal1 dimerization with Circadian Locomotor Output Cycles Kaput (CLOCK) activates downstream target genes, Period (Per1, 2 and 3) and Cryptochrome genes (Cry1 and 2) that in turn inhibits Bmal1/CLOCK activity5. Rev-erbα potently suppresses Bmal1 transcription through a shared response element with retinoid acid receptor-related orphan receptor alpha (RORα), the RevRE/RORE1. Rev-erbα represses, whereas RORα activates Bmal1 gene transcription, and this antagonistic regulation elicit Bmal1 rhythmic oscillation. Interestingly, Rev-erbα itself is a direct target of Bmal1, and the Rev-erbα-Bmal1 regulation constitutes a re-enforcing branch that enhances the robustness of the core clock machinery1,13.
Regenerative myogenesis is a highly orchestrated progression involving myogenic precursor cell (MPC) proliferation, differentiation and fusion to form multinucleated myofibers for muscle repair14,15. Recent studies indicate that the circadian clock regulation is critical for muscle growth and function4. Loss of Bmal1 leads to severe aging-associated sarcopenia9, and Bmal1 and CLOCK are required for myofilament integrity7. We recently demonstrated that Bmal1 promotes myogenic precursor proliferation and differentiation required for skeletal muscle regenerative myogenesis16,17, with its ablation leading to significantly impaired satellite cell proliferative expansion and muscle regeneration following injury. Our findings of Bmal1 modulation of MPC properties implicate potential intrinsic clock control in muscle repair that may require additional clock components. Recent cistrome analysis of Rev-erbα demonstrate an extensive ~28% overlap with Bmal118,19, suggesting that they may exert opposing transcriptional controls of shared targets.
As a ligand-dependent nuclear receptor, Rev-erbα synthetic ligands are currently available to interrogate its biological functions and potential pharmacological interventions of the circadian clock20,21,22. Based on Bmal1 regulation of the myogenic cascade, we hypothesize that the transcription repressor Rev-erbα may inhibit myogenesis to suppress regenerative repair. In the current study, we employed genetic and pharmacological approaches to probe the physiological functions of Rev-erbα in myogenic regulations and muscle regeneration.
Materials and Methods
Animals
Mice were maintained in the Baylor College of Medicine vivarium under a constant 12:12 light dark cycle. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the IACUC committee of Baylor College of Medicine. Global gene-targeted Rev-erbα-null (Rev−/−) mice were generated by the Genetically Engineered Mouse Core at Baylor College of Medicine using targeted embryonic stem cells (Velocigene ES clone 11705A-E7, KOMP), as described previously23. Male mice of 8–10 weeks of age were used for the study. Homozygote Rev−/− mice were obtained through heterozygote breeding, with age-matched littermate WT mice as controls.
Primary myoblast isolation and culture
Primary myoblasts were isolated from hind limb muscle of 4 week-old mice, as described16. Briefly, muscles were minced and subjected to collagenase digestion and seeded on collagen-coated plates, using pre-plating to deplete fibroblasts. Cells were subsequently expanded in myoblast growth media for 6 passages. Purity of myoblasts obtained was confirmed by uniform differentiation into myosin heavy chain (MyHC)-positive myotubes. Proliferative myoblast cultures were maintained in F-10 medium supplemented with 20% FBS and 5 ng/ml bFGF. 2% horse serum supplemented DMEM was used for induction of differentiation. Crushed muscle extract (CME) was obtained as described previously17, from leg muscles from 8–10 week old C57BL/6 mice gently pressed using blunt forceps. Supernatant after 2 hours 4 °C incubation in Tris-buffered saline was obtained and protein concentration determined. Primary myoblast were treated by muscle extract at a protein concentration of 300 μg/ml.
Microarray Analysis
Mouse WG-6 V2.0 whole genome expression arrays were obtained from Illumina. Total RNA from three independent primary myoblast samples were used to synthesize cRNA using Illumina TotalPrep RNA amplification kit (Thermo Scientific). Biotin Labeling and hybridization of cRNA were performed according to manufacturer’s protocol, as described24. Arrays were scanned using BeadArray Reader and the data extracted using Illumina BeadStudio software. Unmodified microarray data were background-subtracted and quantile-normalized using Lumi and analyzed with the limma package within R to test differentially gene expression25. All analyses were corrected for multiple hypothesis testing26, and effects determined as significant with an adjusted p-value ≤ 0.05. The microarray dataset was deposited in NCBI Gene Expression Omnibus accession number GSE110765. KEGG pathway analysis was carried out among significantly regulated genes to identify enriched biological processes.
EdU labelling and proliferation assay in vitro
Primary myoblasts seeded on collagen-coated chamber slides were labeled with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) for 6 hours. EdU incorporation was detected by Click-iT Imaging Kit (Invitrogen) with Alexa 488 Fluorophore and DAPI was used to label nuclei. Total number of EdU+ cells was calculated from 4 representative fields and proliferation rate calculated as percentage of EdU+ to total nuclei.
Quantitative TUNEL assay
Primary myoblasts were plated on collagen-coated 96 well plate. Cells were treated with endonuclease to induce apoptosis as positive control. Terminal deoxynucleotidyl transferase in situ labeling and detection of apoptosis was performed 24 hours after seeding by colorimetric TACS apoptosis detection kit (Trevigen), according to manufacturer’s protocol.
Analysis of Wnt signaling activity
Wnt activity in primary myoblasts, as assessed by β-catenin intracellular localization at basal state without stimulation and after Wnt3a stimulation, was carried out as described16. Briefly, primary myoblasts were treated by unconditioned or conditioned media from L3-Wnt3a (40%) for 10 minutes, and nuclear fraction was collected by ultracentrifugation. β-catenin level in the nuclear and total cellular extracts were analyzed by immunoblot. Ratio of the nuclear fraction to total β-catenin was used to assess Wnt signaling activity after normalization to TBP. TopFlash Wnt luciferase reporter assay was performed as described16. SR9011 and SR8278 (10 nM) were added for 8 hours prior to Wnt3a stimulation overnight. Luciferase activity was measured using Dual-Glo luciferase assay (Promega) 24 hours after treatment and values normalized to Renilla readings.
Muscle Injury Model
Cardiotoxin (CTX, 30 μl of 10 μM solution) was injected into Tibialis Anterior muscle along the longitudinal axis, as described27,28, with PBS-injected animals as controls. Muscles on post-injury days were collected at 4 PM, the time of CTX injection, to ensure comparable circadian time sampling. Tissues were fixed in 10% paraformaldehyde for paraffin sections, or frozen in liquid nitrogen-cooled isopentane for cryosections. Histological analysis was performed in middle region of the Tibialis Anterior. Myofiber cross-section areas were measured in H/E staining sections of five representative fields of (150 myofibers per mice), using the Nikon NIS-Elements software. For EdU in vivo labeling, mice were injected i.p. of EdU (5 mg/Kg) 6 hours prior to sample collection. Click-iT EdU kit (ThemoFisher) was used for EdU detection.
Histology, immunofluorescence staining and morphometric analysis
Muscles were snap frozen in liquid-nitrogen cooled isopentane and embedded in OCT for cryosections. 10 μm cryosections of middle region of the injured TA muscle were used for immunostaining. Sections were fixed in 4% paraformaldehyde and permeabilized by 0.2% Triton X-100, and endogenous IgG blocked prior to primary antibody incubation. The number of Pax7+ satellite cells or EdU+Pax7+ proliferative satellite cells was determined from three representative fields of each mice, and percentage of Pax7+ satellite cells to total nuclei or myofibers were calculated.
Immunoblot analysis
20–40 µg of total protein was resolved on SDS-PAGE gel followed by immunoblotting after nitrocellulose membrane transfer, as described previously (Chatterjee et al., 2013), and developed by chemiluminescence (Supersignal; Pierce Biotechnology). Source and dilution information of primary antibodies are included as Suppl. Table 1. Specific secondary antibodies were used at a dilution of 1:3000.
RNA extraction and quantitative reverse-transcriptase PCR analysis
RNeasy miniprep kits (Qiagen) were used to isolate total RNA from snap-frozen muscle tissues and cells, respectively. cDNA was generated using q-Script cDNA Supermix kit (Quanta Biosciences) and quantitative PCR was performed using a Roche 480 Light Cycler with Perfecta SYBR Green Supermix (Quanta Biosciences). Relative expression levels were determined using the comparative Ct method to normalize target genes to 36B4 internal control. Primer sequences used are listed in Suppl. Table 2.
Chromatin Immunoprecipipation (ChIP)-qPCR Analysis
Immunoprecipitation was performed using a Rev-erbα antibody specific for IP (Proteintech 14506-1-AP), or control rabbit IgG with Magnetic Protein A/G beads, using the Magna ChIP A/G kit (Millipore), similarly as described previously29,30. Briefly, C2C12 myoblasts were fixed using 1% formaldehyde for 10 minutes, lysed and sonicated using Biodisruptor to shear the chromatin. The immunoprecipitated chromatin fragments were eluted, treated with proteinase K and purified using DNA purification column. Real-time PCR using Perfecta SYBR Green Supermix (Quanta Biosciences) was carried out using 1 µl of eluted chromatin for each reaction in triplicates with specific primers for predicted RORE response elements identified on the target promoters or first intron. Known Rev-erbα binding site in Bmal1 first intron was included as positive control and TBP first exon genomic primers as negative control. ChIP rimer sequences used are listed in Supplementary Material Table S3. Values were expressed as fold enrichment normalized to IgG control.
Statistical analysis
Data are presented as mean ± SE. One way ANOVA or two-tailed Student’s t-test were performed as appropriate as indicated. Non-parametric Kruskal–Wallis ANOVA for entire dataset and Mann-Whitney test for individual cross section area categories were used to assess statistical difference in myofiber size distribution. Statistical analyses were carried out using Prism by GraphPad. P < 0.05 was considered statistically significant.
Results
Rev-erbα is highly enriched in skeletal muscle and responds to myogenic stimuli
To study the function of Rev-erbα in skeletal muscle, we examined the abundance of its protein expression among distinct tissues and muscle fiber types at 9 AM and 5 PM. Interestingly, at both times of the day, Rev-erbα protein is more abundant in skeletal muscles as compared to heart, liver and white adipose tissue (Fig. 1A). Among the muscle types, a distinct diurnal pattern of expression with higher expression at 5 PM than 9 AM is observed in slow-twitch Type I soleus and mixed fiber type of Tibialis Anteria, but not evident in the fast-twitch extensor digitalis longus (EDL). The average expression in diaphragm is low as compared to other muscles. Thus the current study, we confined our sample collection to 4–5 PM when Rev-erbα protein, particularly in Tibialis Anterior, is more abundant. Further analysis of clock components in Tibialis Anterior reveal that Rev-erbα mRNA level is nearly 6-fold of that of Rev-erbβ, a closely related gene of Rev-erbα (Fig. 1B). Both Rev-erbα and Rev-erbβ are expressed at higher levels than the core clock gene Bmal1 that is expressed at comparable level with the myogenic factor Myod1. The relative expression of Rev-erbα transcript to Rev-erbβ and Bmal1 is similar in C2C12 myoblasts, although lower than that of the Myod1 (Fig. 1C). To explore potential Rev-erbα involvement in myogenic response in vivo, we examined the dynamics of its expression following cardiotoxin (CTX)-induced muscle injury. CTX elicits regenerative myogenic repair, as indicated by strong myogenin induction at 5 and 8 days post injury, which gradually declines till day 15 (Fig. 1D). In sharp contrast to myogenin induction, Rev-erbα protein was markedly suppressed at day 5, with nearly abolished expression at 8 days after injury and recovered slightly at day 15. This finding suggests muscle damage or its associated myogenic signals may inhibit Rev-erbα. Indeed, treatment of primary myoblasts with crushed muscle extract (CME), obtained from lightly traumatized muscle that is known to contain myogenic stimuli31, results in significant down-regulation of Rev-erbα mRNA (Fig. 1E). The suppression of Rev-erbα occurs early at 6 hours after CME treatment and precedes the induction of myogenic markers at 12 hours, including myogenin and embryonic myosin heavy chain genes (Myhc3). Thus, Rev-erbα inhibition by muscle injury may occur as an early response to myogenic stimuli. One of the known mitogenic component of CME tested, Fgf2, can indeed suppress Rev-erbα protein at either a low or high concentration in primary myoblast cultures (Fig. 1F).
Rev-erbα expression and dynamic regulation in skeletal muscle and myoblasts. (A) Immunoblot analysis of time-dependent expression profile of Rev-erbα protein in distinct tissues at 9 AM and 5 PM. Each lane represents pooled protein sample of n = 5. (B,C) RT-qPCR analysis of comparison of Rev-erbα and related core circadian clock gene expression in mice quadricep (B, n = 6), and C2C12 myoblasts (C, n = 3). (D) Immunoblot analysis of dynamic expression profiles of Rev-erbα and myogenin during 15 days post cardiotoxin-induced injury of Tibialis Anteria (TA) muscle (n = 5–6/time point). (E) RT-qPCR analysis of Rev-erbα and myogenic gene expression in primary myoblasts (n = 4/group) following treatment of crushed muscle extract (CME) at a protein concentration of 300 μg/ml, for 6 and 12 hours respectively. ***P ≤ 0.05 or 0.01 by Student’s t test. (F) Suppression of Rev-erbα protein expression as analyzed by immunoblot analysis in response to myogenic stimuli Fgf2 treatment in primary myoblasts for 24 hours (n = 4/group). Fgf2 concentration: low: 2.5 ng/ml, high: 25 ng/ml.
Gene expression profiling reveals Rev-erbα regulation of Wnt and proliferative pathways
We performed gene expression profiling to explore Rev-erbα functions in MPC using proliferative satellite cell-derived primary myoblasts from WT and Rev−/− mice. Differentially expressed genes, with fold change ≥ 1.5, were subjected to KEGG pathway analysis to identify Rev-erbα-differentially regulated biological functions. Top 15 ranked biological processes were listed (Fig. 2A), with the complete dataset deposited as NCBI GEO GSE110765. Consistent with the established metabolic functions of Rev-erbα, metabolic pathways are shown as top Rev-erbα-regulated processes in these cells, while potentially novel Rev-erbα-modulated functions, such as axon guidance, drug metabolism and protein metabolism, were identified. Notably, Wnt signaling and cancer pathways are ranked as the top two Rev-erbα-regulated pathways. As Wnt pathway is a major developmental signal that drives muscle development and repair32,33,34, Rev-erbα regulation of this cascade implicates its potential functions in myogenenic differentiation. In contrast, Rev-erbα differentially-regulated targets in cancer pathways are critical cell cycle regulators that could be involved in MPC proliferation. Additional pathways are identified that could also modulate myogenesis, including MAPK and actin cytoskeleton regulations35,36. Based on this analysis, we next examined potential Rev-erbα regulation of myogenesis through the Wnt and proliferative pathways.
Rev-erbα transcriptional regulation of the Wnt signaling pathway and cell cycle regulators. (A) KEGG pathway gene enrichment analysis of differentially expressed genes in Rev−/− vs. WT primary myoblasts. Genes that were significantly regulated with fold change ≥ 1.5 and adjusted P ≤ 0.05 from microarray analysis were analyzed (n = 3). Top 10 most significantly regulated biological pathways ranked by adjusted P values are listed. The dataset is deposited as NCBI GSE110765. (B) ChIP-qPCR analysis of Rev-erbα occupation of identified RORE sites in regulatory regions ±1 kb of candidate target genes in Wnt signaling or cell cycle regulators in proliferative C2C12 myoblasts (n = 3). Values are expressed as fold enrichment over IgG and known Rev-erbα binding site in Bmal1 first intron was included as positive control.
Based on Rev-erbα differentially-regulated transcripts revealed by gene expression profiling, we next identified its candidate target genes in the Wnt and proliferative pathways. Following screening for putative consensus Rev-erbα binding sequence ROR-response element (RORE) within the +2 kb promoter region plus 1 kb downstream of transcription start site among these genes, ChIP-qPCR was performed to detect direct Rev-erbα chromatin occupancy on identified regulatory sites (Fig. 2B). Indeed, using the RORE Rev-erbα binding site on its known target Bmal1 first intron as a positive control, robust enrichment of chromatin association comparable to the known Bmal1 site were detected in a number of genes in Wnt signaling cascade as well as cell cycle regulators (c-Met, Cyclin D1 and D2). The striking finding of direct Rev-erbα transcriptional control of distinct steps along the Wnt pathway, including the ligand Wnt10a, receptors (Fzd5, Lrp6, Dvl2), and the transcription mediator (Tcf3), suggests that it may exert robust coordinated regulation of Wnt signaling, whereas its control of multiple proliferative genes indicates potential inhibitory effect on cell cycle.
Ablation of Rev-erbα promotes myogenic gene expression in skeletal muscle and myoblast proliferation
Global Rev-erbα-null (Rev−/−) mice were generated as previously described23. In Rev−/− mutant mice, Rev-erbα transcript in representative mixed fiber type muscle quadriceps is undetectable (Fig. 3A). Due to the loss of Rev-erbα repression activity on its direct target gene, Bmal1 mRNA level is markedly up-regulated ~5-fold as compared to the WT, as expected. The loss of Rev-erbα does not affect muscle development and growth, and histological analysis of quadriceps muscle cross sections of 8-week-old Rev−/− mice did not reveal obvious defects.
Rev-erbα inhibits myoblast proliferation. (A) RT-qPCR analysis of Rev-erbα and Bmal1 mRNA in quadriceps of WT and Rev−/− mice (n = 6/group). (B–G) Genetic loss-of-function of Rev-erbα promotes proliferation of primary myoblasts. (B) RT-qPCR analysis of expression of cell cycle regulators (n = 4). (C) Immunoblot analysis of Cyclin D1 protein level before and during differentiation in WT and Rev−/− primary myoblasts (n = 3/time point). (D) Growth rate of primary myoblasts as assessed by increase in cell numbers monitored for 7 days in culture. Cells were seeded at a starting density of 1 × 106/10 cm plate (n = 4/time point). **p ≤ 0.01 by one-way ANOVA. (E,F) Representative images of EdU labeling with corresponding DAPI image to assess proliferation (E) with quantification normalized to total cell numbers assessed by DAPI (F, n = 6). (G) Analysis of apoptosis by quantitative TUNEL assay in primary myoblasts (n = 5/group). **P < 0.01 Rev−/− vs. WT by Student’s t test. (H,I) Effects of Rev-erbα ligands, agonist SR9011 and antagonist SR8278, on primary myoblast proliferation, as represented by quantification of percent of EdU+ cells/DAPI (H, n = 6/treatment group), with representative images of EdU labelling and DAPI shown (I, 10X). Cells were treated with DMSO (0.1%), SR9011 (10 nM) or SR8278 (10 nM) for 24 hours prior to EdU labelling for 6 hours. ***P ≤ 0.05 or 0.01 treated vs. DMSO by Student’s t test.
As findings from the microarray study suggested, we further validated Rev-erbα transcriptional regulation of cancer pathway genes by RT-qPCR analysis. Rev-erbα-deficient myoblasts display ∼3-fold inductions of Cyclin D1 and c-Met, whereas the cell cycle inhibitor p21 was significantly suppressed (Fig. 3B). Furthermore, Cyclin D1 protein level in proliferating Rev−/− myoblasts is markedly elevated as compared to WT prior to myogenic induction (D0 level prior or differentiation, Fig. 3C), although it can be similarly suppressed upon differentiation. We next tested whether Rev-erbα transcriptional control of cancer/proliferative pathways impacts proliferation, and found that primary myoblasts lacking Rev-erbα display an accelerated growth rate that nearly doubles that of the WT cells after seven days in culture (Fig. 3D). Direct examination of proliferative myoblasts by EdU incorporation revealed a significant 54.8% increase of proliferation rate in Rev−/− cells as compared to WT controls (Fig. 3E,F). In addition, quantitative TUNEL assay to assess apoptotic rate in Rev−/− cells these cells found a ~53% reduction under with basal untreated, or nuclease-treated conditions (Fig. 3G).
The genetic loss-of-function studies indicate that Rev-erbα negatively modulates myoblast proliferation. As the activity of Rev-erbα is amenable to modulation by synthetic ligands21,22, we tested whether pharmacological interventions of Rev-erbα affect myoblast proliferation, and found that activation of Rev-erbα by synthetic agonist SR9011 significantly attenuates, whereas its inhibition by an antagonist SR8278 augments primary myoblast proliferation (Fig. 3H,I). Thus, findings from genetic and pharmacological manipulations collectively demonstrate that Rev-erbα inhibits MPC proliferation.
Rev-erbα suppresses Wnt pathway gene expression and signaling activity
As microarray study indicate that Wnt pathway is a top Rev-erbα-regulated process, we performed RT-qPCR analysis among differentially regulated genes identified and found that multiple components in the canonical Wnt cascade are significantly induced in Rev−/− myoblasts as compared to WT (Fig. 4A). These genes include Wnt ligands, Wnt10a and Wnt3a, the Wnt receptor, Fzd5, and the Wnt signaling transcription mediator Tcf4. Of note, loss of Rev-erbα does not alter the transcript level of Dishevelled 2 (Dvl2) and β-catenin. Expression of the Wnt target gene, Axin2, is also up-regulated, suggesting increased Wnt signaling activity in Rev-erbα-deficient myoblasts. Canonical Wnt signaling is transmitted through the inhibition of GSK-3β activity upon Wnt ligand stimulation and the subsequent nuclear translocation of β-catenin to activate TCF-induced transcription37. We thus determined Wnt signaling activity through analysis of β-catenin nuclear translocation in response to Wnt3a. As shown by immunoblot analysis of nuclear fraction vs. total cellular protein extract in WT cells (Fig. 4B), Wnt activity as assessed by nuclear β-catenin level is low in the absence of Wnt, and Wnt3a stimulation elicited a robust ~6-fold nuclear accumulation, as expected. In contrast, although the basal level of nuclear β-catenin remains low in Rev−/− myoblasts, Wnt3a-stimulated nuclear translocation is markedly augmented, reaching ∼70% higher than that of the WT as shown by quantitative analysis. Using a Wnt-responsive TopFlash-Luciferase reporter to monitor TCF-mediated transcriptional activity16, we further examined whether Rev-erbα inhibition by stable shRNA knockdown (RevKD) affects Wnt signaling in C2C12 myoblasts, generated using clonal selection after transfection as previously16. As shown in Fig. 4C, Wnt signaling as detected by the reporter was significantly higher in RevKD cells, with Wnt3a-stimulated activity increased ~90% as compared to cells with scrambled control shRNA (SC).
Rev-erbα regulation of Wnt pathway gene expression and signaling activity. (A) RT-qPCR analysis of Wnt pathway gene expression in WT and Rev−/− primary myoblasts (n = 4). (B) Wnt signaling activity as assessed by immunoblot analysis of nuclear translocation of β-catenin in WT and Rev−/− primary myoblasts. The ratio of nuclear to cytosolic β-catenin protein levels under basal and Wnt3a-stimulated conditions were calculated after normalization to TBP as loading control. (C) Wnt signaling activity was assessed by TopFlash-Luc reporter under basal and Wnt3a-stimulated conditions in C2C12 myoblasts with stably transfected scrambled control (SC) shRNA or Rev-erba knockdown (RevKD) shRNA (n = 5). **p ≤ 0.05 by Student’s t test. (D) The effects of Rev-erbα ligands, agonist SR9011 and antagonist SR8278, on Wnt signaling activity were assessed by TopFlash luciferase reporter assay in C2C12 myoblasts (n = 5/group). Ligands were added 8 hours prior to overnight Wnt3a (40%) stimulation before luciferase assay. **P < 0.01 vs. DMSO control by Student’s t test. (E,F) The effect of agonist SR9011 (E), and antagonist SR8278 (F) on Wnt signaling pathway gene expression as analyzed by RT-qPCR analysis, at 4 days of C2C12 myoblast differentiation (n = 3). ***P ≤ 0.05 or 0.01 vs. DMSO control.
Using this approach, we further determined the effects of pharmacological modulations of Rev-erbα activity on Wnt signaling. As shown by the Wnt-responsive luciferase reporter activity, Rev-erbα agonist, SR9011, markedly suppressed Wnt signaling at basal and Wnt3a-stimulated conditions (Fig. 4D). In contrast, the Rev-erbα antagonist, SR8278, augmented TopFlash reporter activity by 2-fold at basal state, although not under Wnt3a-stimulated state likely due to the saturated transcriptional activity levels induced by Wnt3a. In line with the observed effects of Rev-erbα ligands on Wnt signaling, SR9011 markedly inhibited the expression of Wnt pathway components, Axin2 and Wnt10 (Fig. 4E). In contrast, SR8278 was able to up-regulate the expression of Axin2, Dvl2, β-catenin and Tcf3 genes within the Wnt cascade (Fig. 4F).
Rev-erbα inhibits myogenic differentiation
As Rev-erbα inhibits Wnt signaling32,33,34, we postulate this regulation may lead to suppression of myogenic differentiation. We thus tested Rev-erbα-deficient primary myoblasts differentiation upon 2% horse serum induction, and found that the loss of Rev-erbα accelerated their morphological transformation toward organized myotubes (Fig. 5A). The advanced differentiation is further validated by a ~60% increase in the number of MyHC+ myofibers by immnostaining (64.4% vs. 42.8%) at 24 hours upon differentiation, together with higher percentage of fused myofibers containing multiple nuclei (Fig. 5B,C). Accompanying the accelerated morphological transformation, Rev−/− myoblasts display elevated expression of Myf5, Myod and myogenin during the differentiation time course as compared to WT cells, particularly at late differentiation stage of day 2 (Fig. 5D).
Loss of Rev-erbα enhances myogenic differentiation of Rev−/− primary myoblasts. (A) Representative phase-contrast images of morphologic progression of WT and Rev−/− primary myoblasts subjected to 2% horse serum-induced myogenic differentiation for 48 hours. (B,C) Representative images of MyHC immunostaining at 24 hours after myogenic induction (B, 10X). Quantification of total MyHC+ and fused myofibers (myofibers containing ≥ 2 nuclei) was shown in (C, 20X, n = 5). (D) RT-qPCR analysis of myogenic transcription factor induction during WT and Rev−/− primary myoblasts differentiation over 2 days (n = 3).
We next examined whether Rev-erbα ligands affects myoblast differentiation. Opposite to the effect of genetic loss-of-function, activation of Rev-erbα by SR9011 markedly suppresses the appearance of myotubes at day 3 of early myoblast differentiation in C2C12 cell line, as compared to the DMSO-treated controls (Fig. 6A). At 6 days post myogenic induction when myotube formation of DMSO-treated myoblasts is largely complete, there is still a significant percentage of cells in SR9011-treated culture remain undifferentiated. Immunostaining of MyHC at 3 days post-differentiation confirmed an evident defect in myotube formation, as SR9011 nearly abolished the mature myocyte marker expression (Fig. 6B,C). Gene expression analysis revealed that SR9011 repressed the expression of known Rev-erbα target genes, Rev-erbα itself and Bmal1, indicating the activation of Rev-erbα transcription activity. Furthermore, myogenic regulatory factors (Myod1, Mrf4, myogenin) and myocyte-specific markers (Myhc3, Myhc7 and MLC1) were markedly down-regulated by SR9011 as compared to control (Fig. 6D), corroborating the impaired differentiation phenotype in SR9011-treated cells.
Activation of Rev-erbα by agonist SR9011 inhibits myogenic differentiation. (A–C) Inhibition of Rev-erbα agonist SR9011 on morphological progression of C2C12 myogenic differentiation for 6 days, as shown by representative phase-contrast images (A). Effect of SR9011 on myogenic differentiation as indicated by immunostaining of MyHC in day 3-differentiated C2C12 on chamber slides (B), with quantification of total MyHC+ and fused myofibers (C, n = 5/group) in the presence of vehicle (DMSO) or SR9011. DMSO (0.1%) or SR9011 (10 nM) were added 8 hours before myogenic induction and maintained throughout differentiation. (D) RT-qPCR analysis of SR9011 effect on suppressing myogenic gene expression at day 4 of C2C12 differentiation (n = 3). ***P ≤ 0.05 or 0.01 SR9011 vs. DMSO by Student’s t test.
In contrast to SR9011, the Rev-erbα antagonist, SR8278, promotes the morphological changes toward mature myotube formation in C2C12 myoblasts relative to DMSO-treated controls (Fig. 7A). SR8278 treatment significantly increased the percentage of MyHC+ myofibers and fused myofiber containing multiple nuclei upon 3 days of myogenic induction (Fig. 7B,C). The accelerated morphological differentiation was accompanied by augmented expression of myogenic regulatory factors (Myf5, Myod1 and Mrf4) and myocyte-specific markers (Myhc3, Myhc7 and MLC1, Fig. 7D). SR8278 effectively blunted the transcription repressor activity of Rev-erbα, as indicated by the induction of Rev-erbα and Bmal1 transcripts. Thus, both genetic and pharmacological findings demonstrate that Rev-erbα inhibits MPC differentiation.
Inhibition of Rev-erbα by antagonist SR8278 promotes myogenic differentiation. (A–C) Advanced morphological progression of C2C12 myoblast differentiation treated by SR8278 as compared to DMSO, as shown by representative phase-contrast image at day 0 to 6 in culture (A). Effect of SR8278 on myogenic differentiation as indicated by MyHC immunostaining in day 3-differentiated C2C12 on chamber slides (B), with quantification of total MyHC+ and fused myofibers (C, n = 5/group), in the presence of vehicle (DMSO) or SR8278. Cells were treated with DMSO (0.1%) or agonist SR8278 (10 nM) 8 hours prior to induction of differentiation and maintained throughout differentiation. (D) RT-qPCR analysis of effect of SR8278 on myogenic gene expression at day 4 of C2C12 differentiation (n = 3). ***P ≤ 0.05 or 0.01 SR8278 vs. DMSO by Student’s t test.
Loss of Rev-erbα augments muscle regeneration and satellite cell proliferative expansion
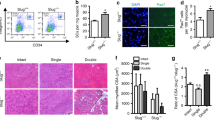
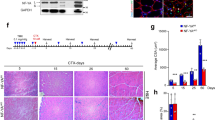
Upon muscle injury, the orderly activation, proliferation, and differentiation of MPCs, satellite cells and its progenies, critically determine the regenerative repair response15,38. Based on our findings of Rev-erbα inhibition of proliferation and differentiation in myoblasts, loss of these activities may promote muscle regeneration in vivo. We thus test the regenerative response of WT and Rev−/− animals to cardiotoxin (CTX)-induced muscle injury CTX was injected throughout the length and thickness of Tibialis Anterior muscle to ensure uniform injury response, as described17, and muscle regenerative repair was monitored for 30 days after injury. Initial histological examination of the regeneration time course suggests enhanced repair in Rev-erbα-null mutants as compared to WT (Fig. 8A). At the peak of new myofiber synthesis at day 5, when nascent myofibers of WT are small, new Rev−/− myofibers appear larger with robust basophilic staining. More advanced maturation of well-formed new myofibers containing central nuclei is evident by day 8 post-injury. At day 30 when the normal morphology of the muscle is largely restored by regenerated myofibers, quantitative analysis of cross-section areas reveal significantly skewed distribution toward larger myofibers in Rev−/− mice as compared to the WT (Fig. 8B). The expression kinetics of key factors involved in MPC proliferative and myogenic response during the regeneration time course was examined, as shown in Fig. 7C. Upon muscle damage, satellite cells are activated to proliferate and expand to provide new myonuclei for regenerative repair. This normal response of satellite cell expansion in WT mice is reflected by the strong induction of the satellite cell marker, Pax7 protein, which peaks at 5 days after injury, gradually declines at day 8 and terminates at 14 days (Fig. 8C). In comparison, Rev-erbα-null mice display elevated Pax7 induction throughout the satellite activation stages at day 3 and 5, with its peak level advanced to day 3. The continued high expression of Pax7 in Rev-erbα-null mice as compared to WT at day 8 post-injury indicates a potentially more sustained satellite cell proliferative expansion phase. In line the expression kinetics of Pax7, Cyclin D1 induction at day 3 and 5 are markedly higher in Rev−/− mice, suggesting increased proliferative activities of cycling myogenic progenitors. Interestingly, myogenin levels in these mice exhibit a similarly augmented kinetics with an early peak expression at day 3, suggesting concomitantly enhanced myogenic maturation of activated progenitors. As the elevated Pax7 protein observed at day 8 near the end of the proliferative expansion suggests more abundant Pax7-expressing satellite cells, we determined satellite cell numbers directly through Pax7 immunofluorescence staining in muscle sections (Fig. 9A). Consistent with the low expression of Pax7 in WT mice at day 8 post injury, the number of Pax7+ satellite cells detected is limited. In contrast, ~50% higher amount of satellite cells was found in regenerating Rev−/− muscle than the WT (Fig. 9B,C). Furthermore, identification of proliferating satellite cell by EdU incorporation in vivo revealed nearly doubling of the number of cells with EdU+/Pax7+ co-staining in Rev−/− muscle as compared to WT (Fig. 9D). These findings demonstrate that, in line with the observed functions of Rev-erbα in suppressing MPC proliferative and myogenic activities, the loss of Rev-erbα augments satellite cell-mediated regenerative repair in vivo.
Loss of Rev-erbα enhances muscle regeneration. (A) Representative images of H/E histological analysis of muscle regeneration at 3, 5, 8 and 30 days. The number of mice used for these analysis are: day 0 (WT n = 6, Rev−/− n = 6), day 3 (WT n = 6, Rev−/− n = 5), day 5 (WT n = 7, Rev−/− n = 6), day 8 (WT n = 5, Rev−/− n = 6), day 14 (WT n = 6, Rev−/− n = 5), day 30 (WT n = 6, Rev−/− n = 8). (B) Quantitative analysis of regenerated myofiber diameter distribution at 30 days after cardiotoxin injury in WT (n = 6) and Rev−/− mice (n = 8). Values are represented as the percentage of number of myofibers within the indicated range over total number of myofibers. Three representative 10X sections with ~450 total myofibers were counted for each mouse in the group. Non-parametric Kruskal–Wallis ANOVA for dataset p-value < 0.0001, and non-parametric Mann-Whitney test are indicated for individual cross section area category: ***P ≤ 0.05 or 0.01 Rev−/− vs. WT. (C) Immunoblot analysis of protein levels of cell cycle regulator and myogenic marker in regenerating TA muscle lysate at indicated time points after CTX injury. Protein samples were pooled from each group of WT or Rev−/− at indicated time point as shown in (A).
Loss of Rev-erbα promotes satellite cell proliferative expansion during muscle regeneration. (A,B) Representative images of Pax7 immunofluorescent staining to identify total and EdU staining to label proliferative satellite cells, in day 8-regenerating TA post CTX injury (A). Arrowheads denote co-localization of Pax7 (red), EdU (green) with DAPI (blue) that identifies proliferating satellite cells. Phase contrast images of corresponding fields were shown in (B). (C–E) Quantification of number of Pax7+ satellite cells expressed as percentage of total nuclei (C), or normalized as the percentage of total number of myofibers (D); and the number of proliferative satellite cells as indicated by co-localized Pax7+ and EdU+ staining (E, n = 4). ***P ≤ 0.05 or 0.01 Rev−/− vs. WT by Student’s t test.
Discussion
Maintenance of proper MPC proliferation and differentiation functions critically determines skeletal muscle growth and repair15,38. Our study identifies that the clock repressor Rev-erbα is an inhibitory factor of MPC proliferative and myogenic properties, and these functions suppress skeletal muscle regeneration in vivo.
The appropriate balance and separation between proliferation and differentiation of MPCs ensures the expansion of the myogenic precursor pool and their subsequent maturation for muscle repair. Manipulations that promote MPC proliferation alone hinder myogenic differentiation, whereas enhancing differentiation limits MPC proliferative growth39,40,41,42. Our current study uncovers an intriguing finding that the loss of Rev-erbα function coordinately facilitates MPC proliferation with myogenic progression. Similar to the known effects of IGF-1 on myoblasts43,44,45 but distinct from typical cell cycle regulators or myogenic factors, these actions likely contributed to the enhanced myogenic repair observed in Rev-erbα-null mice in concert. These inhibitory Rev-erbα functions in regenerative myogenesis could be a mechanism to prevent excessive tissue remodeling in response to injury and maintain tissue homeostatic control. On the other hand, suppressing Rev-erbα activity may have the potential to promote skeletal muscle regenerative capacity. Given that we have demonstrated previously that Bmal1 promotes MPC myogenic activities16,17, Rev-erbα may function in concert with Bmal1 or additional clock regulators in MPC to orchestrate skeletal muscle regenerative myogenesis.
Through gene expression profiling, we uncovered a novel mechanistic link between Rev-erbα and the Wnt signaling pathway. We identified that genes along distinct steps of the Wnt cascade are direct Rev-erbα target genes with RORE consensus binding sites. This raises an intriguing possibility whether RORα, the transcription activator of the RORE elements and critical for the core clock loop1, may exert positive transcriptional control of Wnt and proliferative pathway to modulate myogenesis. Future studies of potential RORα functions in myogenic pathways would lend additional support to circadian clock mechanisms involved. And, as another ligand-modulated nuclear receptor, RORα regulations of myogenic repair may also reveal novel therapeutic targets for muscle diseases46,47,48. Interestingly in previous studies, we found that Bmal1 exerts positive transcriptional control of key signaling nodes of the Wnt pathway to promote myogenesis16. The new findings in our study suggest that it is possible that the Bmal1 activation together with Rev-erbα repression, likely at distinct times in a circadian cycle, may underlie rhythmic expression of Wnt pathway genes, as we demonstrated in synchronized myoblasts in the previous study16. As Wnt pathway is a major developmental signal that drives myogenesis, its modulation by clock regulators may confer an additional layer of control to fine-tune the myogenic cascade. A study by Downes et al. found that ectopic overexpression of a Rev-erbα dominant negative mutant promotes myogenic differentiation49, a finding that is consistent with our current results. Notably in our study, the effect of loss of endogenous Rev-erbα on muscle regeneration in vivo is not as marked as demonstrated by the dominant negative mutant. It is possible that a closely related Rev-erbβ gene may have partially overlapping myogenic functions with Rev-erbα that provides functional redundancy in muscle regeneration18.
An intimate link between circadian clock and cell cycle regulation has long been established50,51. CLOCK-controlled mRNA in growth and differentiation comprise ~15% of all transcripts in skeletal muscle7. Studies of Bmal1 on MPC proliferation and satellite cell expansion16,17 are in line with this notion11,12. Based on the significant overlap of Rev-erbα and Bmal1 genomic targets and their opposing functions in the clock circuit18, Rev-erbα may inhibit MPC proliferation. Although few studies directly examined Rev-erbα function in proliferative pathways, several observations to date supports its involvement. Rev-erbα-deficiency increases hippocampal neuronal progenitor proliferation52, whereas its activation by SR9011 inhibits breast cancer cell proliferative growth53. With our finding of Rev-erbα inhibitory effect on myoblast proliferation, it may modulate proliferative behaviors in a broad array of tissue/cell types. Many of Rev-erbα differentially-regulated genes in cancer/proliferative pathway we identified shares significant overlap with a published ChIP-Seq dataset18. Indeed, our analysis of chromatin association of Rev-erbα among its differentially-regulated genes indicate that several cell cycle regulators are its direct transcriptional targets in proliferative myoblasts, although additional targets of Rev-erbα in proliferative pathway may be uncovered through global ChIP-Seq approaches in future studies.
Rev-erbα expression was strongly suppressed by muscle injury stimuli. This finding suggests that physiological inhibitory effects of Rev-erbα on MPC proliferation and myogenesis may help maintain a quiescent state, and that its relief upon injury facilitates myogenic activation in repair. Possibly due to its dynamic regulation by injury stimuli, no significant effect of Rev-erbα ablation on muscle growth was evident under normal condition. Potential functional redundancy between Rev-erbα and Rev-erbβ with overlapping genomic targets is another possibility. Current findings of Rev-erbα regulation of MPC behavior establish a basis for future in-depth analysis of molecular clock function in muscle stem cells. A growing body of evidence indicates that the circadian clock is required for coordinating stem cell response to external stimuli in tissue growth and remodeling processes54,55,56,57. Rev-erbα regulation of proliferation and differentiation pathways, as we uncovered in this study, may apply to a myriad of stem cell paradigms beyond skeletal muscle repair.
Rev-erbα inhibition of MPC behavior in isolated primary myoblasts suggests the cell-autonomous roles of Rev-erbα. Enhanced satellite cell proliferation in regenerative expansion we observed in Rev-erbα-null mice is consistent with this notion. Nonetheless, although MPC is major cellular source for muscle regeneration, optimal repair requires additional stem cell populations and other cell types including macrophages, fibroblasts, endothelial cells38,58,59,60,61. The global ablation of the Rev-erbα-null in our study therefore cannot exclude its involvement in additional processes of regeneration. Particularly, the known immune-modulatory activities of Rev-erbα may contribute to regenerative repair62,63. Future cell type-selective gene targeting studies are required to dissect the cell-autonomous Rev-erbα functions in MPC vs. other cell types in skeletal muscle repair.
As a “druggable” nuclear receptor, Rev-erbα activity is modulated by synthetic ligands21,22. Our in vitro studies of Rev-erbα antagonist SR8278 in myoblasts provide the evidence and mechanisms by which that pharmacological inhibition of Rev-erbα augments MPC myogenic activities. Consistent with our current findings, a recent study by Welch et al.64 tested SR8278 in mdx mice that resulted in functional improvement of dystrophic pathpophysiology. Thus, as these studies uncovered, there are potential therapeutic utilities of Rev-erbα antagonism in degenerative or dystrophic muscle diseases through stimulation of myogenic growth and repair mechanisms. Ultimately, better mechanistic understandings of Rev-erbα function in myogenic progenitor behavior may lead to the discovery of new strategies to enhance regenerative capacity for the maintenance of muscle mass.
Change history
22 March 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41598-022-08153-9
References
Preitner, N. et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002).
Duez, H. & Staels, B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. Journal of applied physiology 107, 1972–1980, https://doi.org/10.1152/japplphysiol.00570.2009 (2009).
Yin, L., Wu, N. & Lazar, M. A. Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nuclear receptor signaling 8, e001 (2010).
Chatterjee, S. & Ma, K. Circadian clock regulation of skeletal muscle growth and repair. F1000Research 5, 1549, https://doi.org/10.12688/f1000research.9076.1 (2016).
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nature reviews. Genetics 18, 164–179, https://doi.org/10.1038/nrg.2016.150 (2017).
Dibner, C., Schibler, U. & Albrecht, U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology 72, 517–549 (2010).
Andrews, J. L. et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proceedings of the National Academy of Sciences of the United States of America 107, 19090–19095 (2010).
Woldt, E. et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nature medicine 19, 1039–1046 (2013).
Kondratov, R. V., Kondratova, A. A., Gorbacheva, V. Y., Vykhovanets, O. V. & Antoch, M. P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes & development 20, 1868–1873 (2006).
McDearmon, E. L. et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science (New York, N.Y 314, 1304–1308 (2006).
Lucassen, E. A. et al. Environmental 24-hr Cycles Are Essential for Health. Curr Biol 26, 1843–1853, https://doi.org/10.1016/j.cub.2016.05.038 (2016).
Miller, B. H. et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proceedings of the National Academy of Sciences of the United States of America 104, 3342–3347 (2007).
Stratmann, M. & Schibler, U. REV-ERBs: more than the sum of the individual parts. Cell metabolism 15, 791–793 (2012).
Buckingham, M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Current opinion in genetics & development 16, 525–532 (2006).
Le Grand, F. & Rudnicki, M. A. Skeletal muscle satellite cells and adult myogenesis. Current opinion in cell biology 19, 628–633 (2007).
Chatterjee, S. et al. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. Journal of cell science 126, 2213–2224 (2013).
Chatterjee, S., Yin, H., Nam, D., Li, Y. & Ma, K. Brain and muscle Arnt-like 1 promotes skeletal muscle regeneration through satellite cell expansion. Experimental cell research (2014).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485, 123–127 (2012).
Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science (New York, N.Y 338, 349–354 (2012).
Burris, T. P. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Molecular endocrinology (Baltimore, Md 22, 1509–1520 (2008).
Kojetin, D., Wang, Y., Kamenecka, T. M. & Burris, T. P. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS chemical biology 6, 131–134 (2011).
Solt, L. A. et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485, 62–68 (2012).
Nam, D. et al. Novel Function of Rev-erbalpha in Promoting Brown Adipogenesis. Scientific reports 5, 11239, https://doi.org/10.1038/srep11239 (2015).
Cvoro, A. et al. A thyroid hormone receptor/KLF9 axis in human hepatocytes and pluripotent stem cells. Stem cells 33, 416–428, https://doi.org/10.1002/stem.1875 (2015).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 3, Article3 (2004).
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behavioural brain research 125, 279–284 (2001).
Stewart, R., Flechner, L., Montminy, M. & Berdeaux, R. CREB is activated by muscle injury and promotes muscle regeneration. PloS one 6, e24714 (2011).
Abbas, A., Raju, J., Milles, J. & Ramachandran, S. A circadian rhythm sleep disorder: melatonin resets the biological clock. The journal of the Royal College of Physicians of Edinburgh 40, 311–313, https://doi.org/10.4997/JRCPE.2010.406 (2010).
Nam, D. et al. The adipocyte clock controls brown adipogenesis through the TGF-beta and BMP signaling pathways. Journal of cell science 128, 1835–1847, https://doi.org/10.1242/jcs.167643 (2015).
Nam, D., Yechoor, V. K. & Ma, K. Molecular clock integration of brown adipose tissue formation and function. Adipocyte 5, 243–250, https://doi.org/10.1080/21623945.2015.1082015 (2016).
Bischoff, R. A satellite cell mitogen from crushed adult muscle. Developmental biology 115, 140–147 (1986).
Brack, A. S., Conboy, I. M., Conboy, M. J., Shen, J. & Rando, T. A. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell stem cell 2, 50–59 (2008).
Brack, A. S. et al. BCL9 is an essential component of canonical Wnt signaling that mediates the differentiation of myogenic progenitors during muscle regeneration. Developmental biology 335, 93–105 (2009).
Cossu, G. & Borello, U. Wnt signaling and the activation of myogenesis in mammals. The EMBO journal 18, 6867–6872 (1999).
Thompson, J., Dolcet, X., Hilton, M., Tolcos, M. & Davies, A. M. HGF promotes survival and growth of maturing sympathetic neurons by PI-3 kinase- and MAP kinase-dependent mechanisms. Molecular and cellular neurosciences 27, 441–452, https://doi.org/10.1016/j.mcn.2004.07.007 (2004).
Bernet, J. D. et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nature medicine 20, 265–271, https://doi.org/10.1038/nm.3465 (2014).
Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480 (2006).
Charge, S. B. & Rudnicki, M. A. Cellular and molecular regulation of muscle regeneration. Physiological reviews 84, 209–238 (2004).
Skapek, S. X., Rhee, J., Spicer, D. B. & Lassar, A. B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science (New York, N.Y) 267, 1022–1024 (1995).
Rao, S. S., Chu, C. & Kohtz, D. S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol 14, 5259–5267 (1994).
Sabourin, L. A., Girgis-Gabardo, A., Seale, P., Asakura, A. & Rudnicki, M. A. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol 144, 631–643 (1999).
Wen, Y. et al. Constitutive Notch activation upregulates Pax7 and promotes the self-renewal of skeletal muscle satellite cells. Mol Cell Biol 32, 2300–2311 (2012).
Engert, J. C., Berglund, E. B. & Rosenthal, N. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J Cell Biol 135, 431–440 (1996).
Florini, J. R., Ewton, D. Z. & Coolican, S. A. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev 17, 481–517, https://doi.org/10.1210/Er.17.5.481 (1996).
Duan, C., Ren, H. & Gao, S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. General and comparative endocrinology 167, 344–351, https://doi.org/10.1016/j.ygcen.2010.04.009 (2010).
Solt, L. A. & Burris, T. P. Action of RORs and their ligands in (patho)physiology. Trends in endocrinology and metabolism: TEM 23, 619–627, https://doi.org/10.1016/j.tem.2012.05.012 (2012).
Solt, L. A. et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472, 491–494, https://doi.org/10.1038/nature10075 (2011).
Wang, Y. et al. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORalpha and RORgamma. ACS chemical biology 5, 1029–1034, https://doi.org/10.1021/cb100223d (2010).
Downes, M., Carozzi, A. J. & Muscat, G. E. Constitutive expression of the orphan receptor, Rev-erbA alpha, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Molecular endocrinology (Baltimore, Md) 9, 1666–1678 (1995).
Masri, S., Cervantes, M. & Sassone-Corsi, P. The circadian clock and cell cycle: interconnected biological circuits. Current opinion in cell biology 25, 730–734, https://doi.org/10.1016/j.ceb.2013.07.013 (2013).
Sotak, M., Sumova, A. & Pacha, J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann Med 46, 221–232, https://doi.org/10.3109/07853890.2014.892296 (2014).
Schnell, A. et al. The nuclear receptor REV-ERBalpha regulates Fabp7 and modulates adult hippocampal neurogenesis. PloS one 9, e99883 (2014).
Wang, Y., Kojetin, D. & Burris, T. P. Anti-proliferative actions of a synthetic REV-ERBalpha/beta agonist in breast cancer cells. Biochemical pharmacology, https://doi.org/10.1016/j.bcp.2015.06.010 (2015).
Bouchard-Cannon, P., Mendoza-Viveros, L., Yuen, A., Kaern, M. & Cheng, H. Y. The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell reports 5, 961–973 (2013).
Janich, P. et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480, 209–214 (2011).
Karpowicz, P., Zhang, Y., Hogenesch, J. B., Emery, P. & Perrimon, N. The circadian clock gates the intestinal stem cell regenerative state. Cell reports 3, 996–1004 (2013).
Plikus, M. V. et al. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proceedings of the National Academy of Sciences of the United States of America 110, E2106–2115 (2013).
Robertson, T. A., Maley, M. A., Grounds, M. D. & Papadimitriou, J. M. The role of macrophages in skeletal muscle regeneration with particular reference to chemotaxis. Experimental cell research 207, 321–331 (1993).
Mitchell, K. J. et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nature cell biology 12, 257–266 (2010).
Murphy, M. M., Lawson, J. A., Mathew, S. J., Hutcheson, D. A. & Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development (Cambridge, England) 138, 3625–3637 (2011).
Christov, C. et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Molecular biology of the cell 18, 1397–1409 (2007).
Gibbs, J. E. et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America 109, 582–587 (2012).
Lam, M. T. et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515, https://doi.org/10.1038/nature12209 (2013).
Welch, R. D., Billon, C., Valfort, A. C., Burris, T. P. & Flaveny, C. A. Pharmacological inhibition of REV-ERB stimulates differentiation, inhibits turnover and reduces fibrosis in dystrophic muscle. Scientific reports 7, 17142, https://doi.org/10.1038/s41598-017-17496-7 (2017).
Acknowledgements
We thank the Histology Core Laboratory at Baylor College of Medicine for their expert technical assistance. We also thank Dr. Aijun Zhang, Methodist Hospital Research Institute, for help with microarray analysis. This project was supported by grants from Muscular Dystrophy Association 381294, American Heart Association 17GRNT33370012, and National Institute of Health 1R01DK112794 to K.M.; and National Institute of Health grant DK097160-01 to V.Y. K.M. as a faculty member of the City of Hope National Cancer Institute-designated Comprehensive Cancer Center is also supported by National Institute of Health award P30CA33572.
Author information
Authors and Affiliations
Contributions
S.C., H.Y. and W.L. designed, performed experiments and prepared figures; J.L. and V.Y. contributed to the generation of Rev-erbα-null mice and editing of the manuscript; K.M. conceived the project, performed experiments, analyzed results, and prepared figures and manuscript.
Corresponding author
Ethics declarations
Competing Interests
I certify that neither I nor my co-authors have a conflict of interest as described above that is relevant to the subject matter or materials included in this work.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in Figure 8A, where D5 WT was incorrectly duplicated as where D5 WT was incorrectly duplicated as Rev-/- D3.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chatterjee, S., Yin, H., Li, W. et al. The Nuclear Receptor and Clock Repressor Rev-erbα Suppresses Myogenesis. Sci Rep 9, 4585 (2019). https://doi.org/10.1038/s41598-019-41059-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41059-7
This article is cited by
-
Targeted screening and identification of chlorhexidine as a pro-myogenic circadian clock activator
Stem Cell Research & Therapy (2023)
-
Time after time: circadian clock regulation of intestinal stem cells
Cellular and Molecular Life Sciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.