Abstract
The Baltic Sea is a shallow basin of brackish water in which the spatial salinity gradient is one of the most important factors contributing to species distribution. The Baltic Sea is infamous for its annual cyanobacterial blooms comprised of Nodularia spumigena, Aphanizomenon spp., and Dolichospermum spp. that cause harm, especially for recreational users. To broaden our knowledge of the cyanobacterial adaptation strategies for brackish water environments, we sequenced the entire genome of Dolichospermum sp. UHCC 0315, a species occurring not only in freshwater environments but also in brackish water. Comparative genomics analyses revealed a close association with Dolichospermum sp. UHCC 0090 isolated from a lake in Finland. The genome closure of Dolichospermum sp. UHCC 0315 unraveled a mixture of two subtypes in the original culture, and subtypes exhibited distinct buoyancy phenotypes. Salinity less than 3 g L−1 NaCl enabled proper growth of Dolichospermum sp. UHCC 0315, whereas growth was arrested at moderate salinity (6 g L−1 NaCl). The concentrations of toxins, microcystins, increased at moderate salinity, whereas RNA sequencing data implied that Dolichospermum remodeled its primary metabolism in unfavorable high salinity. Based on our results, the predicted salinity decrease in the Baltic Sea may favor toxic blooms of Dolichospermum spp.
Similar content being viewed by others
Introduction
The Baltic Sea is one of the largest brackish water ecosystems in the world. It undergoes substantial spatiotemporal variation in the salinity gradient maintained by uneven saline-water inflow via the Danish Straits and varying amounts of riverine runoff. The surface-water salinity varies from >20 practical salinity units (PSUs) in Kattegat Bay to almost freshwater concentrations in the northernmost Gulf of Bothnia and eastern Gulf of Finland1. Climate change models predicted that the overall future salinity in the Baltic Sea will decrease, due to increased precipitation and freshwater inflow within the catchment area2,3,4. The salinity range is one of the most important factors contributing to the prevailing species diversity and distribution in this area5,6,7,8, and as a result of salinity decrease, the distribution of freshwater species may shift more to the south2. However, due to uneven saltwater pulses, organisms may occasionally become stressed by unfavorably high salt concentrations.
The Baltic Sea is notorious for its annual cyanobacterial blooms composed of Nodularia spumigena, Aphanizomenon flos-aquae, and Dolichospermum spp. (formerly Anabaena spp.)9,10,11. Blooms cause public-health concerns, especially for recreational users, because Nodularia spumigena and Dolichospermum spp. are capable of producing toxins9,12. The hepatotoxins nodularins and microcystins are detected in considerable amounts every summer and, despite general knowledge of their toxicity to mammals, animal poisonings are still reported regularly13.
Dolichospermum spp. is a filamentous and nitrogen-fixing cyanobacterial species that typically forms massive blooms in freshwater environments14,15,16. Furthermore, toxic strains capable of living in intermediate salinities have been found, especially in the less saline Gulf of Finland and coastal regions11,17. Decreased salinity may thus promote the distribution and abundance of Dolichospermum and further favor the formation of toxic Dolichospermum blooms18,19. Due to the genetic and morphological heterogeneity of the genus Anabaena, Dolichospermum was recently distinguished from a benthic Anabaena-type cluster, resulting in the gas vacuole-forming and pelagic morphotype Dolichospermum sp.20. However, Dolichospermum spp. still harbors high genetic and morphological diversity, which results in ambiguity in nomenclatural and phylogenetic clustering. Even mixing with Aphanizomenon spp. and Nostoc sp. has been described21,22. We have previously defined seven Nostocales subgroups (I a–d; II a–c), based on the comparison of average-nucleotide and amino-acid identities of 30 sequenced Nostocales strains23. In this classification, Dolichospermum/Anabaena species were found in four subgroups, implementing a high demand for further taxonomic clarification. However, a latter study suggested even more complex taxonomy due to the close relationship of these species to a third cyanobacterial genus, Aphanizomenon24.
Here, we sequenced the entire genome of Baltic Sea Dolichospermum sp. UHCC 0315 to reinforce the genomic knowledge of filamentous and toxic Baltic Sea cyanobacteria and to gain insight into the niche adaptation strategies of the examined strain. The isolate was obtained from a coastal cyanobacterial bloom in the Helsinki area9. Therefore, we addressed the question of how a slight increase in salinity relevant to the Gulf of Finland can affect the growth and construction of the transcriptomes. Furthermore, comparative genomic analysis was applied to elucidate the ambiguity within the sequenced Dolichospermum/Anabaena species.
Results
Insight into the genome of Dolichospermum sp. UHCC 0315
The whole genome sequence of Dolichospermum sp. UHCC 0315 was obtained using Pacific Biosciences (PacBio) chemistry, and sequencing errors were corrected with Illumina Hiseq2500 reads by two rounds of corrections (Freebayes 217 errors and Pilon 8 errors). The assembly yielded one chromosome of 5.17 Mbp and three plasmids (0.85, 0.53, and 0.94 Mbp). Automatic annotation of the Dolichospermum sp. UHCC 0315 genome revealed a total of 5,362 proteins of which 3,570 (66%) were predicted to be part of 367 metabolic subsystems. Among a wide diversity of functions, several response mechanisms to osmotic, oxidative and heat stress, as well as to antibiotic and toxic heavy metal defense were identified. Furthermore, one possible CRISPR array with 12 spacers and the direct repeat consensus sequence 5′-CTTGCAATTAACCTAATTACTCAAAGCTAATTTCACC-3′ but without any known Cas proteins was detected (Supplementary Fig. S1). Interestingly, spacer 7 matches prophage antirepressor sequences present in the genomes of other Nostocales, Nostoc ‘Peltigera membranacea cyanobiont’ N6 and Nostoc flagelliforme CCNUN1 (Supplementary Fig. S1), supporting the further function of this genetic element as a likely CRISPR repeat spacer array further.
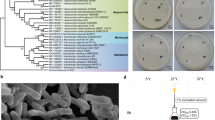
Interestingly, the assembly of the Dolichospermum sp. UHCC 0315 genome unraveled two chromosomal subtypes, A and B, of which subtype A carried an additional five adjacent hypothetical genes (BMF77_1250–1254). Based on the Protein Basic Local Alignment Search Tool (BlastP) search, these five genes encode a putative methyltransferase (BMF77_01250), glycosyltransferase (BMF77_01251; BMF77_01253; BMF77_01254), and a hypothetical protein (BMF77_01252). PacBio coverages of the two identified subtypes were 250 (subtype A) and 70 (subtype B). To ensure that both subtypes were properly distinguished by sequence assembly, we isolated 70 filaments under a microscope and cultivated them in liquid medium. Surprisingly 39 of the cultures were buoyant (subtype A) and 31 grew on the bottom of the flask (subtype B) (Fig. 1a,b), despite both subtypes carried full gvp gene cluster for gas vesicle formation. Differences in cellular morphology under the light microscope were additionally observed and indicate the presence of aerotopes in substrain A (Fig. 1c,d). Presence or absence of the 5-gene region was confirmed by PCR amplification of a segment from gene BMF77_1254 using specific primers (Supplementary Table S1). The amplification product was found only in the buoyant subtype A (Fig. 1e, Supplementary Fig. S2), which further showed that the absence of the BMF77_01250–01254 genes in subtype B was real. Furthermore, the distribution of all three plasmids was tested, using primers amplifying certain regions of the plasmids, and the PCR results indicated that all three plasmids were present in both substrains (Fig. 1f–h, Supplementary Fig. S3).
Identification of two Dolichospermum sp. UHCC 0315 substrains A and B. Subtype A showed buoyant phenotype (a) with rough morphology (presence of aerotopes) under the microscopy (c), whereas subtype B grew on the bottom of the cultivation flask (b) and had smooth morphology (d). The PCR amplification study using specific primers and testing triplicates (e) further showed that the genetic region of BMF77_1250–1254 is present in substrain A (A1–3) but absent in B (B1–3). However, the results indicate that both substrains harbor full sets of three plasmids (f–h). PC = Positive Control; NC = Negative Control. The full-length gel with the PCR amplification of the three plasmids are presented in Supplementary Figs S2 (e) and S3 (f–h).
Comparative genomics
A phylogenomic tree, based on 31 conserved marker genes was constructed to show the placement of the newly sequenced Dolichospermum sp. UHCC 0315 within the cyanobacterial phylum (Fig. 2a). The strain branched together with 16 other Dolichospermum sp., Anabaena sp. and Aphanizomenon flos-aquae with diverse biosynthetic gene clusters, forming a distinct Anabaena/Dolichospermum/Aphanizomenon clade (ADA)24. Other 4 Anabaena strains (PCC 7122, PCC 7108. ATCC 33047 and 4-3) formed different branches in the tree. Furthermore, Average Amino Acid and Nucleotide Identity analyses (Fig. 2b,c, respectively) highlighted the division of the genomes in 6 different subclades (Iα-δ, II and III). Considering the genome of Dolichospermum sp. UHCC 0315, there are five genomes of the genus that are publicly available (Dolichospermum circinale AWQC310F and AWQC131C and Dolichospermum sp. NIES-806 and UHCC 0090). However, while only the UHCC 0315 and UHCC 0090 strains grouped together in a subclade with other 3 Anabaena strains (AL09, LE011-02 and MDT14b) in the genomic analyses, the other ones were scattered in other subclades. A 16S rRNA phylogenetic tree was constructed to further explore the ADA clade (Supplementary Fig. S4).
Maximum likelihood phylogenomic tree based on the concatenated alignment of 31 universal marker genes70 from 75 cyanobacterial genomes. Those analyzed in this study are shown in bold and the presence of gene clusters of natural products are represented (a). Average Amino Acid Identity (AAI) (b), and Average Nucleotide Identity (ANI) (c) heatmaps of genomes in subgroups (Iα-δ, II and III). Subgroups were determined as described earlier23,24.
Orthologous gene clustering was employed to estimate the core and pan-genome of the 21 Anabaena, Dolichospermum and Aphanizomenon genomes (Fig. 3). Whereas the core-genome asymptote is composed of approximately 1,500 genes, the current pan-genome reached more than 12,000 different genes belonging to the ADA clade.
Core and pan-genome plots estimated using the ADA clade (Anabaena sp. WA102, WA93, AL09, LE011-02, MDT14b, WA113 and CRKS33; Aphanizomenon flos-aquae NIES-81, 2012/KM1/D3, LD13, MDT14a and WA102; Dolichospermum sp. NIES-86, UHCC00315, UHCC 0090; and Dolichospermum circinale AWQC310F and AWQC131C plus Anabaena sp. PCC 7122, PCC7108, ATCC 33047 and 4-3. (a) Number of genes in the core-genome size by the number of genomes and (b) number of genes in the pan-genome by the number of genomes.
The closest relative to Dolichospermum sp. UHCC 00315 was the freshwater strain Dolichospermum sp. UHCC 0090 (previously Anabaena sp. 90). Synteny analysis showed a high number of conserved blocks between the two strains in the chromosomes and the pDOL01 and pUHCC0315a plasmids. Nonetheless, the other two plasmids of each strain seemed to be considerably less conserved (Supplementary Fig. S5). The two genomes encode a total of 9,873 proteins of which 7,101 formed shared orthologous clusters (Supplementary Fig. S6). Around 93% of the proteins in those clusters were predicted to be involved in a wide diversity of functions of the cellular primary and secondary metabolisms. On the other hand, 1,816 and 956 proteins were found to be specific to Dolichospermum sp. UHCC 0315 and UHCC 0090, respectively (21.19% and 33,86% of the total number of proteins encoded in each genome). Functions for these two groups of proteins were less successfully predicted compared to the shared genes, pointing to many previously unknown genes among them.
Growth of Dolichospermum sp. UHCC 0315
Dolichospermum sp. UHCC 0315 growth rate was followed at four different salinities (0, 3, 6, or 9 g L−1 NaCl) by measuring the chlorophyll a concentration (Fig. 4a). The highest tested salinity (9 g L−1 NaCl) was deleterious for Dolichospermum sp. UHCC 0315, while 6 g L−1 NaCl inhibited the growth of the culture. Dolichospermum sp. UHCC 0315 was able to grow properly in 3 g L−1 NaCl, but the fastest growth was obtained under freshwater conditions (no added NaCl). Due to the appearance of a yellowish color in the culture, cell counting using the microscope technique was applied to follow the growth (Fig. 4b). The patterns of chlorophyll a and cell counting seemed to correlate, and thus the concentration of chlorophyll a was later used for normalization of the toxin concentration.
Reconstruction of transcriptomes under unfavorable salinities
To understand the cellular response of Dolichospermum sp. UHCC 0315 at moderate salinity, we applied RNA sequencing (RNA-Seq) analysis to compare the normal (0 g L−1 NaCl) and unfavorable conditions (6 g L−1 NaCl). We mapped a total of 4.1 × 108 sequenced paired-end Illumina HiSeq reads for three biological replicates of the control and 4.2 × 108 for three biological replicates of the treatment against the sequenced reference genome. Since the Dolichospermum sp. UHCC 0315 B lacked five genes and the RNA-seq data was generated from the mixture of the subcultures, Dolichospermum sp. UHCC 0315 A was used as a reference genome for mapping and calling of the differentially expressed genes. Using a Log2 fold change (Log2FC) of ±1 and a False-Discovery Rate (FDR) cutoff value < 0.01 provided by DESeq software25, the number of differentially expressed genes for Dolichospermum sp. UHCC 0315 was 348, of which 197 genes were upregulated and 151 were downregulated (Supplementary Table S2). A total of 136 upregulated and 90 downregulated genes were classified as hypothetical proteins or proteins with unknown functions, showing an extensive gap in our knowledge of the metabolism and genetic reservoir of the examined cyanobacteria. Dolichospermum sp. UHCC 0315 was struggling in the unfavorable environment of moderate salinity, suppressing many metabolic processes and protecting cellular biomolecules (Supplementary Fig. S7). The high demand for metabolism remodeling was implemented by adjusting the gene expression of transcription and translation, including 10 upregulated and two downregulated genes (Table 1). Furthermore, the pool of chaperones was reorganized. Nitrogen fixation is energetically expensive for cyanobacteria and pathway seemed to be mainly repressed (BMF77_01942-3; BMF77_01966-67: BMF77_01994-96) despite one nitrogenase gene was upregulated (BMF77_00202). Moreover, the nitrogen requirement was most likely fulfilled by reorganizing existing nitrogen reservoirs e.g. as indicated by the induced expression of BMF77_01290, nitrate reduction (BMF77_3991; 3993), and fine-tuning amino-acid metabolism (BMF77_00644; 02305; 03528). High demand for the reorganization of nitrogen metabolism was also seen among 30 most up and downregulated genes (Fig. 5). A general stress response was implemented, hindering the expression of the genes associated with photosynthesis (BMF77_00624; 00701; 01361; 02844; 02798) and electron transfer (BMF77_00909; 01935; 01997; 02996-7; 03623). In addition, induction of genes involved in modifying cell-wall components or encoding multiple transporters illustrated a need for reorganization of the cell-wall structure. In freshwater cyanobacteria, trehalose and sucrose are the major compatible solutes. Sucrose synthase genes were found in Dolichospermum UHCC 0315 but, surprisingly, there was no differential gene expression detected for the sucrose synthase genes. Hence, the absence of certain genes for compatible solute synthesis and the lacking induction of certain genes are probably important factors limiting the acclimation of Dolichospermum UHCC 0315 to higher salinity.
Biosynthetic potential
Based on antiSMASH analysis, the genome of Dolichospermum sp. UHCC 0315 harbors six known and four unknown gene clusters for the synthesis of naturally bioactive metabolites (Fig. 1a, Supplementary Table S3). Due to the capability of the strain to produce microcystins, our next interest was to follow the toxin concentration at freshwater and moderate salinities. At the latter salinity, the microcystin gene cluster was slightly upregulated (Table 2). Interestingly, the normalized microcystin concentration increased at unfavorably high salinities, reaching the highest amount at day 8 (892.4 ng microcystin/µg chlorophyll, Fig. 6). After that time, the concentration decreased slightly and approached the toxin level of the control cultures during the later phase of the experiment, whereas total quota of the microcystin concentration in the system increased during the experiment (Supplementary Fig. S8a). Combined intra- and extracellular microcystin concentrations were measured every fourth day at 0 and 6 g L−1 NaCl (Supplementary Fig. S8b).
Discussion
Species diversity in brackish water ecosystems is usually relatively narrow, because most of the organisms are adapted to live in either a marine or freshwater environment7. A recent study showed that Nodularia spumigena has remarkable genomic plasticity in acclimation strategies to the brackish water Baltic Sea23. However, in brackish water regions, N. spumigena coexisted with toxic Dolichospermum spp. but the former managed to sense the higher salinities fundamentally better than in freshwater environments23. In the light of projected salinity decreases in the Baltic Sea area, the question of whether freshwater origin Dolichospermum spp. may predominate in future cyanobacterial blooms in the Baltic Sea has arisen. Cyanobacteria from this genus are filamentous, capable of fixing atmospheric nitrogen, usually produce toxins, conditionally buoyant, and form akinetes under unfavorable conditions24,26. The abundance of Dolichospermum in the Baltic Sea is thus a consequence of ecological niche adaptation, for which the genomic inventory and regulatory apparatus provide the genetic underpinning.
Two substrains of Dolichospermum sp. UHCC 0315 (A and B) were distinguished at the genomic and physiological levels. Substrain B lacked a region of five genes, whereas substrain A harbored these genes and was capable of buoyancy. Otherwise, the genomic structure, including the same combination of plasmids, was similar albeit information on single-nucleotide polymorphisms (SNPs) was not gained by the methods used in this study. The association between the genes and the observed buoyancy phenotype is not directly clear, because both subtypes carry the full set of genes for gas vesicle formation (gvpA, gvpC, gvpN, gvpJ, gvpK, gvpF, gvpG, gvpV, gvpW. BMF77_4471–4480), similar to Nostoc sp. PCC7120 and Dolichospermum sp. UHCC 009027,28. Moreover, SNPs may have a great impact on the gene function. Even though the buoyancy of the cyanobacteria can be controlled by mechanisms other than gas vesicles29, the functions tentatively assigned to these five genes point towards the modification of surface properties, so a link, even if circumstantial, with buoyancy cannot be excluded. It is unknown when these changes occurred, during the years in the culture collection or before. However, stable laboratory conditions force cyanobacteria to diminish their energy demand by eliminating functionally redundant and nonessential parts of the genome that enhance fitness in the environment but are unnecessary under stable laboratory conditions28. Moreover, water column vertical migration and co-occurrence of buoyant/non-buoyant populations of cyanobacteria have been documented as result of variations in light incidence and nutrient concentration30. Identification of two substrains in the same culture underpins the importance of whole-genome analysis to elucidate explicit genetic structure and enhance the knowledge of novel genetic elements. The method coupling the short-read Illumina and long-read PacBio sequences used in this study serves as a platform for sequencing plastic genomes with high numbers of repetitive elements.
The genus Anabaena was recently separated into two genera: benthic Anabaena and gas-vesicle-producing planktonic Dolichospermum20. However, the new nomenclature has not yet been fully adopted by the scientific community, and mixing between these two genera and even Aphanizomenon and Nostoc species still occurs9,17. Nonetheless, a recent study indicates that Dolichospermum/Anabaena possess closely related genomic features to Aphanizomenon flos-aquae24. Considering that only Aphanizomenon flos-aquae isolates have been analyzed, future studies with other species, such as A. gracile and A. issatschenkoi, could uncover whether the entire genus is related to Anabaena and Dolichospermum. In the phylogenomic tree, the newly sequenced Dolichospermum sp. UHCC 0315 branched with several other members of the Anabaena/Dolichospermum/Aphanizomenon clade (ADA), while N. spumigena was separated into a single tight cluster in accordance to previous results23. However, 4 Anabaena strains (PCC 7122, PCC 7108. ATCC 33047 and 4-3) genomes formed 2 divergent clades as previously shown23 and might indicate a yet not fully understood taxonomic relationship. In the case of strains ATCC 33047 and 4-3, the taxonomy seems to be even more unclear since 16S rRNA allocated these two strains among Nostoc, Desmonostoc and Aliinostoc. The inclusion of new genomes of these cyanobacteria and further taxonomic studies will possibly indicate whether the ADA clade is polyphyletic or should these four Anabaena genomes be reclassified in a different taxon. As expected, the addition of Dolichospermum sp. UHCC 0315 to the ADA group core-genome analysis did not affect the previously estimated common gene pool of 1,500 genes24. However, the pan-genome of the clade is yet open and calculated to be currently similar to the one for Microcystis (12,000)31. Therefore, future inclusions of new genomes are likely to expand the full complement of genes of the clade.
The genome size of newly sequenced Dolichospermum sp. UHCC 0315 was 5.4 Mbp, which is approximately the same size as its closest counterpart Dolichospermum sp. UHCC 0090 (5.31 Mbp)28. Moreover, the comparison of the two complete genomes indicated a relatively high level of conservation between them and no clear divergence of biosynthetic potential or coding sequences. Therefore, the close genetic association of UHCC 0315 with a freshwater cyanobacteria and the harmfulness of small increases in salinity indicate that Dolichospermum spp. have most probably been transported from freshwater ecosystems to the Baltic Sea.
The most rapid growth of Dolichospermum sp. UHCC 0315 was obtained under freshwater conditions (0 g L−1 NaCl), while higher salinities (>3 g L−1 NaCl) clearly hindered growth. However, slight increases in salinity (3 g L−1 NaCl) were not harmful for the strain, suggesting that the examined cyanobacterium sensed minor increases in salinity in studied environments, which may be caused by water fluctuation and saltwater pulses. Our findings agree well with field studies in the Baltic Sea in which salt concentrations play crucial roles in the abundance and intensity of N. spumigena and Dolichospermum spp. in cyanobacterial blooms19,23. Furthermore, the abundance of Dolichospermum is higher in the low-salinity coastal regions, whereas N. spumigena predominates in the more saline areas9,10.
In general, salt shock inhibits proper functioning of several proteins and metabolic processes such as photosynthesis, central metabolism, and cellular growth, clearly requiring the reconstruction of cellular metabolism32,33,34,35,36,37,38,39,40. For example, nitrogen fixation is an energy-demanding process for diazotrophic cyanobacteria38, and thus reducing expression of the nitrogen fixation (nif) gene cluster decreased the energy requirement of Dolichospermum sp. UHCC 0315 under stressful conditions. The transcript accumulation of chaperones, the molecules that play crucial roles in protein folding, protection, and repair under stressful conditions, were heavily modified. The role of chaperones in maintaining protein integrity under low and high salt stress was previously described35,41, and this study further showed the demand for different types of chaperones in salinity stress. In this study, the expression of group 2 sigma factors was heavily repressed in Dolichospermum UHCC 0315 growing at high salinity. The question of which sigma factors participate in gene expression in changing salinities remains unanswered42.
The genomes of cyanobacteria are highly dynamic entities. One factor affecting genome composition and especially the responses to phage attacks are CRISPR/Cas systems. Therefore, the finding of an unusual CRISPR repeat-spacer array lacking any known cas gene is of interest. Possible orphan repeat-spacer arrays exist in many filamentous cyanobacteria43. However, just next to this array is a gene encoding a 696 amino acid protein with similarity to TnpB transposases of the IS605 family, a feature associated also with CRISPR effector proteins Cas12b (C2c1) and C2c344. The possibility that this array is part of a functional CRISPR system is supported further by our finding that spacer 7 likely targets a phage antirepressor gene present in at least two other Nostocales.
The induction of either general or stress-specific genes and regulatory systems enables cyanobacteria to thrive, even in changing environments. The transcriptional regulatory system of model cyanobacteria for adapting to elevated salt concentrations is induced rapidly31, but complete acclimation of metabolic processes resulting in inhibition of cell division requires longer periods of time33,34. For example, a study of Synechocystis sp. PCC 6803 showed that the number of differentially expressed genes peaked after 30 min of salt shock, and the majority of these genes returned to the control level after acclimation for 24 h35. Relying on the number of differentially expressed genes found after 16 days of incubation in different salt concentrations, we assumed that reconstruction of the transcriptional pattern remained vigorous long enough to minimize the harmfulness of unfavorable salt conditions and fine-tune the metabolic patterns and structural components. Accumulation and production of compatible solutes together with ion exchange through the cell membrane are major salt-stress adaptation strategies for minimizing the harmfulness of increased intracellular ion concentrations36. Of the compatible solutes, low-halotolerance strains are able to produce trehalose and sucrose37. However, only genes for sucrose synthesis were found in the genome of Dolichospermum sp. UHCC 0315 but increased abundance of the transcripts was not identified.
Cyanobacterial genomes harbor a wide variety of gene clusters responsible for the production of biologically active natural products, of which toxins, e.g. microcystins, are the most studied molecules, due to their harmfulness to mammals. A search of the gene clusters responsible for the production of natural products in Dolichospermum sp. UHCC 0315 resulted in similar genomic construction to that found in the genome of Dolichospermum sp. UHCC 009027. However, while the anabaenopeptin gene cluster was absent in the genome of the later, the hassallidin was absent in the former. The concentration of microcystin/cell produced by Dolichospermum sp. UHCC 0315 increased at moderate salinity (6 g L−1 NaCl), which is well in line with previous studies in which high salinity triggered microcystin production in Dolichospermum19. Moreover, salt shock may cause oxidative stress in cyanobacteria which is harmful especially to lipids, and activation of lipid metabolism in the target strain cells was also found45,46. Despite the role of toxins in cyanobacteria being partially unclear, their function as protective elements against oxidative stress has been proposed47,48. Our findings thus further indicated that unfavorably high salinity induces oxidative stress and, together with other cellular stress responses, microcystins may play a role as protective molecules against oxidative stress.
Conclusion
The present study provides new insight into the genetic diversity of the Dolichospermum genus and explores the phylogenetic relationship of this toxic bloom-forming cyanobacterium with other members of the Anabaena and Aphanizomenon flos-aquae taxa. Despite the distinct morphological aspects of these cyanobacteria, the results support previous studies suggesting a demand for an explicit revision of these taxa. Beyond that, the study of Dolichospermum sp. UHCC 0315 genome revealed adaptive responses employed by the strain to explore different salinities. Future alteration in the Baltic Sea water due to climate change will possibly lead to an expansion of blooms dominated by Dolichospermum over Nodularia spumigena.
Methods
Strains, cultivation, and toxin analysis
Microcystin-producing Dolichospermum sp. strain UHCC 0315 (former name Anabaena sp. strain 315) is maintained at the University of Helsinki, Faculty of Agriculture and Forestry, Department of Microbiology Culture Collection (HAMBI, UHCC) under continuous illumination of 3.2–3.7-μmol photons m−2 s−1. The strain was isolated in August 1997 from a cyanobacterial bloom in the coastal Helsinki area9, made axenic, and since isolation cultivated in Z8 medium without nitrogen49. Three replicates were cultivated in four different saline solutions (0, 3, 6, and 9 g L−1 NaCl) at 20 °C under continuous illumination of 3.2–3.7-μmol photons m−2 s−1 for 24 d. Chlorophyll a was measured at 4-d intervals by filtering 1 mL of culture through 21-mm GF/C glass microfiber filters (GE HealthCare, Chicago, IL, USA). Chlorophyll a was extracted using 1 mL of 90% acetone, and absorbance was determined at 664, 647, and 630 nm and calculated as described earlier50. Cell counting was applied accordingly to Coloma et al.51. The combined intra- and extracellular concentrations of microcystins were determined using LC-MS as described earlier23. A standard curve containing a dilution series of known microcystin concentrations was run alongside the toxin samples.
Genome sequencing and annotation
A NucleoBond® anion-exchange (AXG) kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) was used to extract high-molecular-weight genomic DNA from the middle-logarithmic growth phase of Dolichospermum sp. UHCC 0315. The DNA libraries, PacBio RS II sequencing with P6-C4 chemistry, and genome assembly using Hierarchical Genome Assembly Process 3 (HGAP3) protocol with Quiver polishing52 were conducted in the DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki. Paired-end Illumina Hiseq2500 reads (Macrogen Inc., Seoul, South Korea) were mapped twice against the PacBio assembly, using Pilon v. 1.20 software and resulting in correction of 225 sites53.
The genome of Dolichospermum sp. UHCC 0315 was annotated, using Prokka v. 1.1254 and noncoding RNA (ncRNA) by querying the RNA family (rfam) database55, using cmscan from Infernal56. An additional automatic annotation and functional classification in subsystems were performed using the default parameters in the RAST server57 and SEED viewer58. InterProScan 5 was used to polish the protein annotation, followed by manual curation in the genome browser Artemis59. Blast KEGG Orthology And Links Annotation (BlastKOALA) v. 2.160 was used to annotate the KEGG Orthology (KO) orthologs and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Clusters of Orthologous Groups (COG) orthology was annotated by querying the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD)61 with an e-value cutoff of 1e-5, using Reversed Position Specific BLAST (RPSBLAST) 2.2.31+62. BLAST2GO63 was used to annotate the Gene Ontology (GO) term. The putative secondary metabolites and biosynthetic gene clusters were predicted, using the antiSMASH 3.0 online server64. The genome and plasmid sequences were scanned against the Clustered Regularly Interspaced Short Palindromic Repeat database (CRISPRdb), using CRISPRfinder and CRISPRCasFinder to annotate the CRISPR loci65,66,67. The Insertion Sequence (IS) elements were annotated, using the IS Semiautomatic Genomic Annotation (ISsaga) webserver68.
To ensure two genetic substrains, a total of 70 single filaments of Dolichospermum UHCC 0315 were extracted on Z8X agar plates and further cultivated in liquid Z8X medium49. DNA was extracted from three surface-growing and three bottom-growing cultures, using the Macherey-Nagel NucleoSpin kit. The PCR primers were designed to amplify the 300-bp region of the deleted gene BMF77_01254, as well as three regions in plasmids 1–3 (Supplementary Table S1). The PCR mixture contained 1 × DyNAzyme PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.5 µM primers, and 0.5 U DyNAzyme II polymerase in a total volume of 20 µL. As a template, 50–70 ng of genomic DNA was used. DNA from the original Dolichospermum sp. UHCC 0315 culture was used as a positive control and water as a negative control. The PCR program was 94 °C for 3 min; 30 cycles of 94 °C for 1 min, 56 °C for 1 min, and 72 °C for 1 min; and 72 °C for 10 min. PCR amplification products were visualized on 1% agarose gel electrophoresis.
Phylogenetic inference and comparative genomics
A maximum-likelihood phylogenomic tree was constructed by the concatenation of 31 universal marker genes69,70. In brief, the profiles of these marker genes were scanned against the open reading frames (ORFs) of the 76 reference genome sequences and aligned using anvi’o v5.171 workflow for phylogenomics. The protein substitution model LG + I + G was assigned as the best one for the 31 proteins by performing BIC calculation in ProtTest v3.272 (default parameters). A maximum-likelihood tree was estimated with Randomized Axelerated Maximum Likelihood (RAxML) v. 8.2.1073, based on selected partitions and substitution models, with 1000 rapid bootstrap searches. Gloeobacter violaceus PCC 7421 was selected as an outgroup to root the phylogenomic tree. Average Amino Acid and Nucleotide Identity heatmaps were estimate using the average identity matrices protocol in the program GET_HOMOLOGUES74,75 and generated using a seaborn v0.9 library heatmap script76.
The 16S rRNA gene tree was constructed by RAxML v8.0.073 with 1000 bootstrap replicate using 75 cyanobacterial strains. The genes were aligned using MEGA v10.0.5 (default parameters)77 and the evolutionary model GTR + I + G selected as best fitting by jModelTest v2.1.178.
Pan and core-genomes were estimated using the OrthoMCL algorithm and Tettelin function implemented in the program GET_HOMOLOGUES74,75. OrthoVenn with default parameters was employed to generate the Venn diagram of Dolichospermum sp. UHCC 0315 and UHCC 009079. Shared and specific proteins were extracted using the server tools and automatically annotated by the default parameters in the metagenomics analysis server (MG-RAST)80. Pairwise genome alignments and dotplots were performed, using MUMmer v. 3.1.81. Synteny blocks were predicted and visualized, using the jcvi v. 0.7.1 Python library82 and local synteny blocks were visualized, using the genoPlotR v. 0.8.4 package83.
RNA sequencing
On day 16, cells from the cultures containing 0 and 6 g L−1 NaCl were collected on 0.22 µm polycarbonate filters (GE Water and Process Technologies). The filters were frozen in liquid nitrogen and stored at −80 °C. Prior to harvesting, the liquid cultures were fixed by 5% phenol in 10% ethanol. RNA was extracted, using the RNeasy mini kit (Qiagen N.V., Venlo, The Netherlands). For the depletion of genomic DNA, the TURBO DNA-free™ kit (Life Technologies, now ThermoFisher Scientific, Carlsbad, CA, USA) was used, and the RNA was further cleaned and concentrated with the RNA Clean & Concentrator™ kit (Zymo Research Corp., Irvine, CA, USA). RNA integrity and purity were determined, using the Bioanalyzer RNA 6000 Nano Kit (Agilent Technologies Inc., Santa Clara, CA, USA). Ribosomal RNA (rRNA) removal (MICROBExpress™ Bacterial messenger RNA (mRNA) enrichment kit, Life Technologies), complementary DNA (cDNA) library preparation (Bacterial ScrptSeq Complete Kit, Illumina), and paired-end Illumina Hiseq2500 sequencing were carried out at the Institute for Molecular Medicine Finland (FIMM).
The quality of the raw reads was checked using FastQC v. 0.11.4 (Babrahan Bioinformatics, Babrahan Institute, Cambridge, UK)84, and the reads were trimmed with Trimmomatic85. The clean reads were aligned to the reference genomes, using Burrows-Wheeler Aligner-Maximum Exact Match (BWA-MEM) v. 0.7.786 in paired-end mode with default parameters. Sequence Alignment/Map tools (SAMtools) v. 1.287 was applied to convert the resulting SAM format to the Binary Alignment Map (BAM) format and filtered with the BAM filter (Galaxy Version 0.5.7.1) to remove the reads that were unmapped, less than 20 nucleotides long, flagged as secondary alignments, marked as PCR duplicates, or of low quality. The filtered BAM files were sorted with chromosomal coordinates using SAMtools v. 1.1987 and converted to Wiggle format, using the script bam2wig.py from the RNA Sequence Quality Control (RseQC) v. 2.4 package88 in paired-end mode, and normalized to 1000000000 wigsum. The Wiggle format was then converted to Artemis59-compatible genome coverage graphs, using custom scripts available at https://github.com/housw/GRPutils. FeatureCounts v. 1.4.6.p589 was used to quantify unambiguously aligned fragments. Only paired-end reads that constituted a fragment size within 50~600 nucleotides long were considered, and chimeric fragments were excluded. The Galaxy instance of Freiburg University was used for RNA-Seq analysis90,91.
To exclude weakly expressed features, the counts were converted to counts per million (CPM); only features having a higher CPM (sum of CPM across all samples should be more than 3 and at least three samples had a CPM more than 1) were taken for differential expression analysis. Features belonging to rRNA and transfer RNA (tRNA) were excluded in this study. The differentially expressed genes between control and treatment were called, using both edgeR v3.14.092 and DESeq v1.9.1225 following the simple design protocol93. A GO enrichment test was performed for the differentially expressed genes, using GOstats v. 2.40.094 with all the expressed genes as background. Semantic similarities of the enriched GO terms were calculated, using the REVIGO online server95.
Data Availability
The sequences were deposited in the NCBI database under the BioProject accession number PRJNA377208.
References
Zillén, L., Conley, D. J., Andrén, T., Andrén, E. & Björck, S. Past occurrences of hypoxia in the Baltic Sea and the role of climate variability, environmental change and human impact. Earth-Science Rev. 91, 77–92 (2008).
Kjellström, E. & Ruosteenoja, K. Present-day and future precipitation in the Baltic Sea region as simulated in a suite of regional climate models. Clim. Change 81, 281–291 (2007).
von Storch, H., Omstedt, A., Pawlak, J. & Reckermann, M. Introduction and summary. in Second assessment of climate change for the Baltic Sea basin (ed. The BACC II Author Team) 1–22 (Springer International Publishing AG 2015).
Graham, L. P. Climate change effects on river flow to the Baltic Sea. Ambio 33, 235–241 (2016).
Johannesson, K. & André, C. Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol. Ecol. 15, 2013–2029 (2006).
Herlemann, D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579 (2011).
Telesh, I., Schubert, H. & Skarlato, S. Life in the salinity gradient: discovering mechanisms behind a new biodiversity pattern. Estuar. Coast. Shelf Sci. 135, 317–327 (2013).
Celepli, N., Sundh, J., Ekman, M., Dupont, C. L. & Yooseph, S. Meta-omic analyses of Baltic Sea cyanobacteria: diversity, community structure and salt acclimation. Environ. Microbiol. 19, 1–30 (2017).
Halinen, K., Jokela, J., Fewer, D. P., Wahlsten, M. & Sivonen, K. Direct evidence for production of microcystins by Ananbaena strains from the Baltic Sea. Appl. Environ. Microbiol. 73, 6543–6550 (2007).
Sivonen, K. et al. Bacterial diversity and function in the Baltic Sea with an emphasis on cyanobacteria. Ambio 36, 180–185 (2007).
Fewer, D. P. et al. Culture-independent evidence for the persistent presence and genetic diversity of microcystin-producing Anabaena (Cyanobacteria) in the Gulf of Finland. Environ. Microbiol. 11, 855–866 (2009).
Sivonen, K. et al. Occurrence of the hepatotoxic cyanobacterium Nodularia spumigena in the Baltic Sea and structure of the toxin. Appl. Environ. Microbiol. 55, 1990–1995 (1989).
Chen, Y., Shen, D. & Fang, D. Nodularins in poisoning. Clin. Chim. Acta 425, 18–29 (2013).
Laamanen, M. J., Muriel, F. G., Lehitmäki, J. M., Haukka, K. & Sivonen, K. Diversity of toxic and nontoxic Nodularia isolates (Cyanobacteria) and filaments from the Baltic Sea. Appl. Environ. Microbiol. 67, 4638–4647 (2011).
Lyra, C., Laamanen, M., Lehtimäki, J. M., Surakka, A. & Sivonen, K. Benthic cyanobacteria of the genus Nodularia are non-toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia. Int. J. Syst. Bacteriol. 55, 555–568 (2005).
Rippka, R., Iteman, I. & Herdman, M. Form – Anabaena in Bergey’s manual of systematics of archaea and bacteria (ed. Whitman, W. B. et al.), https://doi.org/10.1002/9781118960608.gbm00452 (John Wiley & Sons 2015).
Halinen, K. et al. Genetic diversity in strains of the genus Anabaena isolated from planktonic and benthic habitats of the Gulf of Finland (Baltic Sea). FEMS Microbiol. Ecol. 64, 199–208 (2008).
Vuorinen, I. et al. Scenario simulations of future salinity and ecological consequences in the Baltic Sea and adjacent North Sea areas–implications for environmental monitoring. Ecol. Indic. 50, 196–205 (2015).
Brutemark, A., Vandelannoote, A., Engström-Öst, J. & Suikkanen, S. A less saline Baltic Sea promotes cyanobacterial growth, hampers intracellular microcystin production, and leads to strain-specific differences in allelopathy. PLoS One 10, e0128904, https://doi.org/10.1371/journal.pone.0128904 (2015).
Wacklin, P., Hoffmann, L. & Komárek, J. Nomenclatural validation of the genetically revised cyanobacterial genus Dolichospermum (RALFS ex BORNET et FLAHAULT) comb. nova. Fottea 9, 59–64 (2009).
Lachance, M.-A. Genetic relatedness of heterocystous cyanobacteria by deoxyribonucleic acid-deoxyribonucleic acid reassociation. Int. J. Syst. Bacteriol. 31, 139–147 (1981).
Li, X., Dreher, T. W. & Li, R. An overview of diversity, occurrence, genetics and toxin production of bloom-forming Dolichospermum (Anabaena) species. Harmful Algae 54, 54–68 (2016).
Teikari, J. E., Hou, S., Wahlsten, M., Hess, W. R. & Sivonen, K. Comparative genomics of the Baltic Sea toxic cyanobacteria Nodularia spumigena UHCC 0039 and its response to varying salinity. Front. Microbiol. 9, 358, https://doi.org/10.3389/fmicb.2018.00356 (2018).
Driscoll, C. B. et al. A closely-related clade of globally distributed bloom-forming cyanobacteria within the Nostocales. Harmful Algae 77, 93–107 (2018).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106, https://doi.org/10.1186/gb-2010-11-10-r106 (2010).
Castenholz, R. W. General characteristics of the cyanobacteria in Bergey’s manual of systematics of archaea and bacteria. (ed. Whitman, W. B. et al.), https://doi.org/10.1002/9781118960608.cbm00019 (John Wiley & Sons 2015).
Kaneko, T. et al. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 31, 227–253 (2001).
Wang, H. et al. Genome-derived insights into the biology of the hepatotoxic bloom-forming cyanobacterium Anabaena sp. strain 90. BMC Genomics 13, 613, https://doi.org/10.1186/1471-2164-13-613 (2012).
Chu, Z., Jin, X., Yang, B. & Zeng, Q. Buoyancy regulation of Microcystis flos-aquae during phosphorus-limited and nitrogen-limited growth. J. Plankton Res. 29, 739–745 (2007).
Brookes, J. D. & Ganf, G. G. Variations in the buoyancy response of Microcystis aeruginosa to nitrogen, phosphorus and light. J. Plankton Res. 23, 1399–1411 (2001).
Humbert, J.-F. et al. A tribute to disorder in the genome of the bloom-forming freshwater cyanobacterium Microcystis aeruginosa. PLoS ONE 8, e70747, https://doi.org/10.1371/journal.pone.0070747 (2013).
Billis, K., Billini, M., Tripp, H. J., Kyrpides, N. C. & Mavromatis, M. Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One 9, e109738, https://doi.org/10.1371/journal.pone.0109738 (2014).
Al-Hosani, S., Oudah, M. M., Henschel, A. & Yousef, L. F. Global transcriptome analysis of salt acclimated Prochlorococcus AS9601. Microbiol. Res. 176, 21–28 (2015).
Qiao, J. et al. Integrated proteomic and transcriptomic analysis reveals novel genes and regulatory mechanisms involved in salt stress responses in Synechocystis sp. PCC 6803. Appl. Microbiol. Biotechnol. 97, 8253–8264 (2013).
Marin, K. et al. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant. Physiol. 136, 300–329 (2004).
Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 35, 87–123 (2011).
Klähn, S. & Hagemann, M. Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 13, 551–532 (2011).
Allakhverdiev, S. I. & Murata, N. Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth. Res. 98, 529–539 (2008).
Rai, S., Singh, S., Shrivastava, A. K. & Rai, L. C. Salt and UV-B induced changes in Anabaena PCC 7120: physiological, proteomic and bioinformatic perspectives. Photosynth. Res. 118, 105–114 (2013).
Fernandes, T. A., Iyer, V. & Apte, S. K. Differential responses of nitrogen-fixing cyanobacteria to salinity and osmotic stresses. Appl. Environ. Microbiol. 59, 899–904 (1993).
Fulda, S. et al. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6, 2733–2745 (2006).
Imamura, S. & Asayama, M. Sigma factors for cyanobacterial transcription. Gene Regul. Syst. Biol. 3, 65–87 (2009).
Hou, S. et al. CRISPR-Cas systems in multicellular cyanobacteria. RNA Biology, epub ahead of press. Published online, https://doi.org/10.1080/15476286.2018.1493330 (2018).
Shmakov, S. et al. Discovery and functional characterization of diverse Class 2 CRISPR-Cas systems. Mol. Cell. 60, 385–97 (2015).
Latifi, A., Ruiz, M. & Zhang, C. C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278 (2009).
He, Y.-Y. & Häder, D.-P. Involvement of reactive oxygenspecies in the UV-B damage to the cyanobacterium Anabaena sp. J. Photochem. Photobiol. B 66, 73e80 (2002).
Dziallas, C. & Grossart, H.-P. Increasing oxygen radicals and water temperature select for toxic Microcystis sp. PLoS One 6, e25569, https://doi.org/10.1371/journal.pone.0025569 (2011).
Zilliges, Y. et al. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One 6, e17615, https://doi.org/10.1371/journal.pone.0017615 (2011).
Kótai, J. Instructions for preparation of modified nutrient solution Z8 for algae. NIVA B-11/69 (1972).
Jeffrey, S. W. & Humphrey, G. F. New spectrophotometric equations for determining Chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167, 191–194 (1975).
Coloma, S. E. et al. Newly isolated Nodularia phage influences cyanobacterial community dynamics. Environ. Microbiol. 19, 273–286 (2017).
Chin, C.-H. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods 10, 536–569 (2013).
Walker, B. J. et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963, https://doi.org/10.1371/journal.pone.0112963 (2014).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–4, https://doi.org/10.1093/nar/gki081 (2005).
Nawrocki, E. P., Kolbe, D. L. & Eddy, S. R. Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337 (2009).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 8, 75, https://doi.org/10.1186/1471-2164-9-75 (2008).
Overbeek, R. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–214, https://doi.org/10.1093/nar/gkt1226 (2014).
Carver, T., Harris, S. R., Berriman, M., Parkhill, J. & McQuillan, J. A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469 (2012).
Kanehisa, M., Sato, Y. & Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226, https://doi.org/10.1093/nar/gku1221 (2015).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 1–9 (2009).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Weber, T. et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–43, https://doi.org/10.1093/nar/gkv437 (2015).
Grissa, I., Vergnaud, G. & Pourcel, C. CRISPRFinder: a web tool to identify clustered regularly interspace short palindromic repeats. Nucleic Acids Res. 35, 52–57 (2007).
Couvin, D. et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251, https://doi.org/10.1093/nar/gky425 (2018).
Abby, S. S., Néron, B., Ménager, H., Touchon, M. & Rocha, E. P. C. MacSyFinder: A program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One 9, e110726, https://doi.org/10.1371/journal.pone.0110726 (2014).
Varani, A. M., Siguier, P., Gourbeyre, E., Charneau, V. & Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semiautomatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12, R30, https://doi.org/10.1186/gb-2011-12-3-r30 (2011).
Shih, P. M. et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 110, 1053–1058 (2013).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 30, 1394–1403 (2004).
Eren, A. M. et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319, https://doi.org/10.7717/peerj.1319 (2015).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011).
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Contreras-Moreira, B. & Vinuesa, P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701 (2013).
Vinuesa, P. & Contreras-Moreira, B. Robust identification of orthologues and paralogues for microbial pan-genomics using GET_HOMOLOGUES: a case study of pIncA/C plasmids in Bacterial Pangenomics. Methods in molecular biology. (ed. Mengoni, A., Galardini, M. & Fondi M.) 1231 (Humana Press, New York, NY 2015).
Waskom, M. et al. Mwaskom/Seaborn: V0.9.0 https://github.com/mwaskom/seaborn/releases (2018).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 25, 1253–1256 (2008).
Wang, Y., Coleman-Derr, D., Chen, G. & Gu, Y. Q. OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43, W78–84, https://doi.org/10.1093/nar/gkv487 (2015).
Meyer, F. et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386, https://doi.org/10.1186/1471-2105-9-38 (2008).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biology 5, R1 (2004).
Tang, H., Krishnakumar, V., Li, J. & Zhang, X. jcvi: JCVI utility libraries. Zenodo, https://github.com/tanghaibao/jcvi (2015)
Guy, L., Kultima, J. R. & Andersson, S. G. E. GenoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335 (2010).
Andrews, S. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–30 (2014).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–10, https://doi.org/10.1093/nar/gkw343 (2016).
Cock, P. J. A., Grüning, B. A., Paszkiewicz, K. & Pritchard, L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ 1, e167, https://doi.org/10.7717/peerj.167 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 (2013).
Falcon, S. G. R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One 6, e21800, https://doi.org/10.1371/journal.pone.0021800 (2011).
Acknowledgements
This work, resulting from the BONUS Blueprint project, was supported by BONUS (Art 185) and funded jointly by the E.U. and Academy of Finland to K.S. The bioinformatics analyses were supported by the German Federal Ministry for Education and Research (BMBF) program de.NBI-Partner (Grant 031 L0106B) to WRH and by a China Scholarship Council grant to S.W.H. R.V.P. was supported by an EDUFI fellowship of the Centre for International Mobility (CIMO). Moreover, the authors acknowledge the support of the Freiburg Galaxy Team, funded by BMBF grant 031 A538A RBC (de.NBI). We thank L. Saari for maintenance of the cyanobacterial cultures and the personnel in the sequencing centers at the Institute of Biotechnology, University of Helsinki, FIMM, and Macrogen Ltd.
Author information
Authors and Affiliations
Contributions
J.E.T. and K.S. designed and executed the experiment. M.W. performed LC-MS analysis. R.V.P., J.E.T., S.H. and W.R.H. were responsible for the analysis of the data. All authors participated in the writing and in the discussion of the results.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Teikari, J.E., Popin, R.V., Hou, S. et al. Insight into the genome and brackish water adaptation strategies of toxic and bloom-forming Baltic Sea Dolichospermum sp. UHCC 0315. Sci Rep 9, 4888 (2019). https://doi.org/10.1038/s41598-019-40883-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-40883-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.