Abstract
The FGF receptor signaling pathway is recurrently involved in the leukemogenic processes. Oncogenic fusions of FGFR1 with various fusion partners were described in myeloid proliferative neoplasms, and overexpression and mutations of FGFR3 are common in multiple myeloma. In addition, fibroblast growth factors are abundant in the bone marrow, and they were shown to enhance the survival of acute myeloid leukemia cells. Here we investigate the effect of FGFR stimulation on pediatric BCP-ALL cells in vitro, and search for mutations with deep targeted next-generation sequencing of mutational hotspots in FGFR1, FGFR2, and FGFR3. In 481 primary BCP-ALL cases, 28 samples from 19 unique relapsed BCP-ALL cases, and twelve BCP-ALL cell lines we found that mutations are rare (4/481 = 0.8%, 0/28 and 0/12) and do not affect codons which are frequently mutated in other malignancies. However, recombinant ligand FGF2 reduced the response to prednisolone in several BCP-ALL cell lines in vitro. We therefore conclude that FGFR signaling can contribute to prednisolone resistance in BCP-ALL cells, but that activating mutations in this receptor tyrosine kinase family are very rare.
Similar content being viewed by others
Introduction
Despite generally excellent outcome for children with pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL), therapeutic options are limited for refractory or relapsed cases. Small molecule inhibitors of oncogenic pathways, such as those targeting ABL-, MEK, and JAK kinase families, have shown (pre-)clinical efficacy and may be added to chemotherapy in the future. Here we studied the role of fibroblast growth factor receptor (FGFR) signaling in pediatric BCP-ALL.
FGFs are not only essential in hematopoietic development, but also influence bone formation by regulating expansion and stemness of mesenchymal stromal cells1,2. FGF2 is expressed in the hematopoietic and stromal compartments of the bone marrow1. FGFR fusions have been described in atypical myeloproliferative disorders which may involve lymphoma or present as BCP-ALL3. High FGFR3 expression is a hallmark of t(4;14)-positive multiple myeloma, and activating mutations are recurrently detected in this context4,5,6. Furthermore, FGFR signaling has been shown to contribute to drug resistance in chronic myeloid leukemia7. Thus, oncogenic FGFR signaling contributes to hematologic malignancies, but so far information on its role in pediatric BCP-ALL is limited. We therefore applied a customized targeted sequencing approach for mutational hotspots of FGFR1, FGFR2, and FGFR3 and evaluated the effect of recombinant FGF2 on in vitro prednisolone sensitivity in BCP-ALL cell lines.
Methods
Patient material and cell lines
The study comprised children with BCP-ALL with age at diagnosis ranging from 0 to 18 years. Use of excess materials for research purposes has been approved by the Institutional Review Board of the University Medical Center Rotterdam on August 2nd, 2004, IRB approval file number MEC 2004-203. Written informed consent was obtained from parents or legal guardians and studies were conducted in accordance with the Declaration of Helsinki. Clinical characteristics were kindly provided by the Dutch Childhood Oncology Group (the Hague, Netherlands). Mononuclear cells were isolated using density gradient centrifugation with Lymphoprep (Axis Shield, Norway) as previously described8. All samples contained at least 90% leukemic blasts. Cytogenetic subtypes were determined by karyotype, fluorescence in-situ hybridization (FISH), and/or fusion-gene specific PCR. BCR-ABL1-like cases were identified using microarray gene expression profiling by the means of a 110 probeset classifier9. Leukemic cell lines (697, HAL01, Kasumi-2, LAZ-221, MHH-CALL2, MHH-CALL3, MHH-CALL4, Nalm-6, RCH-ACV, Reh, Sup-B15, Tom-1) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Germany) and routinely validated by DNA fingerprinting. Unless indicated otherwise, all cell lines were cultured in RPMI-1640 supplemented with 10 to 20% fetal calf serum and penicillin, streptomycin and fungizone (all Life technologies).
In vitro cytotoxicity assays
The IC50 of prednisolone was determined in an MTS assay for cell lines and an MTT assay for primary ALL cells based on a six-step range from 250 to 0.008 µg/mL (Nalm6, 697, RCH-ACV, MHH-CALL3) or 1 to 0.003 µg/mL (MHH-CALL2, MHH-CALL4, SupB15, Tom1). For these assays, cells were tested for viability in the prednisolone concentration range with or without 50 ng/mL recombinant human FGF2 (Bio-Rad) and with or without 1 µM AZD4547 (Selleckchem). After four days incubation at 37 °C and 5% CO2, cell viability was evaluated using MTS/MTT. IC50-values were calculated as the prednisolone concentration at which 50% of cells survived relative to the respective prednisolone-free control of each condition. To avoid redundancies with abundant cytokines in fetal calf serum (FCS), these assays were performed under reduced serum conditions (2% FCS for cells usually cultured in 10% FCS, and 4% FCS for those usually cultured with 20% FCS). Sensitivity towards AZD4547 was measured similarly, but at normal serum concentrations (10 µM–0.3 nM).
Sequencing and code availability
DNA was isolated using Trizol reagent (Life Technologies), or in case of three cell lines using the DNeasy Kit (Qiagen). For TruSeq Custom Amplicon sequencing (Illumina, USA), sequencing libraries were prepared from 100–250 ng genomic DNA. Successful library preparation was confirmed using the Labchip GX genomic analyzer (Caliper Life Sciences Benelux N.V., the Netherlands). Samples were then pooled equimolarly and sequenced on an Illumina MiSeq in paired-end reads of 250 bp each. Targeted regions and the analysis script can be provided upon request. Next, sequence reads were aligned to the 1000 genomes human reference sequences (version b37, GATK resource bundle, Broad Institute, USA) using BWA v0.7.1010 and GATK indel realigner v.3.3-0. Single nucleotide variants were called with Freebayes v0.9.18–2411, Varscan v.2.3.712, Bcftools v1.013, and GATK v3.3-014. The resulting variant call format files were annotated using snpEff and snpSift v.4.1a15 and dbNSFP v.2.716. For reliable detection of high-confidence mutations, variants were filtered based on several criteria: For each sample, variants were excluded if they were reported by only one caller, coverage was <100 reads, or <20 reads supported the variant allele. Also variants likely to result from amplification or sequencing errors were excluded, i.e. those that occur repeatedly (n ≥10) but in a low fraction of reads within all samples (variant allele frequency (VAF) within each sample <2%) and those with an unequal distribution between runs. Furthermore, variants were only considered if they were non-synonymous, unlikely to be germline variants and not known SNPs. Mutational hotspots are indicated in Fig. 1c and were obtained from literature and the COSMIC database (v79, released 14th November 2016)6. All data and analysis code can be made available upon request.
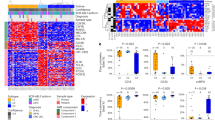
FGFR signaling is not essential in pediatric BCP-ALL cells, but can reduce prednisolone sensitivity. (a) Effect of 50 ng/mL recombinant human FGF2 on ex vivo response to prednisolone in BCP-ALL cell lines and reversal by the FGFR-inhibitor AZD4547 (1 µM). Left panels: prednisolone IC50-concentrations are depicted for each cell line. IC50-values for the combination of FGF2 and AZD4547 were calculated after correction for cytotoxic effects of FGF2 and AZD4547 as single agents on leukemic cell survival (see also panel (b); corresponding dose-response curves are shown in Supplemental Fig. 1b). Bars represent the mean ± SEM of Nalm6 (n = 3), 697 (n = 3), RCH-ACV (n = 3), SupB15 (n = 3), Tom1 (n = 2), MHH-CALL2 (n = 4), MHH-CALL3 (n = 1), and MHH-CALL4 (n = 4). Right panel: mean fold change of prednisolone IC50-values in FGF2-responsive cell lines (Nalm6, 697, RCH-ACV, Tom1, and MHH-CALL3), **p = 0.0012 by Wilcoxon matched pairs test. (b) Ex vivo response of BCP-ALL cell lines to the FGFR-inhibitor AZD4547. Dashed line: 50% survival; Cytogenetic subtypes are indicated in brackets: DUX4: DUX4-rearranged leukemia, TP: TCF3-PBX1-rearranged, BA: BCR-ABL1-rearranged, HD: high hyperdiploid karyotype, CRLF2: high CRLF2 expression, ER: ETV6-RUNX1-rearranged. (c) Schematic representation of protein domains of the FGFR genes and the observed variants. Black bars indicate the regions covered by targeted amplicon sequencing. Variants printed in bold above the scheme represent variants observed in this study, variants below represent those frequently reported in the COSMIC database. Ig: Immunoglobulin-like domain, AD: acidic domain, TMD: transmembrane domain, TKD: tyrosine-kinase domain, A-loop: activation loop. (d) Ex vivo response of primary BCP-ALL cells carrying an FGFR1 D129A mutation.
Results
FGF2 stimulation reduces prednisolone sensitivity in a subset of BCP-ALL cells
FGF2-mediated signaling through FGFR3 has been implicated in therapy resistance in AML7. We therefore evaluated the resistance-inducing effect of FGF2 on prednisolone cytotoxicity, a core component of ALL chemotherapy, using in vitro drug sensitivity assays. To avoid redundancies with abundant cytokines in fetal calf serum (FCS), these assays were performed under reduced serum conditions (2% FCS for cells usually cultured in 10% FCS, and 4% FCS for cells usually cultured with 20% FCS). The IC50 of prednisolone was determined in an MTS assay for eight BCP-ALL cell lines see also Supplemental Fig. 1). In five of the eight tested cell lines exposure to 50 ng/mL recombinant human FGF2 (Bio-Rad, USA) resulted in on average two- to three-fold higher IC50-values for prednisolone (Nalm6, 697, RCH-ACV, Tom1, and MHH-CALL3, Fig. 1a). The resistance induced by the ligand FGF2 was reversed by exposure to 1 µM of FGFR inhibitor AZD4547. AZD4547 as single agent reduced viability in BCP-ALL cell lines only at higher concentrations (5 to 20 µM, Fig. 1b), except for Nalm6 and RCH-ACV cell line (20–40% cell death induced at 1 µM AZD4547). The prednisolone IC50-values were corrected for the cytotoxic effect of AZD4547 as single agent and therefore reflect the synergistic effect of this FGFR inhibitor on the response to prednisolone.
Absence of know oncogenic mutations in the genes FGFR1, -2, and -3
Using the TruSeq Custom Amplicon technique as previously described (ref.17 and Supplementary information), we screened for mutations in 481 primary BCP-ALL cases at initial diagnosis, in 28 BCP-ALL relapse samples (representing 19 unique patients) and in twelve BCP-ALL cell lines. The regions covered by the custom amplicons are indicated in Fig. 1c, and were selected to cover regions of functional relevance (e.g. the tyrosine kinase domain and transmembrane domain) and previously reported mutational hotspots of FGFR1, FGFR2, and FGFR3 (Fig. 1c, hotspots are indicated below each gene). In 481 unique patients we found 4 non-synonymous, likely somatic FGFR variants. These variants were detected in samples taken at initial diagnosis, one FGFR1 (D129A) mutation, two FGFR2 mutations (V274I and D522N), and one FGFR3 (F384I) mutation (Fig. 1c, above gene schematics). No mutations were detected in the B-cell precursor ALL cell line panel or in the 28 relapse samples. The FGFR1 D129A and FGFR2 V274I mutations were predicted to be damaging to the protein function by the SIFT, Polyphen, LRT, and MutationTaster tools18. However, it is important to note that these prediction tools were trained to detect deleterious mutations, and activating mutations could be predicted as benign. Variant allele frequencies (VAF) varied, with 8.4% for the FGFR2 D522N variant, 28.5% for the FGFR1 D129A variant, 40.8% for the FGFR2 V274I variant, and 52.9% for the FGFR3 F384I variant. None of these mutations represents a frequently observed pathogenic mutation, such as those reported in multiple myeloma, according to literature and the COSMIC database (v79 released 14-Nov-16, indicated below each gene in Fig. 1c)19. Their phenotypic effect is unclear, however, all mutations locate within functional domains of the FGFR proteins (colored boxes).
FGFR variants occurred in two ETV6-RUNX1-rearranged cases, one BCR-ABL1-like with a JAK2 translocation, and one high hyperdiploid, and high white blood cell count (≥50 cells/nL) was found in two out of four cases, high minimal residual disease (percentage blasts in bone marrow on day 33 of therapy ≥0.1%) in one out of three cases (Supplemental Table 1 one case without data available). The JAK2-translocated case harboring an FGFR1 D129A mutation experienced a relapse 1 year after diagnosis. In conclusion, FGFR variants occurred in various subtypes and patients did not consistently present with poor or good prognostic factors. Notably, the FGFR inhibitor was not effective in primary leukemic cells carrying the D129A point mutation in FGFR1 (Fig. 1d). Unfortunately no viable leukemic cells were available for other mutated BCP-ALL cases.
Discussion
Here we screened 481 pediatric BCP-ALL cases at initial diagnosis, 19 relapse BCP-ALL cases, and 12 BCP-ALL cell lines for variants in FGFR1, FGFR2, and FGFR3. We found a low frequency of variants among patients at initial diagnosis, and no variants in the relapse cohort or in cell lines. These data indicate that FGFR mutations are very rare in pediatric BCP-ALL. Due to the low incidence and a small relapse cohort, it remains to be determined whether these rare mutations have an effect on the clinical prognosis. With the limited data available, we conclude that this does not seem to be the case.
The observed mutations are located within functional domains of the FGFR proteins: The FGFR1 D129A mutation affects the so-called acid box of the receptor, which has been shown to be essential for auto-inhibition and interaction with adhesion molecules20,21. The FGFR2 mutations V274I and D522N locate in the ligand-binding immunoglobulin-like (Ig) domain and the tyrosine kinase domain (TKD), respectively. They might therefore alter ligand-binding and kinase activation. Last, the FGFR3 F384I mutation is located in the transmembrane domain (TMD), which is frequently affected by mutations that cause aberrant receptor dimerization (e.g. by insertion of a cysteine). Of note, a rare F384L SNP at the same location has been reported and was also found in our screen. No functional consequences have been reported for this SNP, and the high similarity between leucine and isoleucine suggests no effect of the F384I variant on FGFR3 function22.
Given the previous implications of FGFR signaling in resistance, we evaluated its effect on the efficacy of prednisolone in vitro7. Our data indicate that the ligand FGF2 induced resistance to glucocorticoids which could be reversed by inhibition of FGFR. Traer et al. suggested that reactivation of the MAPK pathway by FGFR3 accounts for FGF-mediated resistance of BCR-ABL1-rearranged chronic myeloid leukemia cells to imatinib. In line with this observation, we and others have previously shown that MAPK pathway mediated signaling can also increase resistance towards prednisolone23,24,25. Downstream-located MAPK signaling may therefore be the mediator of FGF-induced glucocorticoid resistance, and could serve as therapeutic target for sensitization towards prednisolone in resistant cases.
Taken together, our data demonstrates that FGFR-activating lesions are rare in newly diagnosed pediatric BCP-ALL, and were not enriched at the time of relapse. Activating mutations which are frequently observed in multiple myeloma or solid cancers were not observed in 481 newly diagnosed nor in 19 relapsed BCP-ALL cases19. As FGFR activating mutations were virtually absent, FGF2 ligand-mediated activation of FGFR-signaling may be more relevant for the sensitivity of leukemic cells to prednisolone.
Data Availability
Data can be made available upon request.
References
Moroni, E., Dell’Era, P., Rusnati, M. & Presta, M. Fibroblast growth factors and their receptors in hematopoiesis and hematological tumors. J Hematother Stem Cell Res 11, 19–32, https://doi.org/10.1089/152581602753448513 (2002).
Coutu, D. L., Francois, M. & Galipeau, J. Inhibition of cellular senescence by developmentally regulated FGF receptors in mesenchymal stem cells. Blood 117, 6801–6812, https://doi.org/10.1182/blood-2010-12-321539 (2011).
Wakim, J. J., Tirado, C. A., Chen, W. & Collins, R. t(8;22)/BCR-FGFR1 myeloproliferative disorder presenting as B-acute lymphoblastic leukemia: report of a case treated with sorafenib and review of the literature. Leukemia research 35, e151–153, https://doi.org/10.1016/j.leukres.2011.05.013 (2011).
Richelda, R. et al. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood 90, 4062–4070 (1997).
Chesi, M. et al. Frequent translocation t(4;14)(p16.3; q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nature genetics 16, 260–264, https://doi.org/10.1038/ng0797-260 (1997).
Chesi, M. et al. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood 97, 729–736 (2001).
Traer, E. et al. FGF2 from Marrow Microenvironment Promotes Resistance to FLT3 Inhibitors in Acute Myeloid Leukemia. Cancer research 76, 6471–6482, https://doi.org/10.1158/0008-5472.CAN-15-3569 (2016).
Den Boer, M. L. et al. Patient stratification based on prednisolone-vincristine-asparaginase resistance profiles in children with acute lymphoblastic leukemia. J Clin Oncol 21, 3262–3268, https://doi.org/10.1200/JCO.2003.11.031 (2003).
van der Veer, A. et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood 122, 2622–2629, https://doi.org/10.1182/blood-2012-10-462358 (2013).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv 1303, 3997 (2013).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv 1207, 3907 (2012).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–576, https://doi.org/10.1101/gr.129684.111 (2012).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993, https://doi.org/10.1093/bioinformatics/btr509 (2011).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43, 491–498, https://doi.org/10.1038/ng.806 (2011).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly 6, 80–92, https://doi.org/10.4161/fly.19695 (2012).
Liu, X., Wu, C., Li, C. & Boerwinkle, E. dbNSFPv3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat 37, 235–241, https://doi.org/10.1002/humu.22932 (2016).
Jerchel, I. S. et al. RAS pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K 32, 931–940, https://doi.org/10.1038/leu.2017.303 (2018).
Liu, X., Jian, X. & Boerwinkle, E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 32, 894–899, https://doi.org/10.1002/humu.21517 (2011).
Greulich, H. & Pollock, P. M. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med 17, 283–292, https://doi.org/10.1016/j.molmed.2011.01.012 (2011).
Kalinina, J. et al. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure 20, 77–88, https://doi.org/10.1016/j.str.2011.10.022 (2012).
Sanchez-Heras, E., Howell, F. V., Williams, G. & Doherty, P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J Biol Chem 281, 35208–35216, https://doi.org/10.1074/jbc.M608655200 (2006).
Kant, S. G. et al. A novel variant of FGFR3 causes proportionate short stature. European journal of endocrinology/European Federation of Endocrine Societies 172, 763–770, https://doi.org/10.1530/EJE-14-0945 (2015).
Rambal, A. A., Panaguiton, Z. L., Kramer, L., Grant, S. & Harada, H. MEK inhibitors potentiate dexamethasone lethality in acute lymphoblastic leukemia cells through the pro-apoptotic molecule BIM. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K 23, 1744–1754, https://doi.org/10.1038/leu.2009.80 (2009).
Aries, I. M. et al. Towards personalized therapy in pediatric acute lymphoblastic leukemia: RAS mutations and prednisolone resistance. Haematologica 100, e132–136, https://doi.org/10.3324/haematol.2014.112995 (2015).
Jones, C. L. et al. MAPK signaling cascades mediate distinct glucocorticoid resistance mechanisms in pediatric leukemia. Blood 126, 2202–2212, https://doi.org/10.1182/blood-2015-04-639138 (2015).
Acknowledgements
This work was supported by the NWO VICI program (grant 016.126.612), the Dutch Cancer Society (grants AMC 2008–4265 and EMCR 2014–6998), and the KIKA Foundation (grants 132 and 161) and the Pediatric Oncology Foundation Rotterdam. The Center for Personalized Cancer Treatment is supported by the NutsOhra Foundation (grant 1102-062). We thank Edwin Cuppen from the Center for Molecular Medicine and Cancer Genomics Netherlands, Division Biomedical Genetics, University Medical Center Utrecht, the Netherlands for access to their sequencing facility, and Ies Nijman and Annelies Smouters of the UMC Utrecht for their help in setting up the analysis pipeline and the Erasmus MC Cancer Computational Biology Center for giving access to their IT-infrastructure and software that was used for the computations and data analysis in this study. We also thank the Dutch Childhood Oncology Group (DCOG) and the German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL) for contributing primary leukemic cells and clinical information.
Author information
Authors and Affiliations
Contributions
I.S.J., M.L.d.B. designed and performed experiments, analyzed and interpreted the data, and wrote the manuscript, A.Q.H. and J.M.B. analyzed and interpreted the data, I.M.A. and M.J.K. designed sequencing experiments, N.J.M.B. assisted in performing sequencing experiments, M.L.d.B. and R.P. conceptualized the study and interpreted the results. All authors read, revised, and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jerchel, I.S., Hoogkamer, A.Q., Ariës, I.M. et al. Fibroblast growth factor receptor signaling in pediatric B-cell precursor acute lymphoblastic leukemia. Sci Rep 9, 1875 (2019). https://doi.org/10.1038/s41598-018-38169-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-38169-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.