Abstract
As part of an ongoing study of new insulin mimetic agents from medicinal plants, the 70% EtOH extract of Symplocos cochinchinensis was found to have a stimulatory effect on glucose uptake in 3T3-L1 adipocyte cells. The intensive targeted isolation of this active extract resulted in ten new hydroxyoleoside-type compounds conjugated with a phenolic acid and monoterpene (1–6 and 8–11), as well as four known compounds (7 and 12–14). The chemical structures of the new compounds were determined based on spectroscopic data analysis (1H and 13C NMR, HSQC, HMBC, NOESY and MS). The absolute configurations of the isolated compounds were determined by electronic circular dichroism (ECD) analysis of derivatives obtained after a series of reactions, such as those with dirhodium (ІІ) tetrakis (trifluoroacetate) and dimolybdenum (ІІ) tetraacetate. In vitro, compounds 3, 7 and 8 moderately increased the 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG) uptake level in differentiated 3T3-L1 adipocytes. For further studies, we evaluated their effects on the expression of glucose transporter-4 (GLUT4), its translocation, protein tyrosine phosphatase 1B (PTP1B) inhibition and expression of phosphorylated Akt. Our results strongly suggest that the traditional uses of this plant can be described as active constituents by hydroxyoleoside-type compounds.
Similar content being viewed by others
Introduction
Globally, the number of people with diabetes mellitus is growing rapidly, and the incidence rate of diabetes is also accelerating, especially as the elderly and obese population increases1. The number of patients with diabetes is expected to increase from 171 million in 2000 to 366 million globally by 2030. According to the American Diabetes Association, the incidence of diabetes is approximately 25.2% of the total elderly population in the United States and 12.0 million seniors suffered from diabetes in 2015. As the increase in diabetic patients is associated with a dramatic increase in the cost of diabetes-related complications, direct healthcare costs and productivity losses in the US alone are estimated to be $ 176 billion and $ 69 billion in 2012, respectively2.
Diabetes is a chronic disease that occurs when the pancreas does not produce enough insulin (type 1 diabetes) or the insulin produced does not function effectively at the site of action (type 2 diabetes, T2D). When insulin does not function properly, the increased blood sugar in the body can cause serious damage to the heart, blood vessels, kidneys, eyes and nerves. Since T2D associated with insulin resistance is the most prevalent3, it is urgently necessary to develop new anti-diabetic agents, especially those of targeting type 2 diabetes.
While many diabetes medications lower blood glucose levels in the short term, they often cause weight gain as a side effect and prolonged use worsens the insulin resistance of diabetic patients3. Insulin mimetics used as oral diabetic agents, which act similar to insulin but do not synthesize fats, have been suggested as a good solution for the treatment for diabetes. Interestingly, food intake and body weight decrease when insulin is selectively delivered to the brain, but not when it is delivered to the whole body4. These results suggest that insulin mimetics that separate glucose-lowering action from the weight gain are a very good pharmacological solution for overcoming insulin resistance as the side effects of diabetes therapies.
Symplocos cochinchinensis (Lour.) S. Moore (www.theplantlist.org) is an evergreen tree that grows up to 35 meters in height and belongs to the Symplocaceae family. This plant is distributed in East Asia, including China, Japan, India, Vietnam and Malaysia5. Ethnobotanical uses of this plant include treatment of diabetes mellitus in traditional “Ayurvedic” Indian medicine6. There are several reports on extracts from this plant showing antidiabetic7,8, antilipidemic and antioxidant activity9, but there have been few studies on the chemical constituents of S. cochinchinensis. The genus Symplocos contains a large amount of seco-iridoid and phenolic compounds10. Recent reports on the antidiabetic activity of oleuropein, which is abundant in olive tree leaves11, led us to isolate active compounds by a special dereplication method aimed at seco-iridoids.
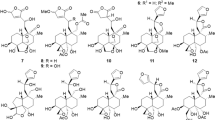
In the search for new insulin mimetics from S. cochinchinensis, the 70% EtOH extract of the plant showed a moderate increase in glucose uptake in differentiated adipocyte cells. Bioassay-guided fractionation resulted in the isolation of ten new hydroxyoleoside-type compounds, including eight phenolic hydroxyoleosides, symplocochinside A-H (1–6, 8 and 9), and two monoterpene-derivatized hydroxyoleosides, symplocochinside I-J (10–11), along with four known compounds, including a megastigmane and triterpene glycosides (Fig. 1). The absolute configurations of the monoterpene attached to hydroxyoleoside (10) and megastigmane (12) were assigned by chemical methods coupled with spectroscopic analysis. All isolates were evaluated for glucose uptake level, GLUT-4 translocation, PTP1B activity and Akt phosphorylation. In this paper, we report the isolation, structural elucidation, determination of absolute configuration and anti-diabetic properties of these isolates.
Results and Discussion
Structural elucidation of new compounds
The 70% EtOH extract of the aerial part of S. cochinchinensis was separated by silica gel, Sephadex LH-20, ODS open column chromatography and preparative HPLC to yield ten new 10-hydroxyoleoside-type seco-iridoids, symplocochinsides A-F (1–6) and G-J (8–11), together with four known compounds (Fig. 1).
Symplocochinside A (1) was obtained as a brownish gum, and the molecular formula was established as C26H30O15 by HREIMS, which showed a ion peak at m/z 605.1478 [M + Na]+ (calcd for C26H30NaO15, 605.1477). The IR spectrum of 1 showed the presence of hydroxyl (3351 cm−1), carboxylic (1692 cm−1) and olefinic (1599 cm−1) functionalities. The 1H NMR spectrum (Table 1) of 1 displayed the typical signals of the 10-hydroxyoleoside skeleton, which is considered an important biosynthetic precursor as one of the major types of seco-iridoids, including one acetal proton of H-1 (δH 5.90, br s), two vinyl protons of H-3 (δH 7.46, br s), one methine group of H-5 (δH 3.88, br d, J = 9.6 Hz), one methylene group of H-6 [(δH 2.68, d, J = 14.4 Hz) and (δH 2.37, dd, J = 15.2, 10.4 Hz)], one olefinic proton of H-8 (δH 6.02, t, J = 6.4 Hz) and oxymethylene protons of H-10 [(δH 4.94, dd, J = 13.6, 8.8 Hz) and (δH 4.80, dd, J = 12.8, 4.8 Hz)]. The corresponding seco-iridoid carbon signals, which consisted of 26 carbon resonances (Table 2) from the 13C NMR spectrum of compound 1, were observed at δC 92.0 (C-1), 152.3 (C-3), 109.0 (C-4), 31.1 (C-5), 40,0 (C-6), 122.3 (C-8) and 133.2 (C-9), as well as one oxygenated methylene carbon at 60.4 (C-10) and two carbonyl carbons at δC 172.8 (C-7) and 167.5 (C-11). Further analysis of the HMBC data of compound 1 (Fig. 2) confirmed the presence of a 10-hydroxyoleoside containing pyran ring moiety by the key correlations from H-8 (δH 6.02, t, J = 6.4 Hz) to C-1/C-5, from H-5 to C-1/C-3, from H-3 to C-1, and from H-10a/H-10b of the oxymethylene to C-8/C-9. In addition, HMBC correlations of H-5 with C-6/C-7/C-11 and of H-3 with C-11 indicated the positions of the carboxylic acids. The relatively large coupling constant (7.2 Hz) for the anomeric proton and six typical carbon resonances at δC 99.1 (C-1″), 73.3 (C-2″), 76.6 (C-3″), 70.0 (C-4″), 77.4 (C-5″) and 61.1 (C-6″) suggested the presence of a β-glucopyranosyl group. The splitting patterns of H-2′ (δH 7.32, d, J = 1.6 Hz), H-5′ (δH 6.79, d, J = 8.0 Hz) and H-6′ (δH 7.12, dd, J = 8.0, 0.8 Hz) in the lower field indicated the presence of a benzene ring with an ABX spin system. The key HMBC correlations from OMe (δH 3.80, s) to δC 148.0 (C-3′) and the carbon resonance at δC 149.4 (C-4′) are indicative of the presence of one methoxy and one hydroxy group, respectively. The large coupling constant between H-7′ (δH 7.55, d, J = 16.0 Hz) and H-8′ (δH 6.49, d, J = 16.0 Hz), the benzene ring system and the distinct UV absorption maximum at 324 nm strongly suggested the presence of a ferulic acid group with α,β-unsaturated ketone moiety. The HMBC correlation from H-10 to C-9′ at δC 166.5 proved that the ferulic acid moiety is linked to C-10 of hydroxyoleoside. The ROESY correlations of compound 1 (Fig. 2) suggested that the C-8 is in the E configuration and H-5 is in the β orientation because H-5 and H-10 become closer to each other, while the H-1/H-1″ and H-1/H-6 are on the same plane. Therefore, the relative configuration could be determined easily as [1S*, 5S*]. The electronic circular dichroism (ECD) spectrum of 1 with oleuropein12 displayed a negative Cotton effect (CE) at 226 nm (Fig. 3A) arising from the 1La band of the 4-carboxy-5,6-dihydro-[1H]-pyrano chromophore13. Since compound 1 assumes a distorted dihydropyran conformation14 and the helicity rule is applicable in this case, the CE with shorter wavelength predicts the 1S and 5S configuration15,16. In addition, chemical shifts of compounds with the 1β configuration generally appeared at approximately 94–96 ppm, while oleonin with the 1α configuration showed a chemical shift of 105.7 ppm in CD3OD17. Thus, compound 1 was determined as 10-O-trans-feruloyl-10-hydroxyoleoside with 1S and 5S (Fig. 1).
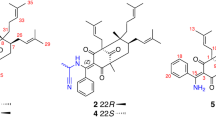
(A) Application of a helicity rule in experimental CD spectra (MeOH) for the absolute configuration of the dihydropyran moiety from all isolated seco-iridoids. (B) Empirical CD method using [Rh2(OCOCF3)4] in CDCl3 of compound 10a. (C) Experimental CD method using [Mo2(OAc)4] in DMSO for compound 12.
Symplocochinside B (2) was purified as a brownish gum, and its molecular formula was established as C26H30O15 from the HRESI mass data, which showed a ion peak at m/z 605.1476 [M + Na]+ (calcd for C26H30NaO15, 605.1477). The 1H NMR data of 2 (Table 1) were similar to those of 1 except for the configuration of the ferulic acid double bond. The J values of H-7′ (δH 6.86, d, J = 13.0 Hz) and H-8′ (δH 5.78, d, J = 13.0 Hz) of compound 1 are indicative of its cis-configuration. Whether the cis form of compound 2 is a plant-derived compound was determined by the retention time and abundance when the partial extract of S. cochinchinensis was co-injected with 2 on LC/MS. Hence, the structure of 2 was characterized as 10-O-cis-feruloyl-10-hydroxyoleoside.
Symplocochinside C (3) (Fig. 1) was isolated as a brownish gum, and its molecular formula was established as C25H28O14 by HRESI mass spectrum, which showed a ion peak at m/z 575.1371 [M + Na]+ (calcd for C25H28NaO14, 575.1371). The distinct UV patterns of 3, which indicate the presence of a cinnamic acid moiety, and the characteristic proton peaks of H-1 (δH 5.90, s) and H-3 (δH 7.46, br s) showed the common features of this seco-iridoid. The 1H and 13C NMR spectra of 3 were similar to those of 1 except for the absence of the methoxy group. Since the trans-cinnamic acid derivative can be converted to the cis-isomer through photoisomerization18, the structure of compound 3 was determined after conversion to the trans form by the reaction with iodine19. Thus, the structure of 3 was determined as 10-O-trans-p-coumaroyl-10-hydroxyoleoside.
Symplocochinside D (4) was obtained as a brownish gum, and its molecular formula was established as C25H28O14 from the HRESI mass spectrum with a peak at m/z 575.1381 [M + Na]+ (calcd for C25H28NaO14, 575.1371). The 1D NMR of 4 (Tables 1 and 2) showed almost same patterns as those of 3 except for the coupling constants of H-7′ (δH 6.87, d, J = 11.2 Hz) and H-8′ (δH 5.77, d, J = 12.8 Hz), indicating that compound 4 is the cis-isomer of compound 3. The pure form of cis-configured compound 4 could be obtained. Hence, the structure of 4 was identified as 10-O-cis-p-coumaroyl-10-hydroxyoleoside.
Symplocochinside E (5) (Fig. 1) was isolated as a brownish gum, and its molecular formula was established as C27H32O15 from the HRESI mass spectrum, which showed a sodium adduct ion peak at m/z 619.1673 [M + Na]+ (calcd for C27H32NaO15, 619.1633). The NMR spectra of 5 (Tables 1 and 2) were similar to those of 1 except for one additional methoxy group. The HMBC correlations (Supplementary Fig. S18) of OMe (δH 3.86, s) with the carbon at δC 152.9 (C-4′), of H-2′/H-6′ with C-4′ and of H-5′ with C-3′ revealed that the benzene ring had two methoxy groups at the para and meta positions. The coupling constants of H-7′ (δH 7.63, d, J = 15.9 Hz) and H-8′ (δH 6.39, d, J = 15.9 Hz) are indicative of the trans-configuration. Accordingly, the structure of 5 was assigned as 10-O-trans-3′,4′-dimethoxycinnamoyl-10-hydroxyoleoside.
Symplocochinside F (6), a brownish gum, had the molecular formula C28H34O16, as deduced from the HRESI mass spectrum, which showed a deprotonated ion peak at m/z 625.1776 [M − H]− (calcd for C28H33O16, 625.1774). The NMR spectra of 6 (Tables 1 and 2) were similar to those of 5 except for one additional methoxy groups, which is supported by the HMBC correlation (Supplementary Fig. S22) of OMe (δH 3.86, s) with the carbon at 154.8 (C-5′). The coupling constants of H-7′ (δH 7.62, d, J = 15.9 Hz) and H-8′ (δH 6.46, d, J = 16.0 Hz) are indicative of the trans-configuration. Thus, the structure of 6 was established as 10-O-trans-3′,4′,5′-trimethoxycinnamoyl-10-hydroxyoleoside.
Symplocochinside G (8) was obtained as a brownish gum, and its molecular formula was established as C23H26O13 from the HRESI mass spectrum with a peak at m/z 533.1292 [M + Na]+ (calcd for C23H26NaO13, 533.1266). In the 1H and 13C spectra of compound 8, chemical shifts and splitting patterns of H-2′/6′ (δH 8.02, d, J = 7.2), H-3′/5′ (δH 7.47, t, J = 7.8) and H-4′ (δH 7.60, t, J = 7.5) in the aromatic regions clearly indicated the presence of a monosubstituted aromatic ring. The key HMBC correlation (Supplementary Fig. S26) of H-10 with C-7′ at δC 167.8 showed that the benzoic acid group is connected to C-10. Thus, the chemical structure of 8 was assigned as 10-O- benzoyl-10-hydroxyoleoside.
Symplocochinside H (9) (Fig. 1) was purified as a brownish gum, and its molecular formula was established as C18H24O13 from the HRESI mass spectrum with a peak at m/z 447.1149 [M − H]− (calcd for C18H23O13, 447.1144). Analysis of the 1H and 13C-NMR data of compound 9 (Tables 1 and 2) showed similar chemical shifts except for the characteristic peaks at δC 170.2 and 20.7 resulting from the acetyl group. The key HMBC correlations (Supplementary Fig. S30) from the acetyl group at δH 2.01 and H-10 [δH 4.81 (dd, J = 13.6, 8.0 Hz) and δH 4.65 (m)] to the carbonyl carbon at δC 170.2 supported that this acetyl group is linked to C-10. Hence, the structure of 9 was elucidated as 10-O-acetyl-10-hydroxyoleoside.
Symplocochinside I (10), a yellowish gum, had the molecular formula C26H38O14, as deduced from the HRESI mass spectrum, which showed a deprotonated ion peak at m/z 573.2200 [M − H]− (calcd for C26H37O14, 573.2189). The three singlet methyl groups of H-8′ (δH 1.56, br s), H-9′ (δH 1.16, s) and H-10′ (δH 1.63, br s) in 1H-NMR spectrum of compound 10 (Table 1) indicated that these methyl groups are bound to quaternary carbons. The methylene protons of H-2′ [(δH 2.42, d, J = 13.6 Hz) and (δH 2.38, d, J = 13.7 Hz)] were geminally coupled with a large coupling constant, suggesting a lack of adjacent protons. The 13C-NMR and HSQC spectral data of compound 10 showed the presence of 10 peaks other than the carbon peaks of 10-hydroxyoleoside (Supplementary Figs S33 and S34), which are three sp3 primary carbons, three sp3 methylene carbons and one sp3 quaternary carbon with a deshielded chemical shift (δC 70.1) implying the presences of one oxygenated quaternary carbon, one carbonyl group, and two olefinic carbons. The HMBC correlations of H-10 with C-1′, H-9′ with C-2′/C-4′, of H-6′ with C-4′/C-5′, of H-5′ with C-3′ and of H-4′ with C-2′/C-5′ suggested the connectivity of the 10-hydroxyoleoside skeleton with 3-hydroxydimethyloctenoic acid20,21, which is a monoterpene also known as 3-hydroxycitronellic acid. The isolation of 3-hydroxycitronellic acid from 10 by selective hydrolysis was not successful due to racemization (data not shown). Therefore, after the reaction with dirhodium (ІІ) tetrakis (trifluoroacetate), the empirical ECD method was employed for the determination of the absolute configuration at C-3′22. The acetate form of compound 10 was purified after the peracetylation reaction (Supplementary Table S1) and was subjected to complexation with [Rh2(OCOCF3)4] in CDCl3. According to the bulkiness rule, the negative Cotton effect at 350 nm (band E) was observed (Fig. 3B), which means that the absolute configuration of C-3′ is 3′R. Thus, compound 10 was elucidated as (3′R)-10-O-3′-hydroxycitronellyl-10-hydroxyoleoside.
Symplocochinside J (11) was obtained as a yellowish gum, and its molecular formula was established as C26H36O13 from the HRESI mass spectrum which showed a deprotonated ion peak at m/z 555.2088 [M − H]− (calcd for C26H35O13, 555.2083). The characteristic peaks of H-1 (δH 5.82, s)/H-3 (δH 7.37, s)/H-10 [(δH 4.82, dd, J = 13.8, 8.2 Hz) and (δH 4.67, dd, J = 13.3, 4.6)] and additional methyl groups at δC 25.49, 18.5 and 17.6 implied it is also an analogue of 10-hydroxyoleoside with a terpene (Tables 1 and 2). Compared to compound 10, the additional double bond carbons of C-2′(δC 114.8)/C-3′(δC 160.5), the lack of any oxygenated quaternary carbon and the same small J-value of H-2′ (δH 5.68, d, J = 0.8)/H-9′ (δH 2.10, d, J = 0.8) indicated that the methyl group of C-9′ is bound to the sp2 quaternary carbon of C-3′. The key HMBC correlation (Supplementary Fig. S38) of H-9′ with C-2′/C-4′ along with the similar pattern of HMBC correlations with 10 signified that 11 is an analogue of 10-hydroxyoleoside substituted with a different monoterpene, namely, geranic acid, which is supported by the comparison with previously reported data23. Thus, the structure of compound 11 was elucidated as 10-O-geranyl-10-hydroxyoleoside.
Compound 12 (Fig. 1) was obtained as a brownish gum, and its molecular formula was established as C13H20O3 from the HRESI mass spectrum with a peak at m/z 225.1484 [M + H]+ (calcd for C13H21O3, 225.1485). The comparison with previously reported NMR data24 showed the compound has the same planar megastigmane structure (Supplementary Table S1). However, the NOESY spectrum (Supplementary Fig. S43) indicated the possibility of different configurations at C-8 and C-9 from those of the known compound 8,9-dihydromegastigmane-4,6-diene-3-one. In the NOESY spectrum, the correlations between H-7 at δH 6.06 (d, J = 9.2 Hz)/H-13 at δH (2.15, d, J = 0.8 Hz) and H-8 at δH (4.68, dd, J = 9.4, 5.3 Hz)/H-9 at δH (3.75, m) indicated that the olefinic carbon of C-7 is in the E configuration and that the relative configuration of C-8 and C-9 is [8R*, 9S*]. The specific rotation of the known compound is +54.0 (c 1.52 MeOH), whereas that of 12 was −88.9 (c 0.2 MeOH). Since the planar structure of the known compound was reported without the absolute configuration, the absolute configuration of compound 12 was determined by the helicity rule using the ECD measurement after derivatization with dimolybdenum tetraacetate25,26. It is possible to apply this method because compound 12 is erythro-1,2-diol and there is a bulkiness difference between the two substituents around the hydroxyl groups. The CD measurement after complexation with [Mo2(OAc)4] showed a negative CE in the band II region (403 nm, −0.15 mdeg) used as the diagnostic band (Fig. 3C). Accordingly, the structure of 12 was determined as (8R,9S)-8,9-dihydromegastigmane-4,6-diene-3-one.
Based on NMR, MS and optical rotation data and comparison with literature values, the known compounds were elucidated as, 10-cinnamoyloxyoleoside (7)27, (8R,9S)-8,9-dihydromegastigmane-4,6-diene-3-one (12)24, nigaichigoside F1 (13)28, and trachelosperoside A1 (14)29 (Fig. 1).
Measurements of insulin mimetic activity with 2-NBDG on differentiated 3T3-L1 adipocytes
Unlike the insulin in the brain, insulin at the periphery acts as an anabolic factor and causes weight gain as a side effect if the energy usage of the patient is not increased. Insulin mimetics have similar characteristics to insulin in terms of the reduction in food intake and body weight in rats when administered intracerebroventricularly. Considering the abovementioned difference, insulin memetics appear to be more advantageous than insulin because of their potential to pass through the blood-brain barrier (BBB), which allows us to find insulin mimetics from natural resources with fewer side effects. To measure for insulin mimetics, the 2-NBDG assay, which is used a fluorescent-tagged glucose analogue for monitoring the glucose uptake in cells, was introduced. All isolates (1–14) were evaluated for 2-NBDG uptake in differentiated 3T3-L1 adipocytes at a concentration of 40 μM (Fig. 4A and Supplementary Fig. S49). Most of the phenolic acid- derivatized seco-iridoid showed activity, whereas compound 9 with an acetyl group and compound 10 with a monoterpene showed weak activity. Comparison of the activities of the compounds with the trans and cis forms showed that the trans isomers had stronger activity than cis. Among these compounds, compounds 3, 7 and 8 showed stronger activities compared to others. Thus, fluorescent signals measurement was performed using a fluorescence microscopy for assessing the transport efficacy of 2-NBDG into cells. Increased signal intensities after treatment of the compounds were more strongly observed in cells treated with 3, 7 and 8 at 40 μM compared to those in cells of the control group (DMSO, Fig. 4B). It was also observed that selected compounds 3, 7 and 8 increased the 2-NBDG uptake in a dose-dependent manner (Fig. 5A,B). Taken together, these results suggest that derivatization of seco-iridoids with trans-configured phenolic acids is relevant to activity. These results are consistent with the ethnopharmacological history of this plant as a diabetes remedy in Ayurvedic formulations.
Enhancement of glucose uptake by compounds 1–14 (40 μM) in 3T3-L1 adipocytes using a fluorescent glucose probe 2-NBDG. (A) After 1 hour of incubation with test compounds, the fluorescence signals were measured and the results were presented as the mean ± SD (n = 3), *p < 0.05 and **p < 0.01, and ***p < 0.001, compared to vehicle. (B) The green fluorescent signals significantly increased in the compounds treated-cells, which indicate that 2-NBDG was successfully transported into the cells.
(A) Stimulation of the glucose uptake by compounds 3, 7, and 8 at different concentrations in 3T3-L1 adipocytes were obtained using the fluorescence microscopy method and (B) the microplate reader detection. The 3T3-L1 adipocytes were incubated with compounds for 1 hour. The cells were then captured for fluorescence images or measured fluorescence intensities. Data were calculated as the mean ± SD (n = 3), *p < 0.05 and **p < 0.01, and ***p < 0.001, compared to negative control. (C) Effect of compounds 3, 7 and 8 on GLUT4 translocation to the plasma membrane of 3T3-L1 adipocytes. The cells were incubated with compounds for 24 hours or insulin for 2 hours. The levels of GLUT4 expression were evaluated using Western blot method. Full length western blots are provided in Supplementary Fig. S56. Results were calculated as the mean ± SD (n = 6), **p < 0.01, compared to negative control. (D) Graphical representation of the type of inhibition for compounds 3 at different concentrations (10, 20 and 40 μM, respectively) on the PTP1B enzyme. (E) Docking simulation of compound 3 into the active site of PTP1B enzyme (PDB code 1Q6T) and its interaction.
GLUT4 translocation by selected compounds in the differentiated 3T3-L1 adipocytes
Concentrations of plasma glucose levels are normally regulated by glucose transporter proteins in muscle and adipose tissue. Among the 14 sugar transporter proteins, GLUT4 is the principal isoform that plays a major role in glucose homeostasis. Intracellular glucose uptake occurs via GLUT4 recruitment to the cell surfaces of muscle and adipose cells. GLUT4 trafficking is stimulated acutely by both insulin via the PI3K/Akt pathway and exercise via the AMPK pathway30. Therefore, the potential of insulin mimetics can be estimated partially by the measurement of GLUT4 translocation or/and expression. Compounds 3, 7 and 8, which showed activity in the 2-NBDG assay, were subjected to the measurement of GLUT4 translocation in the plasma membrane. Western blot analysis showed increased GLUT4 translocation in the plasma membrane of differentiated adipocytes upon treatment with the compounds at 40 μM (Fig. 5C). Compound 3 (40 μM) and insulin (100 nM) clearly increased GLUT4 expression in whole cell lysate, while compounds 7 and 8 showed relatively weak effects on expression of GLUT4 in adipocytes (Supplementary Figs S52 and S57). However, compounds 3, 7 and 8 significantly induce the expression of phosphorylated Akt (Supplementary Figs S61 and S62). These results provide insight into how these molecules stimulate glucose uptake through up-regulation of the Akt pathway. The present data strongly indicate that compound 3 promotes glucose uptake by GLUT4 translocation associated with increased level of GLUT4 expression. Although the mechanism of 3 is still unclear and further studies are in need near future, we propose this compound could regulate the glucose metabolism and insulin sensitivity.
Protein tyrosine phosphatase 1B (PTP1B) inhibitory activity of isolated compounds
Upon insulin binding, glucose uptake is increased by a series of signals including phosphorylation of the insulin receptor (IR), transformation of phosphatidylinositol (4,5)-bisphosphate (PIP2) to phosphatidylinositol (3,4,5)-triphosphate (PIP3) by phosphatidylinositol 3-kinase (PI3K), activation of Akt, and translocation of GLUT4. PTP1B exerts negative regulation of insulin and leptin receptor signalling by dephosphorylation of activated IR and Janus kinase 231. Since some seco-iridoids increased glucose uptake through 2-NBDG and GLUT4 translocation, the PTP1B inhibitory activity, which is important in the relevant signal transduction process, was also assessed32. When all isolated compounds were subjected to the PTP1B assay, compound 3 was found to be a good candidate for use as a PTP1B inhibitor, whereas 7 and 8 showed moderate activity at 50 μM (Supplementary Fig. S53). Among these compounds, the IC50 of 3 is 19.54 ± 0.76 μM, showing moderate activity compared to the positive control tested (Supplementary Fig. S54). Compound 3 was further examined in kinetic experiments and was shown to inhibit PTP1B in a non-competitive manner at different concentrations (10, 20 and 40 μM, Fig. 5D). Molecular docking analysis of compound 3 into the active site of PTP1B (PDB ID code 1Q6T) was performed according to the CDOCKER protocol in the CHARMm-based docking algorithm33. As shown in Fig. 5E, the C-7 carboxylic acid group forms a conventional hydrogen bond and a carbon-hydrogen bond with Asp548 and GLY759, respectively. Moreover, the benzene ring shows affinity for Phe682 via a π-π interaction (Supplementary Fig. S55). These key residues have been proposed as active sites A and B of PTP1B. Additionally, the CDOCKER interaction energy was calculated to be −61.18 kcal/mol. Overall, the observed effects appear to contribute to the inhibitory activity of compound 3 against PTP1B.
Conclusions
In this paper, eight new analogues of phenolic acid-conjugated 10-hydroxyoleoside type (1–6 and 8–9) and two new monoterpene-conjugated compounds (10 and 11), along with one known seco-iridoid (7), one megastigmane (12) and two triterpenoids, (13–14) were isolated from S. cochinchinensis. The absolute configurations of these compounds were determined by ECD analysis, dirhodium (ІІ) tetrakis (trifluoroacetate) reaction or dimolybdenum (ІІ) tetraacetate reaction. Compounds 3, 7 and 8 exhibited 2-NBDG uptake increasing activity in differentiated 3T3-L1 adipocytes by GLUT4 translocation which was evident in Western blot analysis. Compound 3 increased GLUT4 expression level and direct GLUT4 translocation through the PI3K/Akt pathway via a PTP1B inhibition. These results also imply the structure activity relationship to some degree such that derivatization with a phenolic acid and trans configuration contribute to the activity. In conclusion, investigation of new seco-iridoids as ant-diabetic compounds enriches the chemical profile of S. cochinchinensis and provides evidence for the traditional ethnopharmacological uses of this plant.
Methods
General experimental procedures
Optical rotations were recorded on a JASCO P-2000 polarimeter (JASCO International Co. Ltd., Tokyo, Japan). ECD spectra were measured using Chirascan plus (Applied photophysics Ltd., Surrey, United Kingdom). IR data were obtained using a Nicolet 6700 FT-IR spectrometer (Thermo Electron Corp., Waltham, MA, USA). The 1D and 2D NMR spectra were obtained in deuterated solvents using an AVANCE 800 MHz spectrometer (Bruker, Germany). HRESIMS values were obtained using an Agilent Technologies 6530 Q-TOF MS spectrometer (Agilent Technologies, Inc., Santa Clara, CA, USA). Regular column chromatography (CC) was carried out with silica gel (particle size: 63–200 μm, Zeochem, Lake Zurich, Switzerland), RP-C18 (particle size: 75 μm, nacalai tesque, Kyoto, Japan), and Sephadex LH-20 (GE Healthcare, Little Chalfont, UK). Silica gel 60 F254 and RP-18 F254S TLC plates were obtained from Merck (Darmstadt, Germany). A Gilson HPLC purification system was used at a flow of 2 mL/min and UV detection at 205, 254, and 300 nm using an Optima Pak C18 column (10 × 250 mm, 5 μm particle size; RS Tech, Seoul, Korea) and a COSMOSIL 5C18-MS-II column (10 × 250 mm, 5 μm particle size; Nacalai Tesque, Kyoto, Japan). Analytical-grade solvents were used for extraction and isolation.
Plant material
S. cochinchinensis was collected in Kim Boi district, Hoa Binh province, Vietnam and authenticated by Dr. Tran Van On, Head of the Department of Botany, Hanoi University of Pharmacy, Vietnam. A voucher specimen (SCL-02) was deposited at the Korea Bioactive Natural Material Bank, Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul, Republic of Korea.
Extraction and isolation
The stems and leaves of S. cochinchinensis (4 kg) were extracted with 70% EtOH (4 × 11 L, for 4 h each) at 60 °C. The combined extract was concentrated by an evaporator to yield a dried residue (752.6 g). Dried extract was suspended in H2O and then partitioned with n-hexane, EtOAc and n-BuOH successively. The EtOAc portion (57.3 g) was subjected to silica gel CC (8 × 50 cm) and eluted with gradient system of n-hexane/acetone from 5:1 to 0:1 to yield five fractions (F.1–F.5). F.5 (12 g) was subjected to reversed-phase chromatography (5 × 40 cm) with 45% MeOH to obtain six subfractions (F.5.1–F.5.6). F.5.1 (2.93 g) was subjected to Sephadex-20 (2 × 40 cm) with 50% MeOH to obtain two subfractions (F.5.1.1–F5.1.2). Compounds 1 (9 mg), 2 (4.4 mg), 3 (11.2 mg) and 4 (3.5 mg) were purified from F.5.1.2 with ODS silica gel HPLC and eluted with MeCN/H2O (v/v, 19/81 to 21/79). F5.1.1 (566.4 mg) was subjected to reversed-phase chromatography (2 × 50 cm) with 35% MeOH to give six subfractions (F.5.1.1.1–F5.1.1.6). Compound 8 (4 mg) was purified from subfraction F.5.1.1.6 by ODS silica gel HPLC with MeCN/H2O (v/v, 24/76). F.5.1.1.1 was subjected to semi-prep HPLC with MeCN/H2O (v/v, 12:88), resulting in the isolation of compound 9 (16.0 mg). F.5.2 (730 mg) was subjected to reversed-phase chromatography with 40% MeOH to obtain six subfractions (F.5.2.1–F.5.2.6). F.5.2.3 was subjected to semi-prep HPLC with MeCN/H2O (v/v, 23:77), resulting in the isolation of compounds 5 (1.5 mg) and 6 (0.5 mg). F.5.3 was separated by Sephadex LH-20 CC into two subfractions (F.5.3.1–F5.3.2). Subfraction F.5.3.1 was subjected to semi-prep HPLC with MeCN/H2O (v/v, from 28/72 to 32/68), resulting in the isolation of compound 10 (3 mg). Subfraction F.5.3.2 was subjected to semi-prep HPLC with a MeCN/H2O (v/v, 28/72 to 32/68), resulting in the isolation of compound 7 (1.6 mg). Subfraction F.5.5.1 was subjected to Sephadex LH-20 and reversed-phase column sequentially, resulting in the isolation of compound 13 (7.7 mg). Subfraction F.5.6 was subjected to a reversed-phase column (3.5 × 50 cm) and eluted with 10% MeCN and compounds 11 (1.9 mg) and 14 (5.1 mg) were isolated from subfraction F.5.6.1 and F.5.6.2, respectively by semi-prep HPLC with MeOH/H2O (v/v, 55/45 to 61/39). F.2 was subjected to direct semi-prep HPLC with MeCN/H2O (v/v, 19/81 to 21/79), resulting in the isolation of compound 12 (2.2 mg).
10-O-trans-Feruloyl-10-hydroxyoleoside (1)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒211.0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 224 (2.3), 324 (1.9) nm; IR νmax 3350, 2918, 2352, 1692, 1599, 1393, 1161 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 605.1478 [M + Na]+ (calcd for C26H30NaO15, 605.1477).
10-O-cis-Feruloyl-10-hydroxyoleoside (2)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒80.6 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 228 (2.4), 319 (1.9) nm; IR νmax 3424, 2973, 2349, 1705, 1647, 1517, 1164, 1052, 1033 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 605.1476 [M + Na]+ (calcd for C26H30NaO15, 605.1477).
10-O-trans-p-Coumaroyl-10-hydroxyoleoside (3)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒124.9 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 228 (2.6), 312 (2.6) nm; IR νmax 3407, 2980, 2349, 1747, 1658, 1516, 1171, 1052, 678, 9 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 575.1371 [M + Na]+ (calcd for C25H28NaO14, 575.1371).
10-O-cis-p-Coumaroyl-10-hydroxyoleoside (4)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒29.2 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 228 (2.5), 308 (2.3) nm; IR νmax 3420, 2981, 2349, 1680, 1647, 1516, 1397, 1163, 1051, 671 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 575.1381 [M + Na]+ (calcd for C25H28NaO14, 575.1371).
10-O-trans-3′,4′-Dimethoxycinnamoyl-10-hydroxyoleoside (5)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒44.7 (c 0.50, MeOH); UV (MeOH) λmax (log ε) 232 (2.2), 298 (1.9), 324 (2.0) nm; IR νmax 2927, 2373, 1746, 1509 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 619.1673 [M + Na]+ (calcd for C27H32NaO15, 619.1633).
10-O-trans-3′,4′,5′-Trimethoxycinnamoyl-10-hydroxyoleoside (6)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒105.1 (c 0.50, MeOH); UV (MeOH) λmax (log ε) 230 (1.8), 306 (1.6) nm; IR νmax 3419, 2917, 2361, 1716, 1509, 1156 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 625.1776 [M − H]− (calcd for C28H33O16, 625.1774).
10-O-Benzoyl-10-hydroxyoleoside (8)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒159.2 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 236 (2.3) nm; IR νmax 3428, 2972, 1705, 1278, 1055, 1014, 716 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 533.1292 [M + Na]+ (calcd for C23H26NaO13, 533.1266).
10-O-Acetyl-10-hydroxyoleoside (9)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒95.1 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 226 (2.3) nm; IR νmax 3386, 2972, 1712, 1637, 1397, 1254, 1053 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 447.1149 [M − H]− (calcd for C18H23O13, 447.1144).
(3′R)-10-O-3′-Hydroxycitronellyl-10-hydroxyoleoside (10)
Yellowish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒117.9 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 226 (2.4) nm; IR νmax 3405, 2919, 2850, 2349, 1715, 1638, 1435, 1202, 1078, 672 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 573.2200 [M − H]− (calcd for C26H37O14, 573.2189).
Tetraacetate of compound 10 (10a)
Colourless gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{25}\) −113.1 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 233 (2.5) nm; IR νmax 3271, 2898, 1748, 1646, 1508, 1224, 676 cm−1; 1H and 13C NMR, see supporting data; HRESIMS m/z 741.2621 [M − H]− (calcd for C34H45O18, 741.2611).
10-O-Geranyl-10-hydroxyoleoside (11)
Yellowish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{25}\) −91.1 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 222 (2.4) nm; IR νmax 3316, 3121, 2894, 1748, 1646, 1508, 676 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 555.2088 [M − H]− (calcd for C26H35O13, 555.2083).
(8R,9S)-8,9-Dihydromegastigmane-4,6-diene-3-one (12)
Brownish gum; \({[{\rm{\alpha }}]}_{{\rm{D}}}^{20}\) ‒88.9 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 209 (2.3), 285 (2.6) nm; IR νmax 3400, 2972, 2360, 1649, 1387, 1033, 671 cm−1; 1H and 13C NMR, see Tables 1 and 2; HRESIMS m/z 225.1484 [M + H]+ (calcd for C13H21O3, 225.1485).
Determination of absolute configuration of sugars
Compound 9 (1.0 mg) was hydrolysed by 0.5 M HCl (1.0 mL) at 90 °C for 1 hour35. The solvent was neutralized with Na2CO3 and concentrated in vacuo. L-Cysteine methyl ester hydrochloride in anhydrous pyridine (0.5 mg) was added to the resulting residue, followed by heating for 1 hour at 60 °C. Phenyl isothiocyanate (0.1 mL) was added and heated at 60 °C for 1 hour. The solution was then analyzed by reversed-phase HPLC under the following conditions: an INNO C18 column (120 Å, 4.6 × 250 mm, 5 μm); MeCN/H2O mobile phase (27:73, v/v); a diode array detector; a detection wavelength of 254 nm; and a flow rate of 0.6 mL/min. Comparisons of the retention time of the derivative of compound 9 with that of the derivative of an authentic sample of D-glucose (retention time: 23.18 min) proved the D-configuration of the glucose moiety in compound 9.
Absolute configuration of the tertiary alcohol moiety in 10
Compound 10 (13.2 mg) was kept in pyridine/Ac2O 1:1 (4 mL) at room temperature for 19 hours to give peracetylated compound 10a. The mixture was neutralized with NaHCO3 and dried under vacuum. A colorless gum (10.6 mg, 80.3%) was obtained by extraction with EtOAc and the product was further purified to over 99% using prep-HPLC. Compound 10a (0.5 mg) was then dissolved in a dry solution of [Rh2(OCOCF3)4] (1.0 mg) in CDCl3 (600 μL). The resulting mixture was used for CD measurements and the obtained CD spectrum was compared with that of compound 10a for clarity. The Cotton effect at 350 nm (E band) was correlated with the absolute configuration of the tertiary alcohol36.
Preparation of Mo2-complexes of compound 12
The CD spectra were measured at room temperature in DMSO with 1.0 nm/step scans using a 2 mm cell over the range of 250–650 nm according to the Snatzke’s method25. To form complexes, compound 12 (0.18 mg, 1.33 mM/L) was dissolved in a solution of [Mo2(OAc)4] (0.34 mg, 1.33 mM/L) in DMSO at a 1/1 ratio of the molybdenum complex to the diol.
Measurement of glucose uptake using 2-NBDG in differentiated 3T3-L1 adipocytes
To determine the level of glucose uptake into 3T3-L1 adipocytes, a fluorescent derivative of glucose (2-NBDG) (Invitrogen, OR, USA) was used as previously described with slight modifications34,37. First, 3T3-L1 preadipocytes were differentiated by Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, IL, USA) containing 10% fetal bovine serum (FBS) (Gibco, NY, USA), 1 μM dexamethasone (Sigma, MO, USA), 520 μM 3-isobutyl-1-methyl-xanthine (Sigma, MO, USA) and 1 μg/mL insulin (Roche, Germany). After 2 days of incubation, the cells were replaced with fresh media [10% FBS, 1 μg/mL insulin, 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, NY, USA)] and allowed to continue differentiating for 4–6 days. The glucose uptake assay was carried out as follows. 3T3-L1 adipocytes were seeded onto 96 well plates using glucose-free media supplemented with 10% FBS and incubated for 1 day. The cells were then incubated with media containing the test compounds and with or without 2-NBDG. After incubation for 1 hour at 37 °C, the cells were washed with cold PBS, and fluorescence measurements were obtained using a fluorescence microplate reader (Spectra Max GEMINI XPS, Molecular Devices, Sunnyvale, CA, USA) at ex/em = 450/535 nm. To capture bright and fluorescence images, 3T3-L1 adipocytes were maintained on sterilized glass coverslips in glucose-free media containing 10% FBS. The glucose uptake assay was performed as described above (Supplementary Figs S50 and S51). After washing with cold PBS, the cells were replaced with PBS containing 1% BSA (Sigma, MO, USA) and the slides were analysed by fluorescence microscopy (Olympus ix70 Fluorescence Microscope, Olympus Corporation, Tokyo, Japan).
Western blot analysis for GLUT4 translocation and expression
3T3-L1 adipocytes were grown in 6-well plates with DMEM containing 10% FBS (Gibco, NY, USA). The cells were then incubated with the tested compounds for 24 hours or insulin (as a positive control) for 2 hours using serum free media. To evaluate GLUT4 expression in the plasma membrane, the plasma membrane fraction was isolated from the whole cell protein extract as previously described with slight modifications38. The clear isolation of the plasma membrane fraction was verified by comparing the expression of β-actin protein by Western blotting in the whole cell lysates and the plasma membrane fractions (Supplementary Fig. S59). The whole cell lysate was lysed using RIPA buffer containing protease and phosphatase inhibitors. Protein concentrations were measured using a BCA protein assay kit (Bio-Rad Laboratories, Inc., CA, USA). Equal amounts of proteins were then electrophoresed on 12% SDS-polyacrylamide gels and then transferred to polyvinylidene fluoride (PVDF) membranes (PVDF 0.45 µm, Immobilon-P, USA). After blocking with 5% skim milk for 1 hour, the membrane was incubated overnight with different primary antibodies for GLUT4 (Santa Cruz, CA, USA), Na+/K+ ATPase α1 (Cell Signaling, MA, USA) or mouse monoclonal β-actin (Thermo Fisher Scientific, Rockford, IL, USA). The membrane was then incubated with HRP-conjugated secondary antibodies for 2 hours at room temperature. The membrane was detected using a LAS 4000 luminescent image analyzer (Fuji Film, Tokyo, Japan).
Western blot analysis of Akt phosphorylation
3T3-L1 adipocytes were treated with compound 3 for different time points (Supplementary Fig. S60) or test compounds for 2 hours using serum free media. Then, cell lysates were collected using a lysis buffer [50 mM Tris-HCl (pH 7.6), 120 mM NaCl, 1 mM EDTA, 0.5% NP-40, 50 mM NaF]. Western blotting was performed as described above. The membranes were incubated with the primary antibodies for p-Akt (Ser473) and Akt (Cell Signaling, MA, USA) or mouse monoclonal β-actin (Thermo Fisher Scientific, Rockford, IL, USA).
PTP1B inhibition and kinetic assay
PTP1B enzyme (human, recombinant) was purchased from BIOMOL International LP (USA) and the enzyme assay was performed as previously described34. Briefly, the assay was performed in 96-well plate containing 4 mM p-NPP and PTP1B (0.05–0.1 μg) in enzyme buffer [50 mM citrate (pH 6.0), 0.1 M NaCl, 1 mM dithiothreitol (DTT), and 1 mM EDTA] with or without test compounds in triplicate. In addition, an assay was carried out to exclude the fluorescence effect of the test compounds in the absence of PTP1B enzyme. After incubation at 37 °C for 30 minutes, the reaction was terminated with 10 M NaOH solution. The p-nitrophenol product was then measured at 405 nm using an absorbance microplate reader (VersaMaxTM, Randor, PA, USA). The enzyme kinetics were determined using Lineweaver-Burk plots. The PTP1B inhibition mode was examined at different concentrations of the p-NPP substrate (from 1 to 8 mM) in the absence and presence of the test compounds (10, 20 and 40 μM). Half-maximal inhibitory concentration (IC50) values and inhibition type of PTP1B were calculated by Sigma Plot 10.0 (Systat Software Inc., San Jose, CA, USA).
Molecular docking simulation
Docking studies between the structure of the PTP1B protein and compound 3 were successfully performed using Discovery Studio 4.0/CDOCKER software (Accelrys, San Diego, CA). The PTP1B protein structure was obtained from the Protein Data Bank (http://www.pdb.org) (PDB ID code: 1Q6T). Protein-ligand binding affinities were optimized and calculated according to the CDOCKER interaction energy arising from several interacting bonds, such as conventional hydrogen bonds, carbon-hydrogen bond, π-π T-shaped and π-alkyl interactions.
Statistical analysis
Data were calculated as the mean ± SD of three independent experiments. The mean value of difference groups was calculated using ANOVA, which was carried out using SPSS Statistics 23 (SPSS, Inc., Chicago, IL, USA). IC50 values and inhibition modes were determined by Sigma Plot 10.0 software (Systat Software Inc., San Jose, CA, USA). Statistically significant p values were established at *p < 0.05, **p < 0.01, and ***p < 0.001.
Data Availability
All data generated or analysed for this study are included in this published paper (and its Supplementary Information files).
References
Control, C. F. D. National Diabetes Statistics Report, 2017: Estimates of Diabetes and Its Burden in the United States, Centers for Disease Control and Prevention (2017).
Association, A. D. Economic costs of diabetes in the US in 2012. Diabetes care 36, 1033–1046 (2013).
Chatterjee, S., Khunti, K. & Davies, M. J. Type 2 diabetes. Lancet 389, 2239–2251 (2017).
Air, E. L. et al. Small molecule insulin mimetics reduce food intake and body weight and prevent development of obesity. Nat. Med. 8, 179–183 (2002).
Wu, Y. F. & Nooteboom, H. P. Flora of China, Vol. 15 (eds by Wu, Y. F. & Hwang, S. M.), 235–248 (Science Press, Beijing, 1996).
Gangadharan, N. The state of Ayurveda in eighteenth and nineteenth centuries. Indian J. Hist. Sci. 17, 154–163 (1982).
Sunil, C., Ignacimuthu, S. & Agastian, P. Antidiabetic effect of Symplocos cochinchinensis (Lour.) S. Moore. in type 2 diabetic rats. J. Ethnopharmacol. 134, 298–304 (2011).
Antu, K. A. et al. Antidiabetic property of Symplocos cochinchinensis is mediated by inhibition of alpha glucosidase and enhanced insulin sensitivity. PLoS One 9, e105829 (2014).
Sunil, C., Agastian, P., Kumarappan, C. & Ignacimuthu, S. In vitro antioxidant, antidiabetic and antilipidemic activities of Symplocos cochinchinensis (Lour.) S. Moore bark. Food Chem. Toxicol. 50, 1547–1553 (2012).
Sunil, C. & Ignacimuthu, S. In vitro and in vivo antioxidant activity of Symplocos cochinchinensis S. Moore leaves containing phenolic compounds. Food Chem. Toxicol. 49, 1604–1609 (2011).
Jiménez-Sánchez, C. et al. AMPK modulatory activity of olive-tree leaves phenolic compounds: Bioassay-guided isolation on adipocyte model and in silico approach. PLoS One 12, 1–22 (2017).
Lee-Huang, S. et al. Discovery of small-molecule HIV-1 fusion and integrase inhibitors oleuropein and hydroxytyrosol: Part I. Integrase inhibition. Biochem. Biophys. Res. Commun. 354, 872–878 (2007).
Ferreira, D., Marais, J. P. J., Slade, D. & Walker, L. A. Circular Dichroic Properties of Flavan-3,4-diols. J. Nat. Prod. 67, 174–178 (2004).
Valente, E. J. & Eggleston, D. S. Dihydropyran ring conformations. I. Structures of 2-methoxy- and 2-hydroxy-2,4-dimethyl-3,4-dihydro-2H,5H-pyranol[3,2-c][1]benzopyran-5-ones. Acta Cryst. B47, 498–506 (1991).
Yu, Y. et al. Homosecoiridoids from the Flower Buds of Lonicera japonica. J. Nat. Prod. 74, 2151–2160 (2011).
Obied, H. K. et al. Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat. Prod. Rep. 25, 1167 (2008).
Huang, X.-J. et al. Oleonin, the first secoiridoid with 1a-configuration from Ligustrum lucidum. RSC Adv. 3, https://doi.org/10.1039/C3RA40557E (2013).
Karaköse, H., Jaiswal, R., Deshpande, S. & Kuhnert, N. Investigation of the photochemical changes of chlorogenic acids induced by ultraviolet light in model systems and in agricultural practice with Stevia rebaudiana cultivation as an example. J. Agric. Food Chem. 63, 3338–3347 (2015).
Li, Z. J. et al. A highly efficient transformation of cis- to trans-cinnamic acid derivatives by iodine. Tetrahedron Lett. 56, 7197–7200 (2015).
Zhao, J. C., Li, X. M., Gloer, J. B. & Wang, B. G. First total syntheses and antimicrobial evaluation of penicimonoterpene, a marine-derived monoterpenoid, and its various derivatives. Mar. Drugs 12, https://doi.org/10.3390/md12063352 (2014).
Fkyerat, A., Burki, N. & Tabacchi, R. Enantioselective synthesis of 3-hydroxycitronellic acid isolated from Ceratocystis fimbriata sp. platani. Tetrahedron Asymmetry 7, 2023–2028 (1996).
Frelek, J. & Szczepek, W. J. [Rh2(OCOCF3)4] as an auxiliary chromophore in chiroptical studies on steroidal alcohols. Tetrahedron Asymmetry 10, 1507–1520 (1999).
Masuda, T., Odaka, Y., Ogawa, N., Nakamoto, K. & Kuninaga, H. Identification of geranic acid, a tyrosinase inhibitor in lemongrass (Cymbopogon citratus). J. Agric. Food Chem. 56, 597–601 (2008).
Nguyen, T. H. T., Pham, N. K. T., Pudhom, K., Hansen, P. E. & Nguyen, K. P. P. Structure elucidation of four new megastigmanes from Sonneratia ovata Backer. Magn. Reson. Chem. 52, 795–802 (2014).
Snatzke, G., Wagner, U. & Wolff, H. P. Circulardichroism-LXXV cottonogenic derivatives of chiral bidentate ligands with the complex [Mo2(O2CCH3)4]. Tetrahedron 37, 349–361 (1981).
Frelek, J. et al. Configurational assignment of sugar erythro-1,2-diols from their electronic circular dichroism spectra with dimolybdenum tetraacetate. Tetrahedron Asymmetry 19, 1709–1713 (2008).
Murayama, T., Hatakeyama, K. & Shiraiwa, R. A secoiridoid, 10-cinnamoyloxyoleoside, isolated from the leaves of Helwingia japonica. J. Nat. Med. 58, 42–45 (2004).
Chen, J. H. et al. Bioactivity-guided fractionation of physical fatigue-attenuating components from Rubus parvifolius L. Molecules 18, 11624–11638 (2013).
Abe, F. & Tamauchi, T. Trachelosperosides, glycosides of 19α-hydroxyursane-type triterpenoids from Trachelospermum asiaticum. Chem. Pharm. Bull. 35, 1748–1754 (1987).
Huang, S. & Czech, M. P. The GLUT4 Glucose Transporter. Cell Metab. 5, 237–252 (2007).
Elchebly, M. et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 (1999).
Tsou, R. C., Zimmer, D. J., De Jonghe, B. C. & Bence, K. K. Deficiency of PTP1B in leptin receptor-expressing neurons leads to decreased body weight and adiposity in mice. Endocrinology 153, 4227–4237 (2012).
Lee, A. et al. Enhancement of Glucose Uptake by Meso-Dihydroguaiaretic acid through GLUT4 up-regulation in 3T3-L1 adipocytes. Molecules 22, 1–8 (2017).
Yang, J. L., Ha, T. K. Q., Lee, B. W., Kim, J. & Oh, W. K. PTP1B inhibitors from the seeds of Iris sanguinea and their insulin mimetic activities via AMPK and ACC phosphorylation. Bioorganic Med. Chem. Lett. 27, 5076–5081 (2017).
Tanaka, T., Nakashima, T., Ueda, T., Tomii, K. & Kouno, I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. (Tokyo). 55, 899–901 (2007).
Gerards, M. & Snatzke, G. Circular dichroism, XCIII determination of the absolute configuration of alcohols, olefins, epoxides, and ethers from the CD of their “In situ” complexes with [Rh2(O2CCF3)4]. Tetrahedron Asymmetry 1, 221–236 (1990).
Nguyen, P. H. et al. Anthraquinones from Morinda longissima and their insulin mimetic activities via AMP-activated protein kinase (AMPK) activation. Bioorganic Med. Chem. Lett. 27, 40–44 (2017).
Yamamoto, N., Yamashita, Y., Yoshioka, Y., Nishiumi, S. & Ashida, H. Rapid preparation of a plasma membrane fraction: Western blot detection of translocated glucose transporter 4 from plasma membrane of muscle and adipose cells and tissues. Curr. Protoc. Protein Sci. 29, 18.1–18.12 (2016).
Acknowledgements
This work was financially supported in part by grants from the KBNMB (NRF-2017M3A9B8069409) and from the Basic Science Research Program (NRF-2017R1E1A1A01074674) through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Planning.
Author information
Authors and Affiliations
Contributions
B.W.L. performed phytochemical experiments and the determination of absolute configuration and wrote the manuscript. T.K.Q.H. designed and performed biological experiments. H.T.T.P. carried out phytochemical experiments. Q.H.H. and V.O.T. collected and authenticated the plant. W.K.O. provided experimental ideas, designed the research and analyzed the data. W.K.O. contributed to the supervision and discussion and reviewed/edited the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, BW., Ha, T.K.Q., Pham, H.T.T. et al. Hydroxyoleoside-type seco-iridoids from Symplocos cochinchinensis and their insulin mimetic activity. Sci Rep 9, 2270 (2019). https://doi.org/10.1038/s41598-018-38013-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-38013-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.