Abstract
Gsdf is a key gene for testicular differentiation in teleost. However, little is known about the function of Gsdf in Chinese tongue sole (Cynoglossus semilaevis). In this study, we obtained the full-length Gsdf gene (CS-Gsdf), and functional characterization revealed its potential participation during germ cell differentiation in testes. CS-Gsdf transcription was predominantly detected in gonads, while the levels in testes were significantly higher than those in ovaries. During the different developmental stages in male gonads, the mRNA level was significantly upregulated at 86 dph, and a peak appeared at 120 dph; then, the level decreased at 1 and 2 yph. In situ hybridization revealed that CS-Gsdf mRNA was mainly localized in the Sertoli cells, spermatogonia, and spermatids in mature testes. After CS-Gsdf knockdown in the male testes cell line by RNA interference, a series of sex-related genes was influenced, including several sex differentiation genes, CS-Wnt4a, CS-Cyp19a1a and CS-Star. Based on these data, we speculated that CS-Gsdf may play a positive role in germ differentiation and proliferation via influencing genes related to sex differentiation.
Similar content being viewed by others
Introduction
In teleost, several genes that belong to TGF-β signal components have been identified to have sex-determination functions, including Amhy, Amhr2, GsdfY and Gdf6Y1,2,3,4,5. Studies on these genes have focused on Gsdf (gonadal soma-derived factor), including its specific expression in teleosts, such as Danio rerio, Takifugu rubripes and Oryzias latipes2,6,7,8. In general, Gsdf performs vital functions in male germ cell proliferation and testicular differentiation6,8,9, and as a teleost-specific gene, it is predominantly expressed in Sertoli cells and surrounding cells in mature gonads of Oryzias luzonensis and Oncorhynchus mykiss3,10. In addition, in O. luzonensis, a Y-chromosome localized Gsdf was reported to be the male-determining gene3, and its deletion could cause feminization11,12. Gsdf has been shown to be an excellent candidate for understanding the sex-determination events in Anoplopoma fimbria13.
Despite the divergent role, accumulated data now support that autosomal Gsdf functions as a male sex initiator and initiate testicular differentiation, which has been proven to be an early marker in the gonads of males in Oryzias latipes and Oreochromis niloticus6,11,14,15. As Dmy-independent sex-determining gene during sex-chromosome evolution16,17,18, Gsdf could play an important role in sex differentiation by interacting with Dmy or Dmrt1, and affecting oestrogen production19,20. Additional features of the Gsdf gene included the regulation of primordial germ cell proliferation and meiotic germ cell proliferation or differentiation6,10.
Chinese tongue sole (Cynoglossus semilaevis) is an economically important marine flatfish that is widely cultured in China. This species exhibits sexual growth dimorphism and females grow 2–4 times faster than males21; thus, increasing the proportion of females would increase the culturing productivity. However, many limiting factors result in low female ratios in aquaculture; for instance, genetic females (ZW) could sex-reverse to phenotypic males under some conditions22,23. In Chinese tongue sole, several genes have been reported to be involved in male sex determination and differentiation, including Dmrt1, Tesk1, Piwil2 and Neurl324,25,26,27. Among these genes, Dmrt1 was previously demonstrated to be the male-determining gene22,24, while the others were found to be involved in spermatogenesis. More and more reports focused on the relationship between environmental factors and sex differentiation/gonad development28,29. Although, in our lab, epigenomics data in Shao et al. revealed that Gsdf is an important sex-related gene, its expression is mediated by DNA methylation in C. semilaevis30, while little is known about the features and functions of the Gsdf gene.
To investigate the role of Gsdf in fish with the ZW sex-determining system, in this study, we first cloned the full cDNA sequence of CS-Gsdf. We then analysed the Gsdf expression patterns in gonads at different developmental stages by qRT-PCR and its special distribution in gonads via in situ hybridization (ISH). Furthermore, RNA interference (RNAi) of CS-Gsdf was performed in the testicular cell line, and a series of sex-related genes were analysed.
Results
Analysis of CS-Gsdf cDNA sequence
The complete cDNA sequence is 1,244 bp in length, containing a 139 bp 5′ untranslated region (UTR), a 615 bp open reading frame (ORF) and a 470 bp 3′ UTR. The ORF encoded a putative protein with 204 amino acids (GenBank accession number: MG891889) (Figs. 1, S1 and S2). The putative protein has a predicted molecular weight of 22.75 kDa and a theoretical isoelectric point of 5.66.
Multiple alignment of C. semilaevis Gsdf protein sequences with other teleosts. Sequences are aligned using ClustalX and DNAMAN. The presumed TGF-β domain region is indicated by the red box. The abbreviations of protein names used in this section are as follows: CS-Gsdf: Cynoglossus semilaevis Gsdf; SS-Gsdf: Solea senegalensis Gsdf; SM-Gsdf: Scophthalmus maximus Gsdf; DL-Gsdf: Dicentrarchus labrax Gsdf; AL-Gsdf: Acanthopagrus latus Gsdf; HT-Gsdf: Halichoeres trimaculatus Gsdf; OL-Gsdf: Oryzias latipes Gsdf; OM-Gsdf: Oncorhynchus mykiss Gsdf.
Expression patterns of CS-Gsdf in Chinese tongue sole
To determine the tissue distribution of CS-Gsdf in Chinese tongue sole, we analysed the expression levels in 10 different tissues of 1-year post-hatching female and male tongue sole by qRT-PCR. CS-Gsdf was expressed in only the gonads, and the expression level was much higher in the testis than that in the ovary (Fig. 2A).
Expression analysis of CS-Gsdf in C. semilaevis evaluated by qRT-PCR. (A) CS-Gsdf transcription in various tissues of C. semilaevis. (B) CS-Gsdf transcription in the gonads of different sexual genotypes. (C) CS-Gsdf transcription in the male gonads at different developmental stages. F: female, M: male, PM: pseudo-male, TM: triploid male. Dmrt1-knockout: Dmrt1-knockout fish. The transcription levels were normalized using the β-actin levels. The bars represent the triplicate mean ± SEM values from three separate individuals (n = 3). The different letters on bars denote statistical significance (p < 0.05).
Using qRT-PCR, we also examined the expression levels in the gonads of male, female, pseudo-male, triploid male (infertile) and Dmrt1-knockout fish. The highest CS-Gsdf mRNA transcript level was detected in the gonads of males, while the mRNA expression levels of CS-Gsdf were low in the pseudo-male and triploid male. The lowest expression levels were observed in the gonads of the female and Dmrt1-knockout fish (Fig. 2B).
To study the expression of CS-Gsdf during the differentiation and development of male gonads, we measured the expression levels in testes at different developmental stages. As shown in Fig. 2C, the CS-Gsdf transcripts were detected at 20 days post-hatching (dph) and continued to be expressed at very low levels before sharply increasing at 86 dph. At 120 dph, the expression reached its peak in the testis and then declined at 1-year post-hatching (yph) and 2 yph.
Cyto-localization of CS-Gsdf mRNA in the gonads
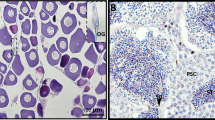
The ISH results demonstrated that CS-Gsdf was mainly expressed in Sertoli cells, spermatogonia and spermatids with intensive hybridization signals at 120 dph and 1 yph (Fig. 3A, B, D and E). In contrast, faint signals were detected in the spermatids of the 2 yph testes (Fig. 3G and H). As a negative control, no positive signal was detected in the hybridization sections with sense probes (Fig. 3C, F and I).
In situ localization of CS-Gsdf mRNA in the gonads of C. semilaevis. (A) Testis of a male at 120 dph. (B) Magnification of the red framed area in A. (C) Testis of a male at 120 dph with sense probes as a control. (D) Testis of a male at 1 yph. (E) Magnification of the red framed area in D. (F) Testis of a male at 1 yph with sense probes as a control. (G) Testis of a male at 2 yph. (H) Magnification of the red framed area in G. (I) Testis of a male at 2 yph with sense probes as a control. Sg: spermatogonia, SC: Sertoli cells, St: spermatids, Bars = 100 μm.
RNAi-mediated CS-Gsdf knockdown and its impact on the mRNA expression of sex-related genes
The RNAi-mediated knockdown in vitro was conducted for CS-Gsdf in a Chinese tongue sole testicular cell line (CSGC). To determine the silencing effects of RNAi, CS-Gsdf expression in CSGC was detected by qRT-PCR 48 h after siRNA transfection. The results revealed that the silencing efficiencies of CS-Gsdf were approximately 79.1%, 70.9% and 71.3% in the si-cse-Gsdf 01, 02 and 03 treatments, respectively (Fig. 4A).
The analysis of Gsdf, Star, Foxl2, Wnt4a and Cyp19a1a expression in cultured testis cells after RNAi. (A) Expression of CS-Gsdf after the transfection of siRNA at 48 h. (B) Expression levels of CS-Star, CS-Foxl2, CS-Wnt4a and CS-Cyp19a1a were determined by qRT-PCR after the transfection of the siRNA for 48 h. NC, si-001, si-002 and si-003 indicate the testis cells transfected with the siRNA of the negative control (NC) interfered with the 001 sites, 002 sites and 003 sites, respectively. Asterisks indicate significant differences (p < 0.05) between the treated group and the control.
To evaluate the effects of CS-Gsdf RNAi-mediated knockdown on the expression of sex-related genes, Foxl2, Star, Wnt4a, and Cyp19a1a were measured. As shown in Fig. 4B, compared with the control, the expression levels of CS-Gsdf, CS-Foxl2, CS-Star, CS-Wnt4a, and CS-Cyp19a1a were detected by qRT-PCR.
Discussion
Gsdf belongs to the transforming growth factor-beta (TGF-beta) family, which is important for the regulation of cell growth, differentiation, and migration31,32. Moreover, recent reports found that members of the TGF-beta family may have a function in the sex-determining process33. To determine the role of CS-Gsdf in tongue sole, we initiated this study.
In this work, we cloned and characterized CS-Gsdf cDNA. The predicted protein contained a highly conserved TGF-beta region, which is characterized by 7 conservative cysteine residues called the conserved cysteine knot motif (Fig. 1), which is not present in trout lacking 6th cysteine residues6. Different from other TGF-β family members, Gsdf lacks glycine residues in the conserved motif34.
The expression analysis revealed that CS-Gsdf was expressed in only gonads, and the expression level was much higher in testes than that in ovaries. The threshold of CS-Gsdf mRNA transcription was observed at 20 dph, which is consistent with the previous reports of its expression in early teleost stages, such as that observed at 5 days post-fertilization (dpf) in O. latipes, 16 dpf in D. rerio6,35 and 30 dpf in O. mykiss10, which are all prior to testicular differentiation6,17. As histological differentiation occurred at 50–65 dph in C. semilaevis, and 70–90 dph were selected to represent appearance of oogonium/spermatogonia, whereas cellular differentiation occurred at 120–150 dph, including oocyte/spermatocyte and so on21,36,37. CS-Gsdf exhibited high expression at 86 dph and peak expression at 120 dph in testes. However, the expression subsequently declined in mature testes which nearly completed cellular differentiation and seminiferous tubule formation20,22,38,39. During germ cells divide rapidly, and Sertoli cells actively regulate the surrounding congregated spermatogonia22,23,40. Thus, the relatively strong signals in Sertoli cells suggested steroidogenesis and spermatogenesis functions41,42,43,44. Furthermore, the lowest CS-Gsdf expression levels were found in Dmrt1-deficient tongue sole (Fig. 2B), and few spermatogonia were observed in this sample. Similar findings in Oreochromis niloticus have been reported45. It has been widely reported that Gsdf exists in Sertoli cells in O. mykiss10, D. rerio35, O. luzonensis3, O. sakaizumii20, Monopterus albus38, and Paralichthys olivaceus39. Given these findings, we speculated that CS-Gsdf probably plays an important role in germ cell differentiation and proliferation.
After the in vitro knockdown of CS-Gsdf in cultured male cells (Fig. 4A), qRT-PCR showed the mRNA levels of the sex-related genes CS-Star, CS-Cyp19a1a, and CS-Wnt4a, which all increased after CS-Gsdf knockdown except CS-Foxl2. While, when CS-Dmrt1 is knocked out, both CS-Cyp19a1a and CS-Foxl2 are significantly upregulated in tongue sole24. Cyp19a1a is both the sex-determining gene and the regulator of steroid hormone synthesis in teleosts, and this gene plays a role in gonadal differentiation. Meanwhile, Star is a rate-limiting step that mediates steroid hormone synthesis46,47,48,49,50,51,52. Therefore, we speculated that CS-Gsdf was involved in sex differentiation through mediating genes related to gonadal hormones. Similar results appeared in two types of medaka (O. latipes and O. sakaizumii), which proved Gsdf affected oestrogen production during sex differentiation19,20. Simultaneously, Wnt4a is a key gene in the Wnt4/β-catenin1 pathway that regulates gonad development/differentiation53,54. CS-wnt4a was also upregulated after RNAi, although the mechanisms remain elusive. Interestingly, CS-Gsdf obviously declined in Dmrt1-deficient gonads, which might be positively regulated by sex-determination genes. In conclusion, CS-Gsdf transcription could affect gene expression related to gonadal hormone genes but exert a negative effect on female Foxl2 gene.
Conclusion
In summary, we cloned and characterized CS-Gsdf from Chinese tongue sole. CS-Gsdf was specifically expressed in gonads, with much higher expression levels in testes than those in ovaries. In testes, the threshold of CS-Gsdf transcription was detected at 20 dph, increased at 86 dph, and peaked at 120 dph, Its mRNA was mainly localized in Sertoli cell and spermatogonia. After in vitro RNAi in the testicular cell line, several sex-related genes were affected. Based on these data, we proposed that CS-Gsdf participates in germ cells differentiation and proliferation of testes, while further studies are needed to elucidate its detailed role.
Materials and Methods
Ethics statement
The handling of experimental fish was approved by the Animal Care and Use Committee of the Chinese Academy of Fishery Sciences, and all protocols were performed in accordance with the guidelines of the Animal Care and Use Committee. To minimize fish suffering, tissues were collected under MS222 anaesthesia.
Experimental fish preparation and sample collection
The Chinese tongue sole used in this study were purchased from the Haiyang High-Tech Experimental Base (Haiyang, Shandong Province, China). Temperature treatments were performed to induce pseudo-males, as previously described55. Ten individuals of each scope (including males, females and pseudo-males) participated in this work. The brain, heart, intestine, gill, kidney, liver, muscle, skin, spleen, and gonads were collected from 1-year-old fish, immediately subjected to liquid nitrogen and then stored at −80 °C until RNA extraction. The gonads at different developmental stages (at 20, 35, 65, 86, 120 dph, 1 and 2 yph) were picked from one side and frozen in liquid nitrogen until RNA extraction. The contralateral gonads were also picked and divided into two sections: one was placed in 4% paraformaldehyde (PFA) for in situ hybridization (ISH), and the other was simultaneously placed in Bouin’s fixative for histological analysis of phenotypic sex. The tail fins of all experimental fish were collected and preserved in 100% ethanol for DNA extraction and subsequent genetic sex determination.
DNA, RNA extraction and cDNA synthesis
The process used to extract genomic DNA followed the standard phenol-chloroform extraction method56, which was then used as a template for subsequent analysis after quantification.
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and then quantified by NanoVue Plus (BiochRom LTD, Cambridge, England). The first-strand cDNA was synthesized using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, City, Country). A total of 800 ng of total RNA from each sample was reverse transcribed into first-strand cDNA and used as the template for qRT-PCR.
Identification of phenotypic and genetic sex
To verify the phenotypic sex of all experimental individuals, gonadal histology was carried out as previously described57. The genetic sex was determined by following the methods of Liu et al.58. The sex-specific simple sequence repeat (SSR) markers scaffold68-2F and scaffold68-2R (Table S1) were designed for PCR amplification as previously described58.
Isolation of CS- Gsdf full-length cDNA
To obtain the full-length cDNA of CS-Gsdf, rapid amplification of cDNA ends (RACE) was performed using the SMART RACE cDNA Amplification Kit (Clontech Inc., Mountain View, CA, USA). RACE-ready first-strand cDNA was synthesized from total RNA according to the manufacturer’s instructions, and the gene-specific primers for outer and nest amplification were designed (Table S1). The outer amplification was performed using touchdown PCR procedures as described by Meng25. PCR products were electrophoresed on a 1.0% agarose gel, and the amplified fragments of expected size were depurated with a Zymo clean Gel DNA Recovery Kit (ZYMO Research, Orange, CA, USA). Purified products were cloned into a pMD18-T vector (TaKaRa, Dalian, China) and sequenced.
Analysis of qPCR
QRT-PCR primers (Table S1) were designed based on the CS-Gsdf cDNA sequence, and their specificity was verified by a single distinct peak obtained in a melting curve analysis. QRT-PCR was conducted using a 7500 ABI real-time PCR system (Applied Biosystems) with SYBR Green Master Mix (TaKaRa). β-actin was used as the internal control59. Three randomly selected individuals were subjected to qRT-PCR, and the experiment was performed in triplicate for each sample.
The relative mRNA expression of target genes was calculated by the 2−ΔΔCt method. All data were tested using one-way ANOVA followed by Duncan multiple comparison tests using SPSS 18.0 (IBM, New York, NY, USA). Significance was accepted only when p < 0.05. All assays in the qRT-PCR complied with the MIQE guidelines60.
In situ hybridization
After dehydration in ethanol, the stored gonad samples were fixed in paraffin wax and sheared as 5 µm sections. A pair of primers (Table S1) for RNA probe synthesis was designed according to the CS-Gsdf ORF sequence. The PCR product was cloned into a pBluescriptSKII plasmid and then linearized with PstI and SalI (TaKaRa). Probes were labelled using DIG RNA Labeling Mix (Roche, Mannheim, Germany). The ISH was performed following a previously described method61 using samples from three different individuals. Images were captured with a Nikon E80i microscope (Nikon, Tokyo, Japan) and then analysed.
In vitro RNAi of Gsdf
The three Gsdf-specific small interfering RNAs (si-cse-Gsdf 01, 02 and 03) were designed and synthesized by RayBiotech C. Ltd. In addition, a nonspecific siRNA negative was used as a control (NC) during the experiment (Guangzhou, Guangdong province, China). The testicular (CSGC) cell line, which was previously created in our laboratory, was employed for RNAi silencing. The CSGC cells were recovered as described by Zhang et al.62, and then transferred to six-well plates. After cultivating at 24 °C for 12 h, the cells completely attached to the plates, and then the labelled siRNAs were transfected into the cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. The calculated average transfection efficiency was approximately 80%. Both the treated groups (using si-cse-Gsdf 01, 02 and 03) and the control group (using NC siRNA) were transfected at a concentration of 30 nM. The collected cells were cultivated at 24 °C for 48 h. The total RNA was extracted from the cells, and cDNA was synthesized as described above. The relative expression levels of the genes related to sex differentiation, such as Star, Cyp19a1a, Foxl2 and Wnt4a, were evaluated by qRT-PCR, and all experiments were performed in triplicate.
References
Hattori, R. S. et al. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci USA 109, 2955–2959 (2012).
Kamiya, T. et al. A trans-species missense SNP in Amhr2 is associated with sex deter-mination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8, e1002798 (2012).
Myosho, T. et al. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191, 163–170 (2012).
Reichwald, K. et al. Insights into Sex Chromosome Evolution and Aging from the Genome of a Short-Lived Fish. Cell 163, 1527–1538 (2015).
Li, M. et al. A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 11, e1005678 (2015).
Shibata, Y. et al. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns 10, 283–289 (2010).
Chen, S. X. et al. A progestin (17α, 20β-dihydroxy-4-pregnen-3-one) stimulates early stages of spermatogenesis in zebrafish. Gen. Comp. Endocrinol. 185, 1–9 (2013).
Kobayashi, T. et al. Estrogen alters gonadal soma-derived factor (Gsdf)/Foxl2 expression levels in the testes associated with testis-ova differentiation in adult medaka, Oryzias latipes. Aquat. Toxicol. 191, 209–218 (2017).
Crespo, B., Gómez, A., Mazón, M. J., Carrilo, M. & Zanuy, S. Isolation and characterization of Ff1 and Gsdf family genes in European sea bass and identification of early gonadal markers of precocious puberty in males. Gen. Comp. Endocrinol. 191, 155–167 (2013).
Sawatari, E., Shikina, S., Takeuchi, T. & Yoshizaki, G. A novel transforming growth factor-beta superfamily member expressed in gonadal somatic cells enhances primordial germ cell and spermatogonial proliferation in rainbow trout (Oncorhynchus mykiss). Dev. Biol. 301, 266–275 (2007).
Zhang, X. et al. Autosomal Gsdf acts as a male sex initiator in the fish medaka. Sci. Rep. 6, 19738 (2016).
Imai, T., Saino, K. & Matsuda, M. Mutation of Gonadal soma-derived factor induces medaka XY gonads to undergo ovarian development. Biochem. Biophys. Res. Commun. 467, 109–114 (2015).
Rondeau, E. B. et al. Genomics of sablefish (Anoplopoma fimbria): expressed genes, mitochondrial phylogeny, linkage map and identification of a putative sex gene. BMC Genomics 14, 452 (2013).
Schartl, M. A comparative view on sex determination in medaka. Mech. Dev. 121, 639–645 (2004).
Kaneko, H. et al. Gonadal soma-derived factor (Gsdf), a TGF-beta superfamily gene, induces testis differentiation in the teleost fish Oreochromis niloticus. Mol. Cell. Endocrinol. 415, 87–99 (2015).
Volff, J. N., Kondo, M. & Schartl, M. Medaka dmY/dmrt1Y, is not the universal primary sex-determining gene in fish. Trends Genet. 19, 196–199 (2003).
Kikuchi, K. & Hamaguchi, S. Novel sex‐determining genes in fish and sex chromosome evolution. Dev. Dyn. 242, 339–353 (2013).
Masuyama, H. et al. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 20, 163–176 (2012).
Chakraborty, T., Zhou, L. Y., Chaudhari, A., Iguchi, T. & Nagahama, Y. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 6, 19480 (2016).
Horie, Y. et al. Androgen induces gonadal soma-derived factor, Gsdf, in XX gonads correlated to sex-reversal but not Dmrt1 directly, in the teleost fish, northern medaka (Oryzias sakaizumii). Mol. Cell. Endocrinol. 436, 141–149 (2016).
Chen, S. L. et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nature Genet 46, 253–260 (2014).
Chen, S. L. et al. Artificial gynogenesis and sex determination in tongue sole (Cynoglossus semilaevis). Mar Biotechnol 11, 243–251 (2009).
Hu, Q. M. et al. Differences in sex reversion and growth between normal and neomale stock in half-smooth tongue sole, Cynoglossus semilaevis. Aquaculture Int 22, 1437–1449 (2014).
Cui, Z. K. et al. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 7, 42213 (2017).
Meng, L. et al. Cloning and characterization of tesk1, a novel spermatogenesis-related gene, in the tongue sole (Cynoglossus semilaevis). PloS One 9, e107922 (2014).
Zhang, L. Y. et al. Cloning, expression and methylation analysis of piwil2 in half-smooth tongue sole (Cynoglossus semilaevis). Mar Genomics 18, 45–54 (2014).
Xu, W. T. et al. Ubiquitin ligase gene neurl3 plays a role in spermatogenesis of half-smooth tongue sole (Cynoglossus semilaevis) by regulating testis protein ubiquitination. Gene. 592, 215–220 (2016).
Santos, D., Luzio, A. & Coimbra, A. M. Zebrafish sex differentiation and gonad development: A review on the impact of environmental factors. Aquatic Toxicology. 191, 141–163 (2017).
Orban, L., Sreenivasan, R. & Olsson, P. E. Long and winding roads: testis differentiation in zebrafish. Molecular & Cellular Endocrinology. 312, 35–41 (2009).
Shao, C. W. et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24, 604–615 (2014).
Hughes, D. E. et al. Estrogen promotes apoptosis of murine osteoclasts mediated by TGF-beta. Nat. Med. 2, 1132–1136 (1996).
Josso, N. & Clemente, N. Transduction pathway of anti-Mullerian hormone, a sex-specific member of the TGF-beta family. Trends Endocrinol. Metab. 14, 91–97 (2003).
Herpin, A. & Schartl, M. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16, 1260–1274 (2015).
Vitt, U., Hsu, S. & Hsueh, A. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol Endocrinol. 5, 681–694 (2001).
Gautier, A., Sohm, F., Joly, J. S., Le Gac, F. & Lareyre, J. J. The proximal promoter region of the zebrafish Gsdf gene is sufficient to mimic the spatio-temporal expression pattern of the endogenous gene in Sertoli and granulosa cells. Biol. Reprod. 85, 1240–1251 (2011).
Li, H. L. et al. Characterization and expression pattern of r-spondin1 in Cynoglossus semilaevis. Journal of Experimental Zoology Part B Molecular & Developmental Evolution. 8, 772–780 (2017).
Li, H. L. et al. Two Figla homologues have disparate functions during sex differentiation in half-smooth tongue sole (Cynoglossus semilaevis). Scientific Reports. 6, 28219 (2016).
Zhu, Y., Wang, C., Chen, X. & Guan, G. Identification of gonadal soma-derived factor involvement in Monopterus albus, (protogynous rice field eel) sex change. Mol. Biol. Rep. 43, 629–637 (2016).
Liu, Y. et al. Sexually dimorphic expression in developing and adult gonads shows an important role of gonadal soma-derived factor during sex differentiation in olive flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 210, 1–8 (2017).
Deng, S. P. & Chen, S. L. cDNA cloning, tissues, embryos and larvae expression analysis of Sox10 in half-smooth tongue-sole, Cynoglossus semilaevis. Mar. Genomics 1, 109–114 (2008).
Le Gac, F., Loir, M., le Bail, P. Y. & Ollitrault, M. Insulin‐like growth factor (IGF‐I) mRNA and IGF‐I receptor in trout testis and in isolated spermatogenic and Sertoli cells. Mol. Reprod. Dev. 44, 23–35 (1996).
Berishvili, G., D’Cotta, H., Baroiller, J. F., Segner, H. & Reinecke, M. Differential expression of IGF-I mRNA and peptide in the male and female gonad during early development of a bony fish, the tilapia Oreochromis niloticus. Gen. Comp. Endocrinol. 146, 204–210 (2006).
Reinecke, M., Schmid, A., Ermatinger, R. & Loffing-Cueni, D. Insulin-like growth factor I in the teleost Oreochromis mossambicus, the tilapia: gene sequence, tissue expression, and cellular localization. Endocrinology. 138, 204–210 (2006).
Viñas, J. & Piferrer, F. Stage-specific gene expression during fish spermatogenesis as determined by laser-capture microdissection and quantitative-PCR in sea bass (Dicentrarchus labrax) gonads. Bio. Reprod. 79, 738–747 (2008).
Jiang, D. N. et al. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 83, 497–508 (2016).
Wang, K. L., Zhang, H., Hu, Q. M., Shao, C. W. & Chen, S. L. Expression and purification of half-smooth tongue sole (Cynoglossus semilaevis) CSDAZL protein. Protein Expr. Purif. 102, 8–12 (2014).
Dong, X. L., Chen, S. L., Ji, X. S. & Shao, C. W. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Ocean. Sin. 30, 68–77 (2011).
Trant, J. M., Gavasso, S., Ackers, J., Chung, B. C. & Place, A. R. Developmental expression of cytochrome P450 aromatase genes (CYP19a and CYP19b) in zebrafish fry (Danio rerio). J. Exp. Zool. 290, 475–483 (2001).
Bauer, M. P., Bridgham, J. T., Langenau, D. M., Johnson, A. L. & Goetz, F. W. Conservation of steroidogenic acute regulatory (StAR) protein structure and expression in vertebrates. Mol. Cell. Endocrinol. 168, 119–125 (2000).
Arukwe, A. Modulation of brain steroidogenesis by affecting transcriptional changes of steroidogenic acute regulatory (StAR) protein and cholesterol side chain cleavage (P450scc) in juvenile Atlantic salmon (Salmo salar) is a novel aspect of nonylphenol toxicity. Environ. Sci. Technol. 39, 9791–9798 (2005).
Arukwe, A. Steroidogenic acute regulatory (StAR) protein and cholesterol side-chain cleavage (P450scc)-regulated steroidogenesis as an organ-specific molecular and cellular target for endocrine disrupting chemicals in fish. Cell. Biol. Toxicol. 24, 527–540 (2008).
Aluru, N., Renaud, R., Leatherland, J. F. & Vijayan, M. M. Ah Receptor-mediated impairment of interrenal steroidogenesis involves StAR protein and P450scc gene attenuation in rainbow trout. Toxicol. Sci. 84, 260–269 (2005).
Zhu, Y. et al. Identification and analysis of the β-catenin1 gene in half-smooth tongue sole (Cynoglossus semilaevis). PLoS One 12, e0176122 (2017).
Hu, Q. M., Zhu, Y., Liu, Y., Wang, N. & Chen, S. L. Cloning and characterization of wnt4a gene and evidence for positive selection in half-smooth tongue sole (Cynoglossus semilaevis). Sci. Rep. 4, 7167 (2014).
Hu, Q. M. et al. Cloning, genomic structure and expression analysis of ubc9 in the course of development in the half-smooth tongue sole (Cynoglossus semilaevis). Comp Biochem Physiol Part B 165, 181–188 (2013).
Chen, S. L. et al. Induction of mitogynogenetic diploids and identification of WW super-female using sex-specific SSR markers in tongue sole (Cynoglossus semilaevis). Mar Biotechnol 14, 120–128 (2012).
Liang, Z., Chen, S. L., Zhang, J., Song, W. T. & Liu, S. S. Gonadal development process observation of half-smooth tongue sole in rearing population. J Southern Agric (In Chinese) 43, 2074–2078 (2012).
Liu, Y. et al. SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semilaevis). J Agric Biotech (In Chinese) 22, 787–792 (2014).
Li, Z. et al. β-Actin is a useful internal control for tissue-specific gene expression studies using quantitative real-time PCR in the half-smooth tongue sole Cynoglossus semilaevis challenged with LPS or Vibrio anguillarum. Fish Shellfish Immuno 29, 89–93 (2010).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Kobayashi, T., Kajiura-Kobayashi, H. & Nagahama, Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mech. Dev. 99, 139 (2000).
Zhang, B. et al. Establishment and characterization of a testicular cell line from the half-smooth tongue Sole, Cynoglossus semilaevis. Int. J. Biol. Sci. 7, 452–459 (2011).
Acknowledgements
We would like to thank Yingming Yang at Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences for sample collection. This work was supported by grants from the National Natural Science Foundation of China (31730099, 31402293, 31472269, 31722058), Special Scientific Research Funds for Central Non-profit Institutes, Yellow Sea Fisheries Research Institute (20603022017003), A Shan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2017ASTCP-OS15), Taishan Scholar Climbing Project of Shandong.
Author information
Authors and Affiliations
Contributions
Y.Z., L.M., S.C. conceived and designed the experiments. Y.Z., W.X., Z.C., N.Z. and H.G. performed the experiments. Y.Z., N.W. and C.S. analyzed the data. Y.Z. L.M. wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, Y., Meng, L., Xu, W. et al. The autosomal Gsdf gene plays a role in male gonad development in Chinese tongue sole (Cynoglossus semilaevis). Sci Rep 8, 17716 (2018). https://doi.org/10.1038/s41598-018-35553-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-35553-7
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.